Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals
Abstract
:1. Introduction
2. Comparative Analysis of the Primary Structure of Defensins Isolated from Wild and Cultivated Cereals
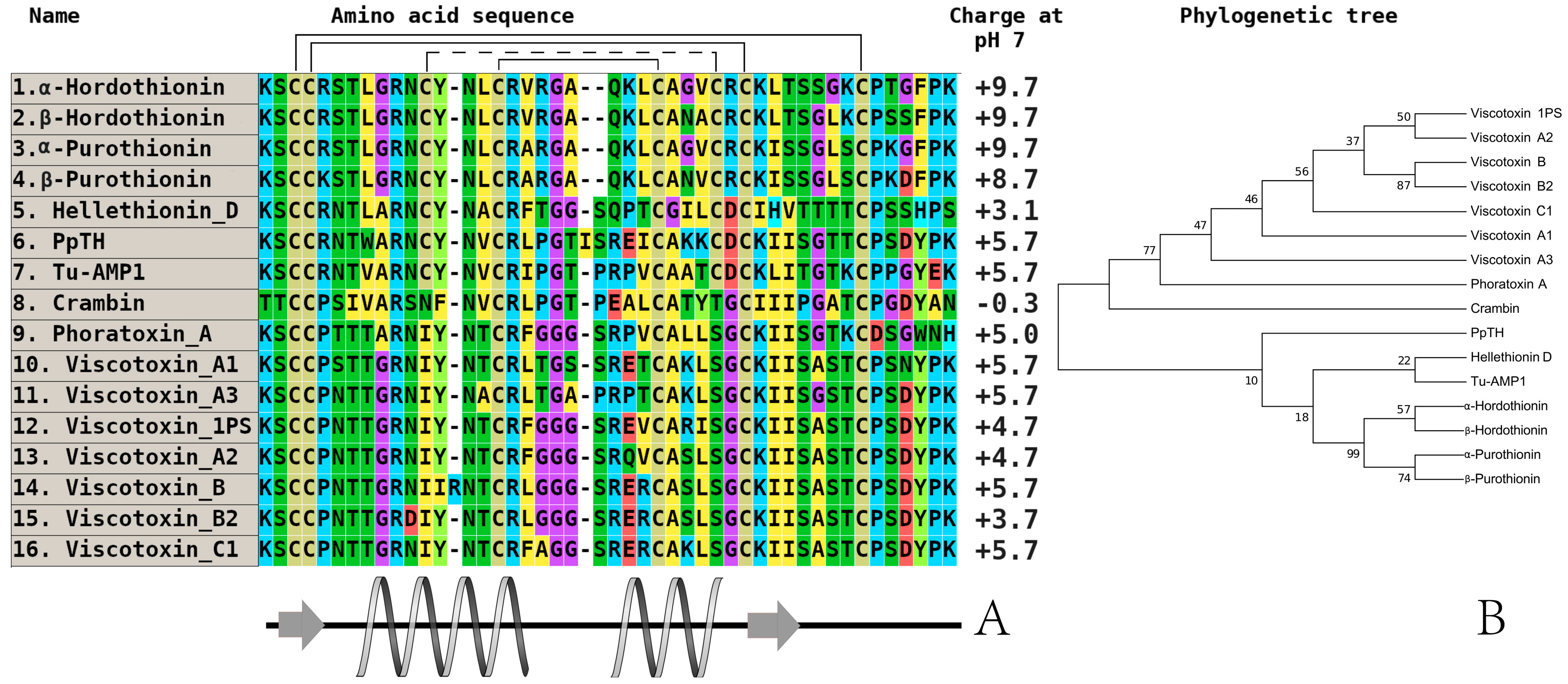
3. Investigation of Structural Determinants of Other AMPs (Thionins, Hevein-Like Peptides, and Alpha-Hairpinins), Which Provide Higher Antifungal Activity to Wild Cereals
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, F.; Cheng, Z. Research Progress on the use of Plant Allelopathy in Agriculture and the Physiological and Ecological Mechanisms of Allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Iqbal, J.; Ullah, A.; Yang, G.; Yousaf, M.; Fahad, S.; Tanveer, M.; Hassan, W.; Tung, S.A.; Wang, L.; et al. Allelopathic potential of oil seed crops in production of crops: A review. Environ. Sci. Pollut. Res. Int. 2016, 15, 14854–14867. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Baek, K.H. Physiological and biochemical perspectives of non-salt tolerant plants during bacterial interaction against soil salinity. Plant Physiol. Biochem. 2017, 116, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, U.C.; Bohra, A.; Jha, R. Breeding approaches and genomics technologies to increase crop yield under low-temperature stress. Plant Cell Rep. 2017, 36, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Mesihovic, A.; Iannacone, R.; Firon, N.; Fragkostefanakis, S. Heat stress regimes for the investigation of pollen thermotolerance in crop plants. Plant Reprod. 2016, 29, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burrit, D.J.; Tran, L.P. The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- de Souza Cândido, E.; Sousa, D.A.; Viana, J.C.; de Oliveira-Júnior, N.G.; Miranda, V.; Franco, O.L. The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides 2014, 55, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yuan, S.S.; Jiang, L.L.; Ye, X.J.; Ng, T.B.; Wu, Z.J. Plant antifungal proteins and their applications in agriculture. Appl. Microbiol. Biotechnol. 2015, 99, 4961–4981. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.S.; Prodhan, Z.H.; Biswas, S.K.; Le, C.F.; Sekaran, S.D. Antimicrobial peptides from different plant sources: Isolation, characterisation, and purification. Phytochemistry 2018, 154, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.M.; Liu, P.Q.; Xu, Y.J.; Xiao, S. Protein trafficking during plant innate immunity. J. Integr. Plant Biol. 2016, 58, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Andreev, Y.A.; Korostyleva, T.V.; Slavokhotova, A.A.; Rogozhin, E.A.; Utkina, L.L.; Vassilevski, A.A.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. Genes encoding hevein-like defense peptides in wheat: Distribution, evolution, and role in stress response. Biochimie 2012, 94, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Rogozhin, E.A.; Musolyamov, A.K.; Andreev, Y.A.; Oparin, P.B.; Berkut, A.A.; Vassilevski, A.A.; Egorov, T.A.; Grishin, E.V.; Odintsova, T.I. Novel antifungal α-hairpinin peptide from Stellaria media seeds: Structure, biosynthesis, gene structure and evolution. Plant Mol. Biol. 2014, 84, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Lobo, D.S.; Pereira, I.B.; Fragel-Madeira, L.; Medeiros, L.N.; Cabral, L.M.; Faria, J.; Bellio, M.; Campos, R.C.; Linden, R.; Kurtenbach, E. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 2007, 46, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.A.; Silverstein, K.A.; Cannon, S.B.; VandenBosch, K.A. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004, 135, 1179–1197. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, A.K.; Brunstedt, J.; Nielsen, J.E.; Mikkelsen, J.D.; Roepstorff, P.; Nielsen, K.K. Processing, disulfide pattern, and biological activity of a sugar beet defensin, AX2, expressed in Pichia pastoris. Protein Exp. Purif. 1999, 16, 377–387. [Google Scholar] [CrossRef] [PubMed]
- de Zélicourt, A.; Letousey, P.; Thoiron, S.; Campion, C.; Simoneau, P.; Elmorjani, K.; Marion, D.; Simier, P.; Delavault, P. Ha-DEF1, a sunflower defensin, induces cell death in Orobanche parasitic plants. Planta 2007, 226, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Do, H.M.; Lee, S.C.; Jung, H.W.; Sohn, K.H.; Hwang, B.K. Differential expression and in sutu localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum anuum. Plant Sci. 2004, 166, 1297–1305. [Google Scholar]
- Fujiumura, M.; Ideguchi, M.; Minami, Y.; Watanabe, K.; Tadera, K. Purification, characterization and sequencing of novel antimicrobial peptides Tu-AMP1 and Tu-AMP2 from bulbs of tulip (Tulipa gesneriana L.). Biosci. Biotechnol. Biochem. 2004, 63, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.E.; Bassett, C.L.; Artlip, T.S.; Webb, R.P.; Janisiewicz, W.J.; Norelli, J.L. Characterization of a defensin from bark and fruit tissues of peach and antimicrobial activity of a recombinant defensin in the yeast, Pichia pastoris. Physiol. Plant. 2003, 119, 563–572. [Google Scholar] [CrossRef]
- Bloch, C.J.; Richardson, M.A. A new family of small (5 kDa) protein inhibitors of insect α-amylases from seeds of sorghum (Sorghum bicolor (L.) Moebch.) have sequence homologies with wheat γ-purothionins. FEBS J. 1991, 279, 101–104. [Google Scholar] [CrossRef]
- Mendez, E.; Moreno, A.; Colilla, F.; Pelaez, F.; Limas, G.G.; Mendez, R. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, γ-hordothionin, from barley endosperm. Eur. J. Biochem. 1990, 194, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Mendez, E.; Rocher, A.; Calero, M.; Girbes, T.; Citores, L.; Soriano, F. Primary structure of ω-hordothionin, a member of a novel family of thionins from barley endosperm, and its inhibition of protein synthesis in eukaryotic and prokaryotic cell-free systems. Eur. J. Biochem. 1996, 239, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.; Moreno, M.; Molina, A.; Garcia-Olmedo, F. Novel defensin subfamily from spinach (Spinacea oleracea). FEBS Lett. 1998, 435, 159–162. [Google Scholar] [CrossRef]
- Sharma, P.; Lönneborg, A. Isolation and characterization of a cDNA encoding a plant defensin-like protein from roots of Norway spruce. Plant Mol. Biol. 1996, 31, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-H.; Jian, G.-L.; Zhang, Y.-T.; Ai, T.-M. Bacterial expression of a Trichosanthes lirilowii defensin (TDEF1) and its antifungal activity on Fusarium oxysporum. Appl. Microbiol. Biotechnol. 2007, 28, 62–75. [Google Scholar]
- Colilla, F.J.; Rocher, A.; Mendez, E. Gamma-purothionins: Amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 1990, 270, 191–194. [Google Scholar] [CrossRef]
- Vriens, K.; Cammue, B.P.; Thevissen, K. Antifungal plant defensins: Mechanisms of action and production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.; Osborn, R.W. Plant defensins: Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995, 108, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.F.; Vasconcelos, E.A.; Pelegrini, P.B.; Grossi de Sa, M.F. Antifungal defensins and their role in plant defense. Front. Microbiol. 2014, 5, 116. [Google Scholar] [CrossRef] [PubMed]
- Thevissen, K.; Ferket, K.K.; François, I.E.; Cammue, B.P. Interactions of antifungal plant defensins with fungal membrane components. Peptides 2003, 24, 1705–1712. [Google Scholar] [CrossRef] [PubMed]
- Thevissen, K.; Warnecke, D.C.; François, I.E.; Leipelt, M.; Heinz, E.; Ott, C.; Zähringer, U.; Thomma, B.P.; Ferket, K.K.; Cammue, B.P. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 2004, 279, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Egorov, T.A.; Odintsova, T.I.; Pukhalsky, V.A.; Grishin, E.V. Diversity of wheat antimicrobial peptides. Peptides 2005, 26, 2064–2073. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.I.; Egorov, T.A.; Musolyamov, A.K.; Odintsova, M.S.; Pukhalsky, V.A.; Grishin, E.V. Seed defensins from T. kiharae and related species: Genome localization of defensin-encoding genes. Biochimie 2007, 89, 605–612. [Google Scholar] [PubMed]
- Odintsova, T.I.; Rogozhin, E.A.; Baranov, Y.; Musolyamov, A.K.; Yalpani, N.; Egorov, T.A.; Grishin, E.V. Seed defensins of barnyard grass Echinochloa crusgalli (L.) Beauv. Biochimie 2008, 90, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Osborn, R.W.; De Samblanx, G.W.; Thevissen, K.; Goderis, I.; Torrekens, S.; Van Leuven, F. Isolation and characterization of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett. 1995, 368, 257–262. [Google Scholar] [CrossRef]
- Terras, F.R.G.; Schofs, H.M.E.; de Bolle, M.F.C.; Van Leuven, F.; Rees, S.B. In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid lipid transfer proteins. Plant Physiol. 1992, 100, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- De Samblanx, G.W.; Goderis, I.J.; Thevissen, K.; Raemaekers, R.; Fant, F.; Borremans, F.; Acland, D.P.; Osborn, R.W.; Patel, S.; Broekaert, W.F. Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J. Biol. Chem. 1997, 272, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jin, X.; Zhao, Y.; Dong, Q.; Jiang, H.; Ma, Q. Evolution of the defensin-like gene family in grass genomes. J. Genet. 2016, 95, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Shelenkov, A.A.; Odintsova, T.I. Prediction of Leymus arenarius (L.) antimicrobial peptides based on de novo transcriptome assembly. Plant Mol. Biol. 2015, 89, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Bagnaresi, P.; Biselli, C.; Orru’, L.; Amaral Carneiro, G.; Siciliano, I.; Valé, G.; Gullino, M.L.; Spadaro, D. Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genomics 2016, 17, 608. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, P.; Ma, L.; Wang, H.; Borhan, M.H. Genome-wide transcriptomic analyses provide insights into the lifestyle transition and effector repertoire of Leptosphaeria maculans during the colonization of Brassica napus seedlings. Mol. Plant Pathol. 2016, 17, 1196–1210. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.S.; Rajagopalan, N.; Risseeuw, E.P.; Surpin, M.; Ball, F.J.; Barber, C.J.; Buhrow, L.M.; Clark, S.M.; Page, J.E.; Todd, C.D.; et al. Characterization of Triticum aestivum Abscisic Acid Receptors and a Possible Role for These in Mediating Fusairum Head Blight Susceptibility in Wheat. PLoS ONE 2016, 11, e0164996. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Shelenkov, A.A.; Korostyleva, T.V.; Rogozhin, E.A.; Melnikova, N.V.; Kudryavtseva, A.V.; Odintsova, T.I. Defense peptide repertoire of Stellaria media predicted by high throughput next generation sequencing. Biochimie 2017, 135, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Alamillo, J.M.; Garcia-Olmedo, F. Processing of thionin precursors in barley leaves by a vacuolar proteinase. Eur. J. Biochem. 1997, 243, 202–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florack, D.E.; Stiekema, W.J. Thionins: Properties, possible biological roles and mechanisms of action. Plant Mol. Biol. 1994, 26, 25–37. [Google Scholar] [CrossRef] [PubMed]
- García-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodríguez-Palenzuéla, P. Plant defense peptides. Biopolymers 1998, 47, 479–491. [Google Scholar] [CrossRef]
- Bruix, M.; Jiménez, M.A.; Santoro, J.; González, C.; Colilla, F.J.; Méndez, E.; Rico, M. Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: A structural motif common to toxic arthropod proteins. Biochemistry 1993, 32, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Stec, B.; Rao, U.; Teeter, M.M. Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 2: Structure of beta-purothionin at 1.7 A resolution. Acta Crystallogr. D Biol. Crystallogr. 1995, 51, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Kim, E.; Teeter, M.M.; Suh, S.W.; Stec, B. Crystal structure of alpha-hordothionin at 1.9 Angstrom resolution. FEBS Lett. 2005, 579, 2301–2306. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Debreczeni, J.E.; Sevvana, M.; Gruene, T.; Kahle, B.; Zeeck, A.; Sheldrick, G.M. Structures of viscotoxins A1 and B2 from European mistletoe solved using native data alone. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Reimann-Philipp, U.; Schrader, G.; Martinoia, E.; Barkholt, V.; Apel, K. Intracellular thionins of barley. A second group of leaf thionins closely related to but distinct from cell wall-bound thionins. J. Biol. Chem. 1989, 264, 8978–8984. [Google Scholar] [PubMed]
- Stec, B. Plant thionins--the structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Gheysen, G.; Ullah, C.; Verbeek, R.; Shang, C.; Vleesschauwer, D.D.; Höfte, M.; Kyndt, T. The role of thionins in rice defence against root pathogens. Mol. Plant Pathol. 2015, 16, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Zasukhina, G.D.; Odintsova, T.I.; Shulenina, L.V.; Ushenkova, L.N.; Mikhailov, V.F.; Shagirova, Z.M.; Vedernikov, A.N.; Gromov, S.P.; Alfimov, M.V. Antimutagens (β-purothionin and crown compound) as modulators of expression of genes involved in carcinogenesis in human cells. Dokl. Biochem. Biophys. 2012, 446, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Zasukhina, G.D.; Vasilyeva, I.M.; Kadnikov, I.A.; Voronin, M.V.; Odintsova, T.I.; Korostileva, T.V.; Pukhalskii, V.A. Antimutagenic activity of wheat polypeptides in human cells exposed to cadmium chloride. Bull. Exp. Biol. Med. 2013, 155, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Zasukhina, G.D.; Shagirova, J.M.; Babintsev, M.V.; Vasilyeva, I.M.; Rogozhin, E.A.; Odintsova, T.I.; Mikhailov, V.F.; Gromov, S.P.; Vedernikov, A.I.; Alfimov, M.V. Modulation of gene expression by antimutagens in human cells differing in the sensitivity to mutagens. Dokl. Biochem. Biophys. 2013, 453, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.I.; Vasil’eva, I.M.; Korostyleva, T.V.; Utkina, L.L.; Slavokhotova, A.A.; Rogozhin, E.A.; Shiian, A.N.; Pukhal’skiĭ, V.A.; Zasukhina, G.D. Antimutagenic activity of wheat beta-purothionin Tk-AMP-BP. Russ. J. Genet. 2011, 47, 1267–1270. [Google Scholar] [CrossRef]
- Pelegrini, P.B.; Franco, O.L. Plant gamma-thionins: Novel insights on the mechanism of action of a multi-functional class of defense proteins. Int. J. Biochem. Cell. Biol. 2005, 37, 2239–2253. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, V.F.; Shishkina, A.A.; Vasilyeva, I.M.; Shulenina, L.V.; Raeva, N.F.; Rogozhin, E.A.; Startsev, M.I.; Zasukhina, G.D.; Gromov, S.P.; Alfimov, M.V. Comparative analysis of natural and synthetic antimutagens as regulators of gene expression in human cells under exposure to ionizing radiation. Russ. J. Genet. 2015, 51, 147–155. [Google Scholar] [CrossRef]
- Kul’ko, A.B.; Kisil’, O.V.; Sadykova, V.S.; Mikhailov, V.F.; Vasilyeva, I.M.; Shulenina, L.V.; Zasukhina, G.D.; Rogozhin, E.A. Investigation of thionins from blackseed (Nigella sativa L.) possess cytotoxic, regulatory and antifungal activity. Antibiotiki I khimioterapiya 2016, 61, 8–16. (In Russian) [Google Scholar]
- Vasilchenko, A.S.; Smirnov, A.N.; Zavriev, S.K.; Grishin, E.V.; Vasilchenko, A.V.; Rogozhin, E.A. Novel thionins from black seed (Nigella sativa L.) demonstrate antimicrobial activity. Int. J. Pept. Res. Ther. 2017, 23, 171–180. [Google Scholar] [CrossRef]
- Giudici, A.M.; Regente, M.C.; Villalaín, J.; Pfüller, K.; Pfüller, U.; De La Canal, L. Mistletoe viscotoxins induce membrane permeabilization and spore death in phytopathogenic fungi. Physiol. Plant. 2004, 121, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Rodríguez, J.J.; Ochoa-Zarzosa, A.; López-Gómez, R.; López-Meza, J.E. Plant antimicrobial peptides as potential anticancer agents. Biomed. Res. Int. 2015, 2015, 735087. [Google Scholar] [CrossRef] [PubMed]
- Teeter, M.M.; Ma, X.Q.; Rao, U.; Whitlow, M. Crystal structure of a protein-toxin alpha 1-purothionin at 2.5A and a comparison with predicted models. Proteins 1990, 8, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Loeza-Ángeles, H.; Sagrero-Cisneros, E.; Lara-Zárate, L.; Villagόmes-Gόmez, E.; Lόpez-Meza, J.E.; Ochoa-Zarzosa, A. Thionin Thi2.1 from Arabidopsis thaliana expressed in endothelial cells shows antibacterial, antifungal and cytotoxic activity. Biotechnol. Lett. 2008, 10, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Van Parijs, J.W.F.; Broekaert, J.; Goldstein, I.J.; Peumans, W.J. Hevein: An antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta 1991, 183, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Raikhel, N.V.; Lee, H.-I. Structure and function of chitin-binding proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 591–615. [Google Scholar] [CrossRef]
- Beintema, J.J. Structural features of plant chitinases and chitin-binding proteins. FEBS Lett. 1994, 350, 159–163. [Google Scholar] [CrossRef] [Green Version]
- De Bolle, M.F.; David, K.M.; Rees, S.B.; Vanderleyden, J.; Cammue, B.P.; Broekaert, W.F. Cloning and characterization of a cDNA encoding an antimicrobial chitin-binding protein from amaranth, Amaranthus caudatus. Plant Mol. Biol. 1993, 22, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Mariën, W.; Terras, F.R.; De Bolle, M.F.; Proost, P.; Van Damme, J.; Dillen, L.; Claeys, M.; Rees, S.B.; Vanderleyden, J.; et al. Antimicrobial peptides from Amaranthus caudatus seeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry 1992, 31, 4308–4314. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, A.; Anisimova, V.; Nikonorova, A.; Babakov, A.; Krause, E.; Bienert, M.; Grishin, E.; Egorov, T. An antimicrobial peptide Ar-AMP from amaranth (Amaranthus retroflexus L.) seeds. Phytochemistry 2005, 66, 2426–2431. [Google Scholar] [CrossRef] [PubMed]
- Rogozhin, E.A.; Slezina, M.P.; Slavokhotova, A.A.; Istomina, E.A.; Korostyleva, T.V.; Smirnov, A.N.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. A novel antifungal peptide from leaves of the weed Stellaria media L. Biochimie 2015, 116, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Komakhin, R.A.; Vysotskii, D.A.; Shukurov, R.R.; Voblikova, V.D.; Komakhina, V.V.; Strelnikova, S.R.; Vetchinkina, E.M.; Babakov, A.V. Novel strong promoter of antimicrobial peptides gene pro-SmAMP2 from chickweed (Stellaria media). BMC Biotechnol. 2016, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.H.; Xiang, Y.; Liu, X.Z.; Zhang, Y.; Hu, Z.; Wang, D.C. Two novel antifungal peptides distinct with a five-disulfide motif from the bark of Eucommia ulmoides Oliv. FEBS Lett. 2002, 521, 87–90. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Vassilevski, A.A.; Slavokhotova, A.A.; Musolyamov, A.Kh.; Finkina, E.I.; Khadeeva, N.V.; Rogozhin, E.A.; Korostyleva, T.V.; Pukhalsky, V.A.; Grishin, E.V.; et al. A novel antifungal hevein-type peptide from Triticum kiharae seeds with a unique 10-cysteine motif. FEBS J. 2009, 275, 4266–4275. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Vassilevski, A.A.; Slavokhotova, A.A.; Odintsova, T.I.; Grishin, E.V.; Egorov, T.A.; Arseniev, A.S. Solution structure of a defense peptide from wheat with a 10-cysteine motif. Biochem. Biophys. Res. Commun. 2011, 411, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Utkina, L.L.; Zhabon, E.O.; Slavokhotova, A.A.; Rogozhin, E.A.; Shiian, A.N.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I.; Pukhal’skiĭ, V.A. Heterologous expression of a synthetic gene encoding a novel hevein-type antimicrobial peptide of Leymus arenarius in Escherichia coli cells. Russ. J. Genet. 2010, 46, 1645–1651. [Google Scholar] [CrossRef]
- Naumann, T.A. Modification of recombinant maize ChitA chitinase by fungal chitinase-modifying proteins. Mol. Plant Pathol. 2011, 12, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-Like Antimicrobial Peptides of Plants. Biochemistry 2017, 82, 1659–1674. [Google Scholar] [CrossRef] [PubMed]
- Naumann, T.A.; Wicklow, D.T.; Price, N.P. Identification of a chitinase-modifying protein from Fusarium verticillioides: Truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 2011, 286, 35358–35366. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Naumann, T.A.; Price, N.P.; Rogozhin, E.A.; Andreev, Y.A.; Vassilevski, A.A.; Odintsova, T.I. Novel mode of action of plant defense peptides—Hevein-like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 2014, 281, 4754–4764. [Google Scholar] [CrossRef] [PubMed]
- Istomina, E.A.; Korostyleva, T.V.; Rozhnova, N.A.; Rogozhin, E.A.; Pukhalski, V.A.; Odintsova, T.I. Genes encoding hevein-like antimicrobial peptides WAMPs: Expression in response to phytohormones and environmental factors. Russ. J. Genet. 2016, 52, 1176–1185. [Google Scholar] [CrossRef]
- Duvick, J.P.; Rood, T.; Rao, A.G.; Marshak, D.R. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 1992, 267, 18814–18820. [Google Scholar] [PubMed]
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, X.X.; Xia, H.C.; Zeng, R.; Hu, W.G.; Li, Z.; Zhang, Z.C. Purification and characterization of Luffin P1, a ribosome-inactivating peptide from the seeds of Luffa cylindrical. Peptides 2003, 24, 799–805. [Google Scholar] [CrossRef]
- Park, S.S.; Abe, K.; Kimura, M.; Urisu, A.; Yamasaki, N. Primary structure and allergenic activity of trypsin inhibitors from the seeds of buckwheat (Fagopyrum esculentum Moench). FEBS Lett. 1997, 400, 103–107. [Google Scholar] [CrossRef]
- Yamada, K.; Shimada, T.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J. Biol. Chem. 1999, 274, 2563–2570. [Google Scholar] [CrossRef] [PubMed]
- Conners, R.; Konarev, A.V.; Forsyth, J.; Lovegrove, A.; Marsh, J.; Joseph-Horne, T.; Shewry, P.; Brady, R.L. An unusual helix-turn-helix protease inhibitory motif in a novel trypsin inhibitor from seeds of Veronica (Veronica hederifolia L.). J. Biol. Chem. 2007, 282, 27760–27768. [Google Scholar] [CrossRef] [PubMed]
- Oparin, P.B.; Mineev, K.S.; Dunaevsky, Y.E.; Arseniev, A.S.; Belozersky, M.A.; Grishin, E.V.; Egorov, T.A.; Vassilevski, A.A. Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides. Biochem. J. 2012, 446, 69–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolde, S.B.; Vassilevski, A.A.; Rogozhin, E.A.; Barinov, N.A.; Balashova, T.A.; Samsonova, O.V.; Baranov, Y.V.; Feofanov, A.V.; Egorov, T.A.; Arseniev, A.S.; et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 2011, 286, 25145–25153. [Google Scholar] [CrossRef] [PubMed]
- Rogozhin, E.A.; Ryazantsev, D.Y.; Grishin, E.V.; Egorov, T.A.; Zavriev, S.K. Defense peptides from barnyard grass (Echinochloa crusgalli L.) seeds. Peptides 2012, 38, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ryazantsev, D.Y.; Rogozhin, E.A.; Dimitrieva, T.V.; Drobyazina, P.E.; Khadeeva, N.V.; Egorov, T.A.; Grishin, E.V.; Zavriev, S.K. A novel hairpin-like antimicrobial peptide from barnyard grass (Echinochloa crusgalli L.) seeds: Structure-functional and molecular-genetics characterization. Biochimie 2014, 99, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Utkina, L.L.; Andreev, Y.A.; Rogozhin, E.A.; Korostyleva, T.V.; Slavokhotova, A.A.; Oparin, P.B.; Vassilevski, A.A.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. Genes encoding 4-Cys antimicrobial peptides in wheat Triticum kiharae Dorof. et Migush.: Multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 2013, 280, 3594–3608. [Google Scholar] [CrossRef] [PubMed]
- Vasilchenko, A.S.; Yuryev, M.; Ryazantsev, D.Y.; Zavriev, S.K.; Feofanov, A.V.; Grishin, E.V.; Rogozhin, E.A. Studying of cellular interaction of hairpin-like peptide EcAMP1 from barnyard grass (Echinochloa crusgalli L.) seeds with plant pathogenic fungus Fusarium solani using microscopy techniques. Scanning 2016, 38, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. Mol. Life Sci. 2013, 70, 3545–3570. [Google Scholar] [CrossRef] [PubMed]




| Peptide/Microbe | α 1 -Purothionin | α -Hordothionin | γ -1-H-Hordothionin | γ -1-P-Purothionin | γ -1-Zeathionin | Tk-AMP-BP1 | Tk-AMP-BP2 |
|---|---|---|---|---|---|---|---|
| Bipolaris sorokiniana | 3.2 | 5.0 | Not tested | Not tested | Not tested | 5.6 | 32.0 |
| Botrytis cinerea | Not tested | 20.0 | Not tested | Not tested | Not tested | Not tested | 32.0 |
| Fusarium oxysporum | 3.9 | 5.0 | 4.0 | 7.6 | 7.0 | 6.0 | Not tested |
| F. solani | Not tested | 5.0 | Not tested | Not tested | Not tested | Not tested | Not tested |
| F. verticillioides | 1.9 | 2.7 | 2.2 | 3.5 | 3.0 | 4.5 | Not tested |
| Neurospora crassa | Not tested | 10.0 | Not tested | Not tested | Not tested | Not tested | Not tested |
| Peptide/Microbe | WAMP1a (+R) | WAMP1b (−R) | WAMP2a (A34K) | WAMP3a (A34E) | WAMP4a (A34N) | LAMP-1a | Ar-AMP |
|---|---|---|---|---|---|---|---|
| Bipolaris sorokiniana | 3.2 | 5.0 | Not tested | Not tested | Not tested | 5.6 | 32.0 |
| Botrytis cinerea | Not tested | 20.0 | Not tested | Not tested | Not tested | Not tested | 32.0 |
| Fusarium oxysporum | 3.9 | 5.0 | 4.0 | 7.6 | 7.0 | 6.0 | Not tested |
| F. solani | Not tested | 5.0 | Not tested | Not tested | Not tested | Not tested | Not tested |
| F. verticillioides | 1.9 | 2.7 | 2.2 | 3.5 | 3.0 | 4.5 | Not tested |
| Neurospora crassa | Not tested | 10.0 | Not tested | Not tested | Not tested | Not tested | Not tested |
| Peptide/Microbe | EcAMP1 | EcAMP2 | EcAMP3 | Tk-AMP-X1 | Tk-AMP-X2 | MBP-1 | Sm-AMP-X |
|---|---|---|---|---|---|---|---|
| Alternaria alternata | 16.0 | >32.0 | 19.8 | Not tested | 28.8 | Not tested | 14.8 |
| Aspergillus niger | >32.0 | >32.0 | 22.4 | Not tested | >32.0 | Not tested | 4.0 |
| B. sorokiniana | 18.2 | >32.0 | 15.0 | Not tested | Not tested | Not tested | >32.0 |
| C. graminicola | >10 | Not tested | Not tested | >30.0 | >30.0 | Not tested | Not tested |
| D. maydis | >10 | Not tested | Not tested | 30.0 | 17.0 | Not tested | Not tested |
| F. graminearum | 4.5 | >32.0 | 5.5 | 7.5 | 7.5 | 4.0 | Not tested |
| F. oxysporum | 8.8 | >32.0 | 9.6 | Not tested | 13.5 | Not tested | 6.8 |
| F. solani | 4.0 | >32.0 | 4.8 | Not tested | 8.5 | Not tested | 8.0 |
| F. verticillioides | 8.1 | >32.0 | 5.2 | 15.0 | 10.0 | Not tested | Not tested |
| P. infestans | 16.3 | >32.0 | 14.0 | Not tested | 25.4 | Not tested | >32.0 |
| P. ultimum | 14.4 | Not tested | Not tested | Not tested | Not tested | Not tested | >32.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rogozhin, E.; Ryazantsev, D.; Smirnov, A.; Zavriev, S. Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals. Plants 2018, 7, 74. https://doi.org/10.3390/plants7030074
Rogozhin E, Ryazantsev D, Smirnov A, Zavriev S. Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals. Plants. 2018; 7(3):74. https://doi.org/10.3390/plants7030074
Chicago/Turabian StyleRogozhin, Eugene, Dmitry Ryazantsev, Alexey Smirnov, and Sergey Zavriev. 2018. "Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals" Plants 7, no. 3: 74. https://doi.org/10.3390/plants7030074
APA StyleRogozhin, E., Ryazantsev, D., Smirnov, A., & Zavriev, S. (2018). Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals. Plants, 7(3), 74. https://doi.org/10.3390/plants7030074





