LEAFY and Polar Auxin Transport Coordinately Regulate Arabidopsis Flower Development
Abstract
:1. Introduction
2. Results and Discussion
2.1. Loss-of-LFY Function Enhances Floral Primordium Initiation Defects of pin1 and pid Mutants

2.2. Mutations in LFY Enhance Floral Organ Initiation and Growth Defects of pin1 Mutants

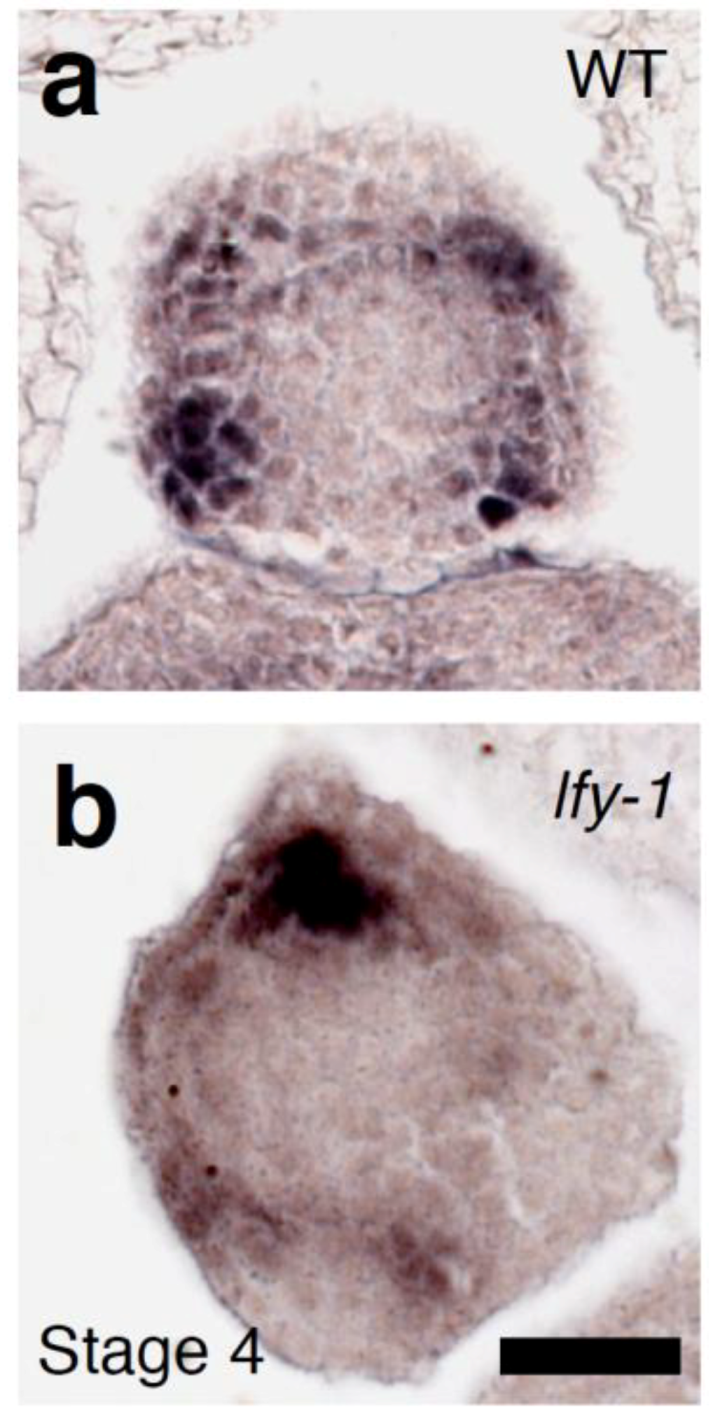
2.3. The Relationship between LFY and Auxin-Mediated Sepal Initiation

2.4. A Link between LFY and Polar Auxin Transport in Sepal Boundary Formation


2.5. A Link between LFY and Auxin Transport in Gynoecium Development

3. Experimental Section
3.1. Plant Materials and Growth Conditions
3.2. Statistical Tests
3.3. Genetics and PCR Genotyping
| Primer name | Sequence |
|---|---|
| Genotyping | |
| lfy-1/-6-FW | AAGCAGCCGTCTGCGGTGTCAGCAGCTGTT |
| lfy-1/-6-RV | CTGTCAATTTCCCAGCAAGACAC |
| pid-16-LP | TCCGTCATAGACAACCTCACC |
| pid-16-RP | GAGTAAGCGTACGAATGAGCG |
| pin1-8-LP | AACTGGCTTCACAGCAGAAAG |
| pin1-8-RP | TCAACAAAAAGGGCATTGTTC |
| LBb1.3 | ATTTTGCCGATTTCGGAAC |
| Primer name | Sequence |
| qRT-PCR | |
| CUC2-FW | GGAAGAGCTCCGAAAGGAGA |
| CUC2-RV | TCCGGTGCTAGCTAAAGTGG |
| ETT-FW | CAGGGACATTTGGAACAAGC |
| ETT-RV | CAGGAAGAAGAGAGACTTGAGCA |
| EIF4-FW | AAACTCAATGAAGTACTTGAGGGACA |
| EIF4-RV | TCTCAAAACCATAAGCATAAATACCC |
| In situ hybridization | |
| CUC2-FW | CGGAATTCATGGACATTCCGTATTACCA |
| CUC2-RV | CCACTAGTTCAGTAGTTCCAAATACAGT |
3.4. Confocal Microscopy
3.5. Quantitative RT-PCR
3.6. Screenshot
3.7. In situ Hybridization
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef]
- Benkova, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jurgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Reinhardt, D.; Pesce, E.R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Rovertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Robert, H.S.; Grones, P.; Stepanova, A.N.; Robles, L.M.; Lokerse, A.S.; Alonso, J.M.; Weijers, D.; Friml, J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013, 23, 2506–2512. [Google Scholar] [CrossRef]
- Friml, J.; Yang, X.; Michniewicz, M.; Weijers, D.; Quint, A.; Tietz, O.; Benjamins, R.; Ouwerkerk, P.B.; Ljung, K.; Sandberg, G.; et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 2004, 306, 862–865. [Google Scholar] [CrossRef]
- Michniewicz, M.; Zago, M.K.; Abas, L.; Weijers, D.; Schweighofer, A.; Meskiene, I.; Marcus, G.H.; Ohno, C.; Zhang, J.; Huang, F.; et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007, 130, 1044–1056. [Google Scholar] [CrossRef]
- Cheng, Y.; Qin, G.; Dai, X.; Zhao, Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18825–18829. [Google Scholar] [CrossRef]
- Cheng, Y.; Qin, G.; Dai, X.; Zhao, Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 21017–21022. [Google Scholar] [CrossRef]
- Furutani, M.; Nakano, Y.; Tasaka, M. MAB4-induced auxin sink generates local auxin gradients in Arabidopsis organ formation. Proc. Natl. Acad. Sci. USA 2014, 11, 1198–1203. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenase controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef]
- Bennett, S.R.M.; Alvarez, J.; Bossinger, G.; Smyth, D.R. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995, 8, 505–520. [Google Scholar]
- Przemeck, G.K.; Mattsson, J.; Hardtke, C.S.; Sung, Z.R.; Berleth, T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 1996, 200, 229–237. [Google Scholar]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef]
- Weijers, D.; Schlereth, A.; Ehrismann, J.S.; Schwank, G.; Kientz, M.; Jurgens, G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 2006, 10, 265–270. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Wu, M-F.; Winter, C.; Berns, M.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.; Wagner, D. A molecular framework for auxin-mediated initiation of floral promordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef]
- Weigel, D.; Nilsson, O. A developmental switch sufficient for flower initiation in diverse plants. Nature 1995, 377, 495–500. [Google Scholar] [CrossRef]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.J.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar] [CrossRef]
- Krizek, B.A. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 1999, 25, 224–236. [Google Scholar] [CrossRef]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef]
- Krizek, B.A. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 2009, 150, 1916–1929. [Google Scholar] [CrossRef]
- Krizek, B.A. Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PHETHORA (AIL/PLT) family. J. Exp. Bot. 2011, 62, 3311–3319. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Sieber, P.; Wellmer, F.; Gheyselinck, J.; Riechmann, J.L.; Meyerowitz, E.M. Redundancy and specillization among plant microRNAs: Role of the MIR164 familiy in developmental robstness. Development 2007, 134, 1051–1060. [Google Scholar] [CrossRef]
- Li, W.; Zhou, Y.; Liu, X.; Yu, P.; Cohen, J.D.; Meyeriwitz, E.M. LEAFY controls auxin response pathways in floral primordium formation. Sci. Signal. 2013, 6, ra23. [Google Scholar]
- Kawamura, E.; Horiguchi, H.; Tsukaya, H. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 2010, 62, 429–441. [Google Scholar] [CrossRef]
- Schuetz, M.; Berleth, T.; Mattsson, J. Multiple MONOPTEROS-dependent pathways are involved in leaf initiation. Plant Physiol. 2008, 148, 870–880. [Google Scholar] [CrossRef]
- Weigel, D.; Meyerowitz, E.M. Activation of floral homeotic genes in Arabidopsis. Science 1993, 261, 1723–1726. [Google Scholar]
- Winter, C.M.; Austin, R.S.; Blanvillain-Baufume, S.; Reback, M.A.; Monniaux, M.; Wu, M.F.; Sang, Y.; Yamaguchi, A.; Yamaguchi, N.; Parker, J.E.; et al. LEAFY Target Genes Reveal Floral Regulatory Logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 2011, 20, 430–443. [Google Scholar] [CrossRef]
- Wu, M.F.; Sang, Y.; Bezhani, S.; Yamaguchi, N.; Han, S.K.; Li, Z.; Su, Y.; Slewinski, T.L.; Wagner, D. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 2012, 109, 3576–3581. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Kilinc, A.; Smyth, D.R. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J. 2012, 71, 724–735. [Google Scholar] [CrossRef]
- Moyroud, E.; Minguet, E.G.; Ott, F.; Yant, L.; Pose, D.; Monniaux, M.; Blanchet, S.; Bastien, O.; Thevenon, E.; Weigel, D.; et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 2011, 23, 1293–1306. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Soowal, L.N.; Lee, I.; Weigel, D. LFY expression and flower initiation in Arabidopsis. Development 1998, 121, 3835–3844. [Google Scholar]
- Takada, S.; Hibara, K.; Ishida, T; Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Wagner, D.; Sablowski, R.W.; Meyerowitz, E.M. Transcriptional activation of APETALA1 by LEAFY. Science 1999, 285, 582–584. [Google Scholar] [CrossRef]
- Massay, F.J. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951, 46, 68–78. [Google Scholar] [CrossRef]
- Long, J.A.; Barton, M.K. The development of apical embryonic pattern in Arabidopsis. Development 1998, 125, 3027–3035. [Google Scholar]
- Wu, M.F.; Wagner, D. RNA in situ hybridization in Arabidopsis. Methods Mol. Biol. 2012, 883, 75–86. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
Appendix
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yamaguchi, N.; Wu, M.-F.; Winter, C.M.; Wagner, D. LEAFY and Polar Auxin Transport Coordinately Regulate Arabidopsis Flower Development. Plants 2014, 3, 251-265. https://doi.org/10.3390/plants3020251
Yamaguchi N, Wu M-F, Winter CM, Wagner D. LEAFY and Polar Auxin Transport Coordinately Regulate Arabidopsis Flower Development. Plants. 2014; 3(2):251-265. https://doi.org/10.3390/plants3020251
Chicago/Turabian StyleYamaguchi, Nobutoshi, Miin-Feng Wu, Cara M. Winter, and Doris Wagner. 2014. "LEAFY and Polar Auxin Transport Coordinately Regulate Arabidopsis Flower Development" Plants 3, no. 2: 251-265. https://doi.org/10.3390/plants3020251
APA StyleYamaguchi, N., Wu, M.-F., Winter, C. M., & Wagner, D. (2014). LEAFY and Polar Auxin Transport Coordinately Regulate Arabidopsis Flower Development. Plants, 3(2), 251-265. https://doi.org/10.3390/plants3020251




