Dead Pericarps of Dry Fruits Function as Long-Term Storage for Active Hydrolytic Enzymes and Other Substances That Affect Germination and Microbial Growth
Abstract
:1. Introduction
2. Methodology
2.1. Plant Material and Growth Conditions
2.2. In-Gel Nuclease Assay
2.3. In-Gel Chitinase Assay
2.4. In-Gel Protease Assay
2.5. Proteome Analysis
2.6. Nutrient Analysis
2.7. Antibacterial Assay
2.8. Seed Germination Assays
3. Results
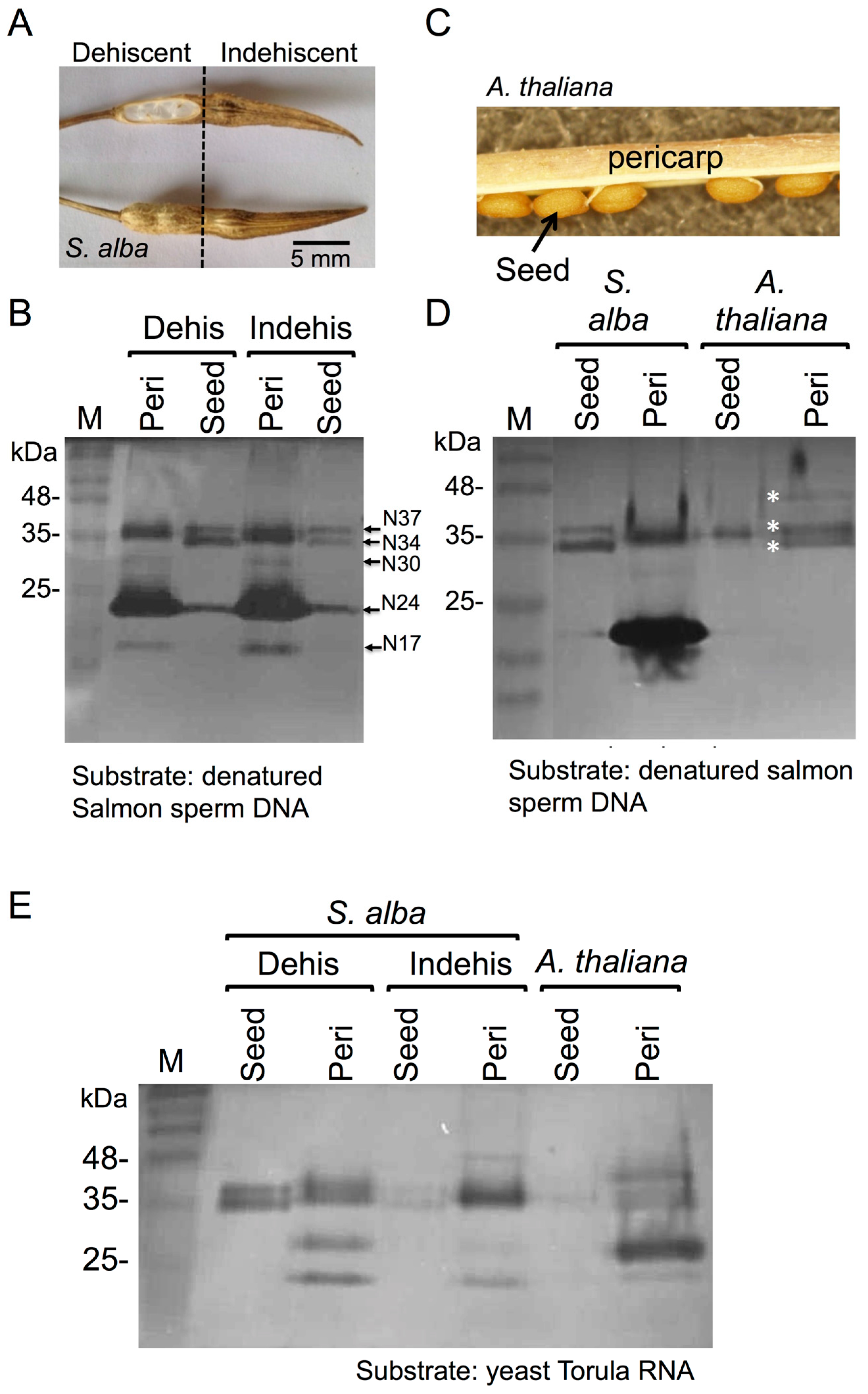
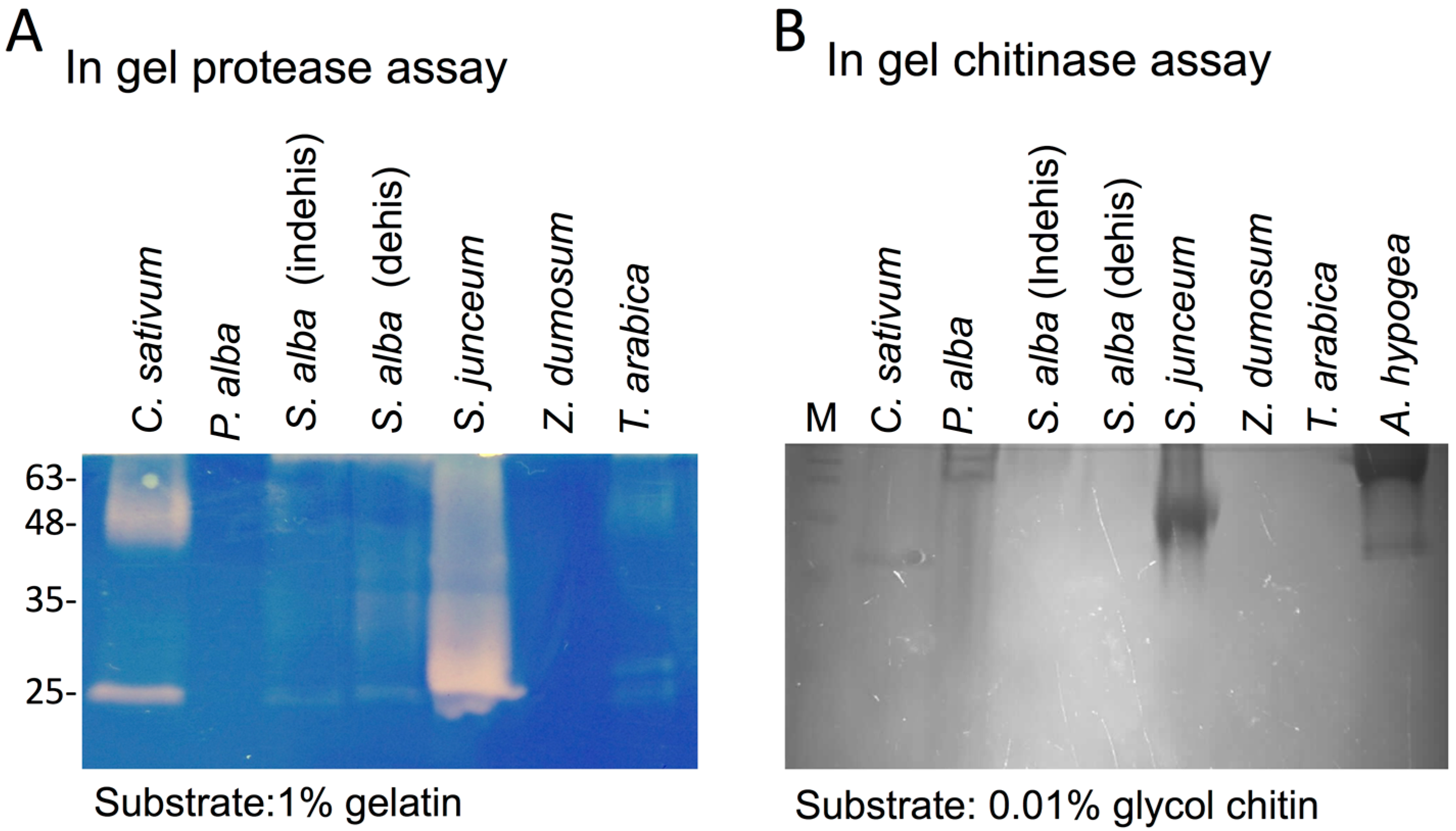
3.1. The Dead Pericarps of Dry Fruits Accumulate Active Hydrolytic Enzymes
3.2. Proteome Analysis of S. alba Indehiscent Pericarps
3.3. The Dead Pericarps of Arabidopsis and Sinapis Accumulate Nutrients
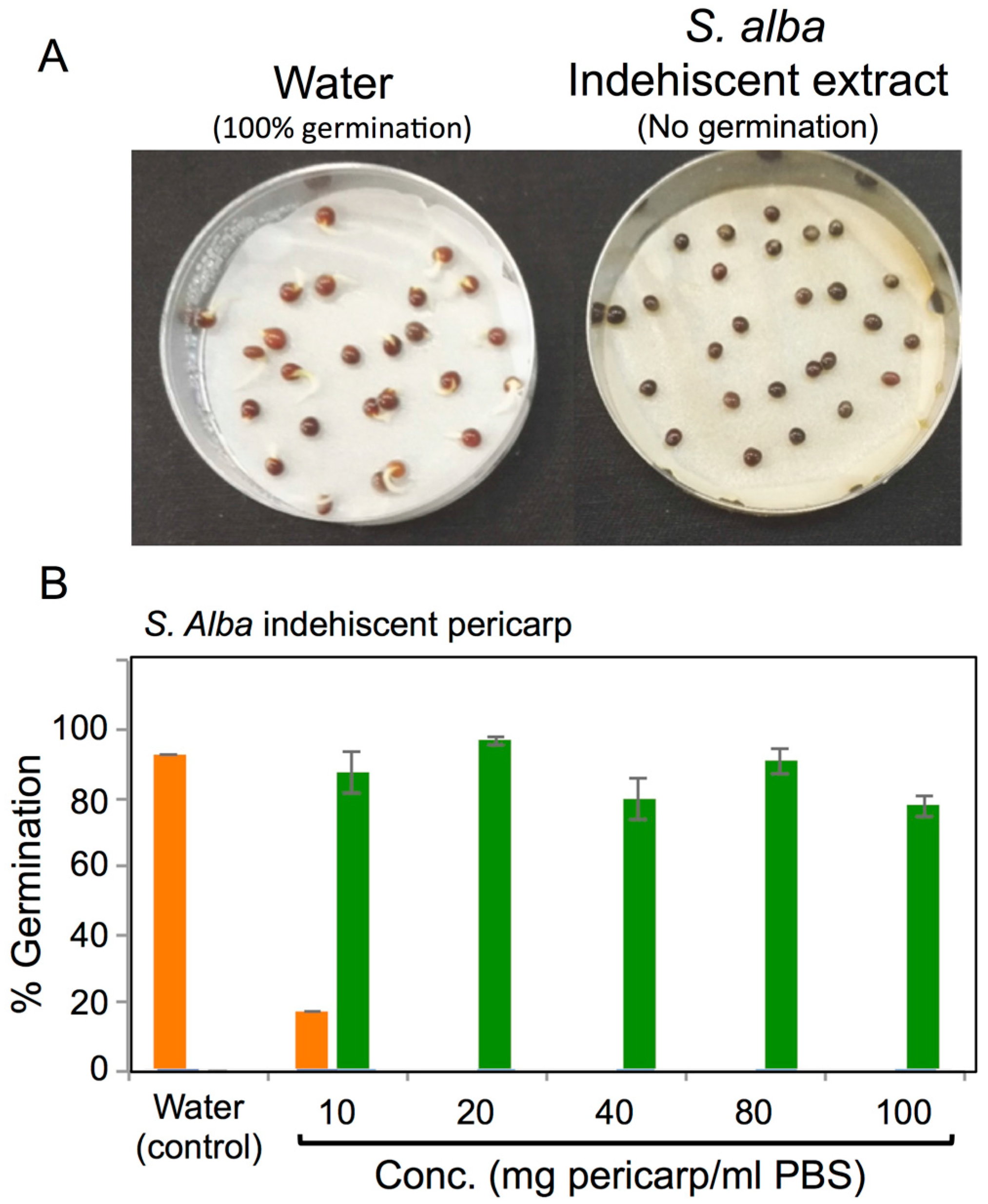
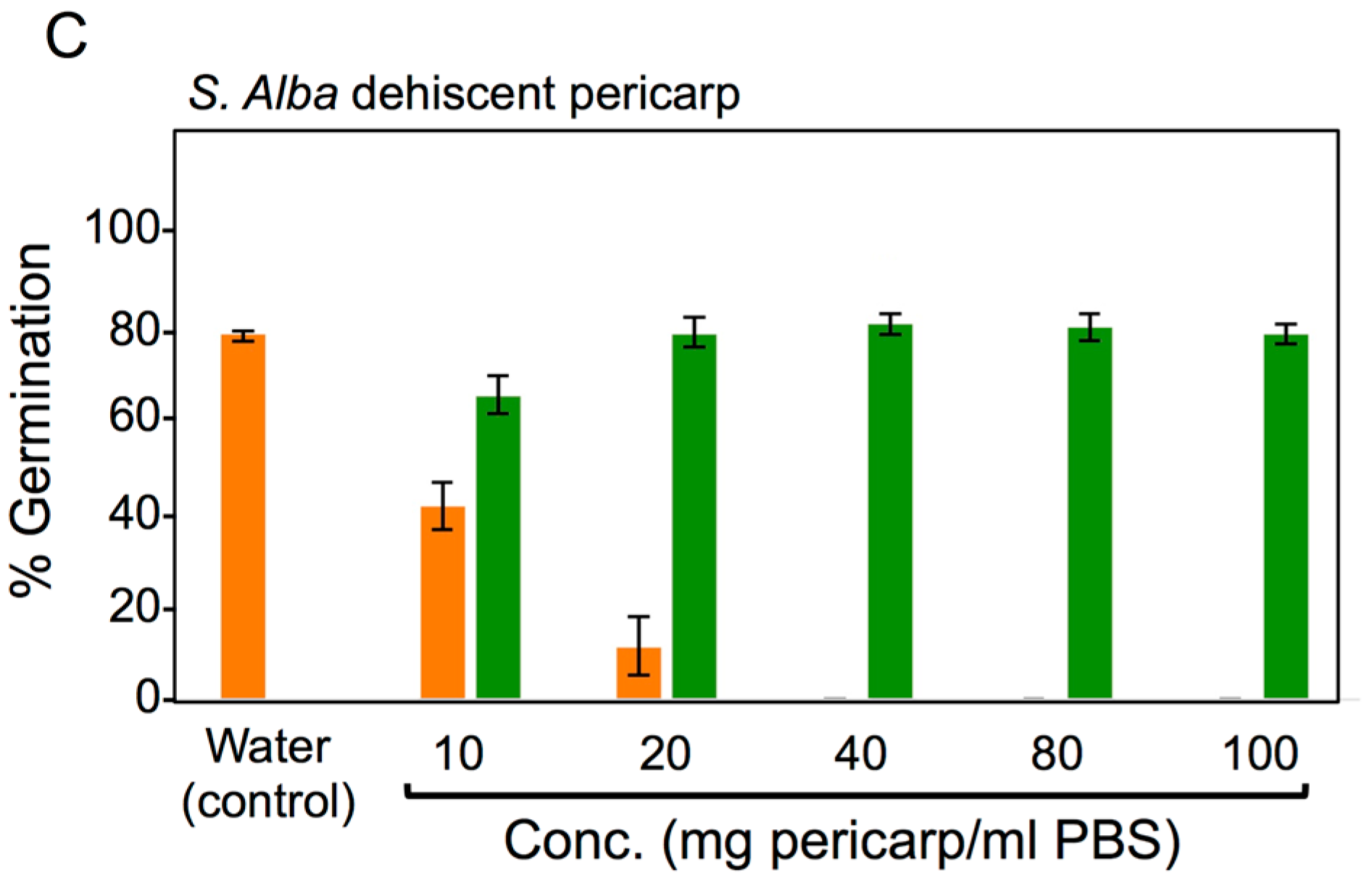
3.4. Effects of Pericarps on S. alba Seed Germination
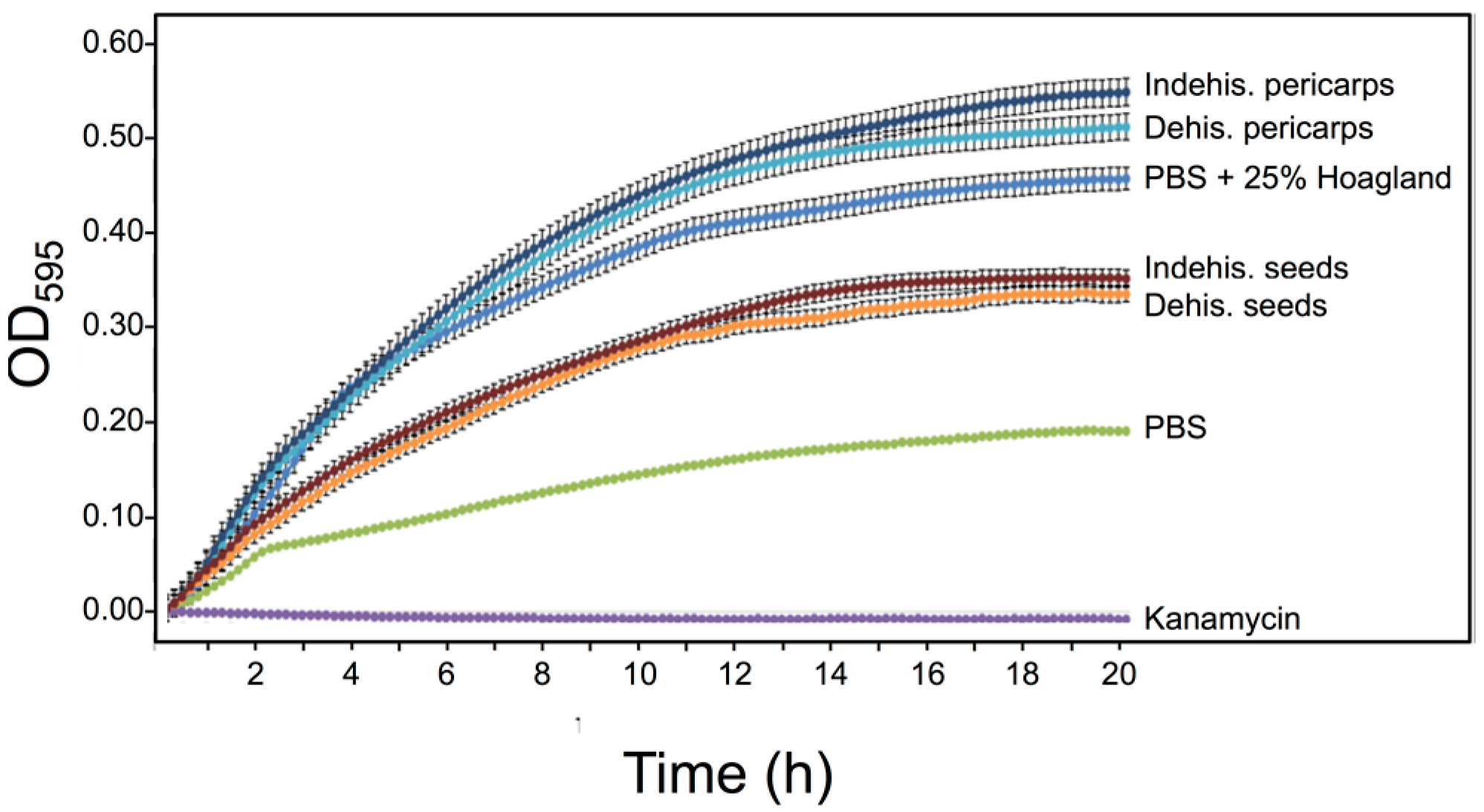
3.5. S. alba Pericarp Extracts Promote Bacterial Growth
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Erikkson, O. Evolution of seed size and biotic seed dispersal in angiosperms: Paleoecological and neoecological evidence. Int. J. Plant Sci. 2008, 169, 863–870. [Google Scholar] [CrossRef]
- Howe, H.F.; Smallwood, J. Ecology of seed dispersal. Ann. Rev. Ecol. Syst. 1982, 13, 201–228. [Google Scholar] [CrossRef]
- Charles, B.B. An Introduction to Plant Structure and Development; Cambridge University Press: Cambridge, UK, 2010; pp. 377–380. [Google Scholar]
- Dominguez, F.; Cejudo, F.J. Programmed cell death (PCD): An essential process of cereal seed development and germination. Front. Plant Sci. 2014, 5, 366. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.M. Seed Dormancy in Grasses; Cambridge University Press: New York, NY, USA, 1990. [Google Scholar]
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Ogawa, K.; Iwabuchi, M. A mechanism for promoting the germination of Zinnia elegans seeds by hydrogen peroxide. Plant Cell Physiol. 2001, 42, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Sari, A.L.I.O.; Oguz, B.; Bilgic, A. Breaking seed dormancy of laurel (Laurus nobilis L.). New For. 2006, 31, 403–408. [Google Scholar] [CrossRef]
- Takos, I.A. Seed dormancy in bay laurel (Laurus nobilis L.). New For. 2001, 21, 105–114. [Google Scholar] [CrossRef]
- Valio, I.F.M. Inhibition of germination of coffee seeds (Coffea arabica L. cv. Mundo novo) by the endocarp. J. Seed Technol. 1980, 5, 32–39. [Google Scholar]
- Thapliyal, R.C.; Naithani, K.C. Inhibition of germination in Nyctanthes arbortristis (Olaceae) by pericarp. Seed Sci. Technol. 1996, 24, 67–73. [Google Scholar]
- Hu, X.W.; Wang, Y.R.; Wu, Y.P. Effects of the pericarp on imbibition, seed germination, and seedling establishment in seeds of Hedysarum scoparium Fisch. et Mey. Ecol. Res. 2009, 24, 559–564. [Google Scholar] [CrossRef]
- Lu, J.J.; Zhou, Y.M.; Tan, D.Y.; Baskin, J.M.; Baskin, C.C. Seed dormancy in six cold desert Brassicaceae species with indehiscent fruits. Seed Sci. Res. 2015, 25, 276–285. [Google Scholar] [CrossRef]
- Chacón, P.; Bustamante, R.O. Effect of seed size and pericarp on seedling recruitment and biomass in Cryptocarya alba (Lauraceae) under two contrasting moisture regimes. Plant Ecol. 2001, 152, 137–144. [Google Scholar] [CrossRef]
- Maruta, E.; Kamitani, T.; Okabe, M.; Ide, Y. Desiccation-tolerance of Fagus crenata Blume seeds from localities of different snowfall regimes in central Japan. J. For. Res. 1997, 2, 45–50. [Google Scholar] [CrossRef]
- Iluz, D. Zoochory: The dispersal of plants by animals. In All Flesh Is Grass: Plant—Animal Interrelationships; Seek, J., Dubinsky, Z., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 201–214. [Google Scholar]
- Raviv, B.; Granot, G.; Chalifa-Caspi, V.; Grafi, G. The dead, hardened floral bracts of dispersal units of wild wheat function as storage for active hydrolases and in enhancing seedling vigor. PLoS ONE 2017, 12, e0177537. [Google Scholar] [CrossRef] [PubMed]
- Raviv, B.; Aghajanyan, L.; Granot, G.; Makover, V.; Frenkel, O.; Gutterman, Y.; Grafi, G. The dead seed coat functions as a long-term storage for active hydrolytic enzymes. PLoS ONE 2017, 12, e0181102. [Google Scholar] [CrossRef] [PubMed]
- Blank, A.; Sugiyama, R.H.; Dekker, C.A. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate-polyacrylamide gel electrophoresis: Use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 1982, 120, 267–275. [Google Scholar] [CrossRef]
- Trudel, J.; Asselin, A. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal. Biochem. 1989, 178, 362–366. [Google Scholar] [CrossRef]
- Solomon, M.; Belenghi, B.; Delledonne, M.; Menachem, E.; Levine, A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 1999, 11, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Patton, T.; Barrett, J.; Brennan, J.; Moran, N. Use of a spectrophotometric bioassay for determination of microbial sensitivity to manuka honey. J. Microbiol. Methods 2006, 64, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Kaeppler, S.M.; Lynch, J.P. Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 2005, 270, 299–310. [Google Scholar] [CrossRef]
- Bhatt, A.; Santo, A. Germination and recovery of heteromorphic seeds of Atriplex canescens (Amaranthaceae) under increasing salinity. Plant Ecol. 2016, 217, 1069–1079. [Google Scholar] [CrossRef]
- Sroelov, R. On germination inhibitors. IV. Germination inhibitors of Sinapis alba and other seeds when enclosed in their fruit. Palestine J. Bot. 1940, 2, 33–45. [Google Scholar]
- Harper, S.H.T.; Lynch, J.M. Microbial effects on the germination and seedling growth of barley. New Phytol. 1980, 84, 473–481. [Google Scholar] [CrossRef]
- Leśniewicz, K.; Pieńkowska, J.; Poręba, E. Characterization of nucleases involved in seedling development of cauliflower. J. Plant Physiol. 2010, 167, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Bambara, R.A. Flap endonuclease 1. Annu. Rev. Biochem. 2013, 82, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Granot, G.; Morgenstern, Y.; Khan, A.; Rapp, Y.G.; Pesok, A.; Nevo, E.; Grafi, G. Internucleosomal DNA fragmentation in wild emmer wheat is catalyzed by S1-type endonucleases translocated to the nucleus upon induction of cell death. Biochim. Biophys. Acta 2015, 1849, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Givaty-Rapp, Y.; Yadav, N.S.; Khan, A.; Grafi, G. S1-type endonuclease 2 in dedifferentiating Arabidopsis protoplasts: Translocation to the nucleus in senescing protoplasts is associated with de-glycosylation. PLoS ONE 2017, 12, e0170067. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Ito, J.; Aoyagi, S.; Fukuda, H. Endonucleases. Plant Mol. Biol. 2000, 44, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Beard, P.; Morrow, J.F.; Berg, P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J. Virol. 1973, 12, 1303–1313. [Google Scholar] [PubMed]
- Grafi, G.; Larkins, B.A. Activity of single-stranded DNA endonucleases in mung bean is associated with cell division. Plant Mol. Biol. 1995, 29, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Gartemann, K.H.; Kirchner, O.; Engemann, J.; Gräfen, I.; Eichenlaub, R.; Burger, A. Clavibacter michiganensis subsp. michiganensis: First steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J. Biotechnol. 2003, 106, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Galiana, E.; Bonnet, P.; Conrod, S.; Keller, H.; Panabières, F.; Ponchet, M.; Poupet, A.; Ricci, P. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol. 1997, 115, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Hugot, K.; Ponchet, M.; Marais, A.; Ricci, P.; Galiana, E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Mol. Plant Microbe Interact. 2002, 15, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Bertini, L.; Caporale, C.; Testa, M.; Proietti, S.; Caruso, C. Structural basis of the antifungal activity of wheat PR4 proteins. FEBS Lett. 2009, 583, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sharma, K.P.; Gaur, R.K.; Gupta, V.K. Role of chitinase in plant defense. Asian J. Biochem. 2011, 6, 29–37. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Ceasar, S.A.; Ignacimuthu, S. Genetic engineering of crop plants for fungal resistance: Role of antifungal genes. Biotechnol. Lett. 2012, 34, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Müntz, K.; Belozersky, M.A.; Dunaevsky, Y.E.; Schlereth, A.; Tiedemann, J. Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. J. Exp. Bot. 2001, 52, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoorn, R.A. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Liberman, L.M. Beneficial Microbes Affect Endogenous Mechanisms Controlling Root Development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 2004, 14, 93–107. [Google Scholar] [CrossRef]
- Richards, S.L.; Wilkins, K.A.; Swarbreck, S.M.; Anderson, A.A.; Habib, N.; Smith, A.G.; McAinsh, M.; Davies, J.M. The hydroxyl radical in plants: From seed to seed. J. Exp. Bot. 2015, 66, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Jeevan Kumar, S.P.; Rajendra Prasad, S.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; Barba-Espín, G.; Hernández, J.A. Elucidating hormonal/ROS networks during seed germination: Insights and perspectives. Plant Cell Rep. 2013, 32, 1491–1502. [Google Scholar] [CrossRef] [PubMed]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef] [PubMed]
- Kohli, P.; Kalia, M.; Gupta, R. Pectin Methylesterases: A Review. J. Bioprocess. Biotech. 2015, 5, 227. [Google Scholar]
- Wurzburger, J.; Leshem, Y. Physiological action of the germination inhibitor in the husk of Aegilops kotschyi boiss. New Phytol. 1969, 68, 337–341. [Google Scholar] [CrossRef]
- Booth, D.T. Seedbed ecology of winterfat: Cations in diaspore bracts and their effect on germination and early plant growth. J. Range Manag. 1989, 42, 178–182. [Google Scholar] [CrossRef]
- Drew, M.C. Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol. 1975, 75, 479–490. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 2000, 51, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitraterich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef] [PubMed]
- Monaco, T.A.; Mackown, C.T.; Johnson, D.A.; Jones, T.A.; Norton, J.M.; Norton, J.B.; Redinbaugh, M.G. Nitrogen effects on seed germination and seedling growth. J. Range Manag. 2003, 56, 646–653. [Google Scholar] [CrossRef]
- Shim, S.I.; Moon, J.C.; Jang, C.S.; Raymer, P.; Kim, W. Effect of potassium nitrate priming on seed germination of seashore paspalum. Hortscience 2008, 43, 2259–2262. [Google Scholar]
- Bian, L.; Yang, L.; Wang, J.A.; Shen, H.L. Effects of KNO3 pretreatment and temperature on seed germination of Sorbus pohuashanensis. J. For. Res. 2013, 24, 309–316. [Google Scholar] [CrossRef]
- Song, W.; Liu, S.; Meng, L.; Xue, R.; Wang, C.; Liu, G.; Dong, C.; Wang, S.; Dong, J.; Zhang, Y. Potassium deficiency inhibits lateral root development in tobacco seedlings by changing auxin distribution. Plant Soil 2015, 396, 163–173. [Google Scholar] [CrossRef]
- Leigh, R.A.; Wynn Jones, R.G. A hypothesis relating critical potassium concentrations for growth and distribution and functions of this ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Tisdale, S.L.; Nelson, W.L.; Beaton, J.D.; Havlin, J.L. Soil Fertility and Fertilizers; Macmillan: New York, NY, USA, 1993; pp. 176–229. [Google Scholar]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godwin, J.; Raviv, B.; Grafi, G. Dead Pericarps of Dry Fruits Function as Long-Term Storage for Active Hydrolytic Enzymes and Other Substances That Affect Germination and Microbial Growth. Plants 2017, 6, 64. https://doi.org/10.3390/plants6040064
Godwin J, Raviv B, Grafi G. Dead Pericarps of Dry Fruits Function as Long-Term Storage for Active Hydrolytic Enzymes and Other Substances That Affect Germination and Microbial Growth. Plants. 2017; 6(4):64. https://doi.org/10.3390/plants6040064
Chicago/Turabian StyleGodwin, James, Buzi Raviv, and Gideon Grafi. 2017. "Dead Pericarps of Dry Fruits Function as Long-Term Storage for Active Hydrolytic Enzymes and Other Substances That Affect Germination and Microbial Growth" Plants 6, no. 4: 64. https://doi.org/10.3390/plants6040064



