Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis
Abstract
:1. Introduction
2. Results and Discussion
2.1. 13-LOX Expression Patterns in Unwounded Rosettes
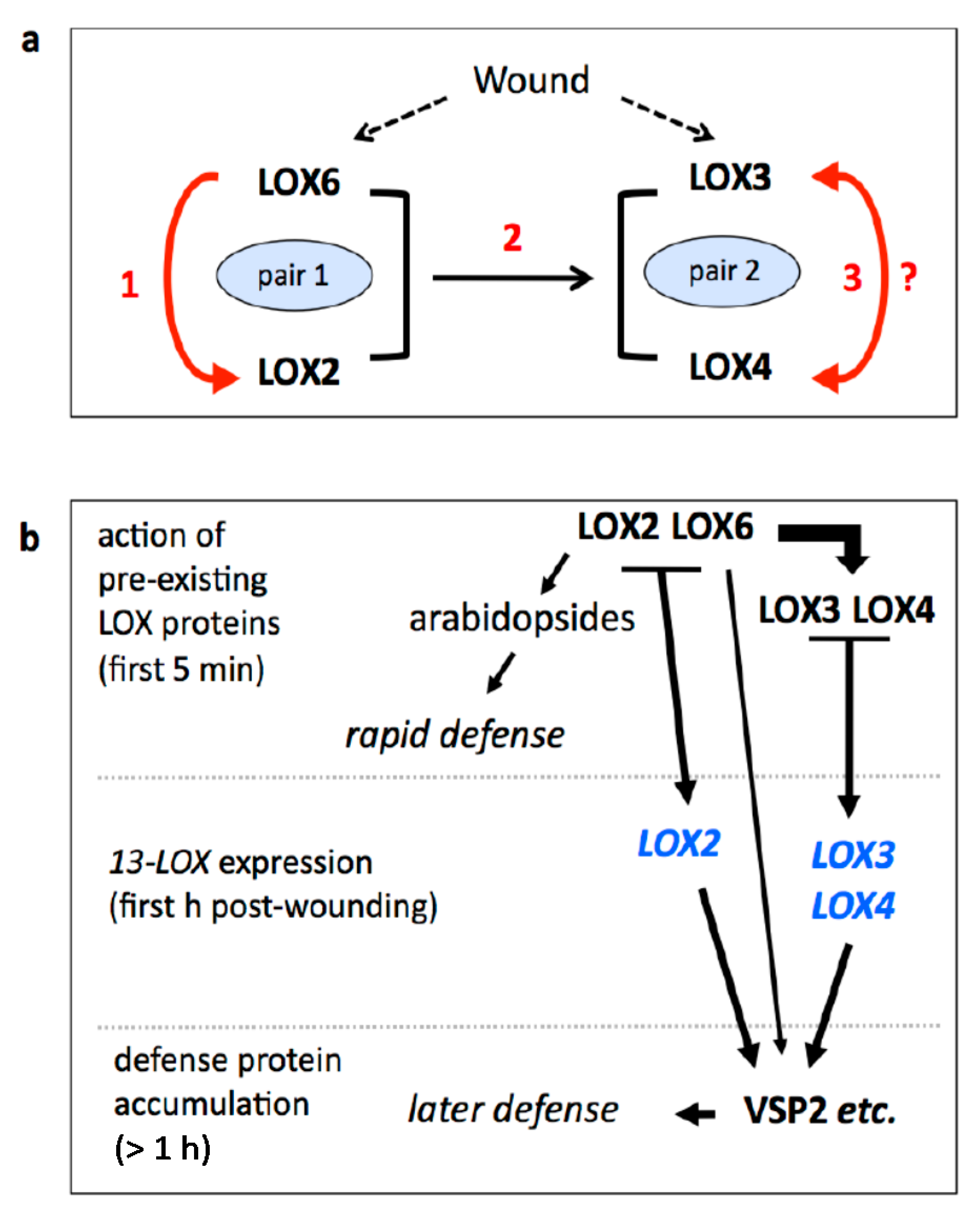
2.2. LOX2 and LOX6 Regulate LOX3 and LOX4 Expression
2.3. 13-LOXs Contribute Differentially to Inducible VSP2 Defense Gene Expression
2.4. Comparison of LOX3, LOX4 and VSP2 Expression in the WT and the lox2 lox6 Double Mutant
2.5. All 13-LOXs Act in Defense against a Lepidopteran Herbivore
2.6. Functional Pairs of 13-LOXs in Leaf Defense
3. Experimental Section
3.1. Plant Material and Growth Conditions
3.2. Insect Feeding Assays
3.3. Gene Expression
3.4. GUS Staining and Light Microscopy
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix


References
- Haeggstrom, J.Z.; Funk, C.D. Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chem. Rev. 2011, 111, 5866–5898. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.; Feussner, I. Lipoxygenases—Structure and reaction mechanism. Phytochemistry 2009, 70, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Bannenberg, G.; Martinez, M.; Hamberg, M.; Castresana, C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 2009, 44, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Ann. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Dong-Sun, L.; Nioche, P.; Hanberg, M.; Raman, C.S. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 2008, 455, 363–368. [Google Scholar]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis—Structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, D.; Chauvin, A.; Acosta, I.F.; Kurenda, A.; Wolfender, J.-L.; Stolz, S.; Chételat, A.; Farmer, E.E. Axial and radial oxylipin transport. Plant Physiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Royo, J.; Leon, J.; Vancanneyt, G.; Albar, J.P.; Rosahi, S.; Ortego, F.; Castanera, P.; Sanchez-Serrano, J.J. Antisense-mediated depletion of a potato lipoxygenase reduces wound induction of proteinase inhibitors and increases weight gain of insect pests. Proc. Natl. Acad. Sci. USA 1999, 96, 1146–1151. [Google Scholar] [CrossRef] [PubMed]
- Halitschke, R.; Baldwin, I.T. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003, 36, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.-B.; et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013, e1003964. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Qi, J.; Ren, N.; Cheng, J.; Erb, M.; Mao, B.; Lou, Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009, 60, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Nemchenko, A.; Borrego, E.; Murray, I.; Sobhy, I.S.; Bosak, L.; DeBlasio, S.; Erb, M.; Robert, C.A.M.; Vaughn, K.A.; et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013, 74, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, A.; Caldelari, D.; Wolfender, J.-L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.; Creelman, R.A.; Mullet, J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 8675–8679. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Dubugnon, L.; Mousavi, S.A.R.; Rudaz, S.; Wolfender, J.-L.; Farmer, E.E. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J. Biol. Chem. 2009, 284, 34506–34513. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Grata, E.; Dubugnon, L.; Rudaz, S.; Farmer, E.E.; Wolfender, J.-L. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 2008, 283, 16400–16407. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, M.A.; Stingl, N.E.; Lautenschlaeger, J.K.; Krischke, M.; Mueller, M.J.; Berger, S. Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 2010, 152, 1940–1950. [Google Scholar] [CrossRef] [PubMed]
- Zoeller, M.; Stingl, N.; Krischke, M.; Fekete, A.; Walker, F.; Berger, S.; Mueller, M.J. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: Biogenesis of pimelic and azelaic acid. Plant Physiol. 2012, 160, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.K.; Fahlberg, P.; Ellerstrom, M.; Andersson, M.X. Oxo-phytodienoic acid (OPDA) is formed on fatty acids esterified to galactolipids after tissue disruption in Arabidopsis thaliana. FEBS Lett. 2012, 586, 2483–2487. [Google Scholar] [CrossRef] [PubMed]
- Vellosillo, T.; Martinez, M.; Lopez, M.A.; Vicente, J.; Cascon, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007, 19, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, V.M.; Chételat, A.; Majcherczyk, P.; Farmer, E.E. Chloroplastic phosphoadenosine phosphosulfate (PAPS) metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 2010, 152, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.; Hause, G.; Kutter, C.; Miersch, O.; Wasternack, C. Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant Cell Physiol. 2003, 44, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Grebner, W.; Stingl, N.E.; Oenel, A.; Mueller, M.J.; Berger, S. Lipoxygenase6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013, 161, 2159–2170. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.; Mitchell-Olds, T.; Stotz, H.U. Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana. Physiol. Plant 2002, 114, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Allmann, S.; Halitschke, R.; Schuurink, R.C.; Baldwin, I.T. Oxylipin channelling in Nicotiana attenuata: Lipoxygenase 2 supplies substrates for green leaf volatile production. Plant Cell Environ. 2010, 33, 2028–2040. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Huang, M.-Y.; Palacio, K.; Schuler, M.A. Variations in CYP74B2 (Hydroperoxide lyase) gene expression differentially affect hexenal signaling in the Columbia and Landsberg erecta ecotypes of Arabidopsis. Plant Physiol. 2005, 139, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Yoa, C.; Henderson, Z.; Kim, S.; Braam, J. Arabidopsis touch-induced morphogenesis is jasmonate-mediated and protects against pests. Curr. Biol. 2012, 22, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Bearenfaller, K.; Loscos, J.; Chetelat, A.; Baginsky, S.; Farmer, E.E. Jasmonate controls polypeptide patterning in undamaged tissue in wounded Arabidopsis leaves. Plant Physiol. 2011, 156, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Ozalvo, R.; Cabrere, J.; Escobar, C.; Christensen, S.A.; Borrega, E.J.; Kolomiets, M.V.; Castresana, C.; Iberkleid, I.; Horowitz, S.B. Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol. Plant Pathol. 2014, 15, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Ren, N.; Qi, J.; Lu, J.; Xiang, C.; Ju, H.; Cheng, J.; Lou, Y. The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol. Plant 2014, 152, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Melan, M.A.; Dong, X.; Endara, M.E.; Davis, K.R.; Ausubel, F.; Peterman, T.K. An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 1993, 1001, 441–450. [Google Scholar] [CrossRef]
- Farmer, E.E.; Gasperini, D.; Acosta, I. The squeeze cell hypothesis for the activation of jasmonate synthesis in response to wounding. New Phytol. 2014, 204, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2011, 75, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Halitschlke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B.; Wolkenfelt, H.; Terlouw, M.; Lawson, E.; Dean, C.; Weisbeek, P. Embryonic Origin of the Arabidopsis Primary Root and Root-Meristem Initials. Development 1994, 120, 2475–2487. [Google Scholar]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chauvin, A.; Lenglet, A.; Wolfender, J.-L.; Farmer, E.E. Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis. Plants 2016, 5, 16. https://doi.org/10.3390/plants5020016
Chauvin A, Lenglet A, Wolfender J-L, Farmer EE. Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis. Plants. 2016; 5(2):16. https://doi.org/10.3390/plants5020016
Chicago/Turabian StyleChauvin, Adeline, Aurore Lenglet, Jean-Luc Wolfender, and Edward E. Farmer. 2016. "Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis" Plants 5, no. 2: 16. https://doi.org/10.3390/plants5020016
APA StyleChauvin, A., Lenglet, A., Wolfender, J.-L., & Farmer, E. E. (2016). Paired Hierarchical Organization of 13-Lipoxygenases in Arabidopsis. Plants, 5(2), 16. https://doi.org/10.3390/plants5020016





