Multiple Pathways in the Control of the Shade Avoidance Response
Abstract
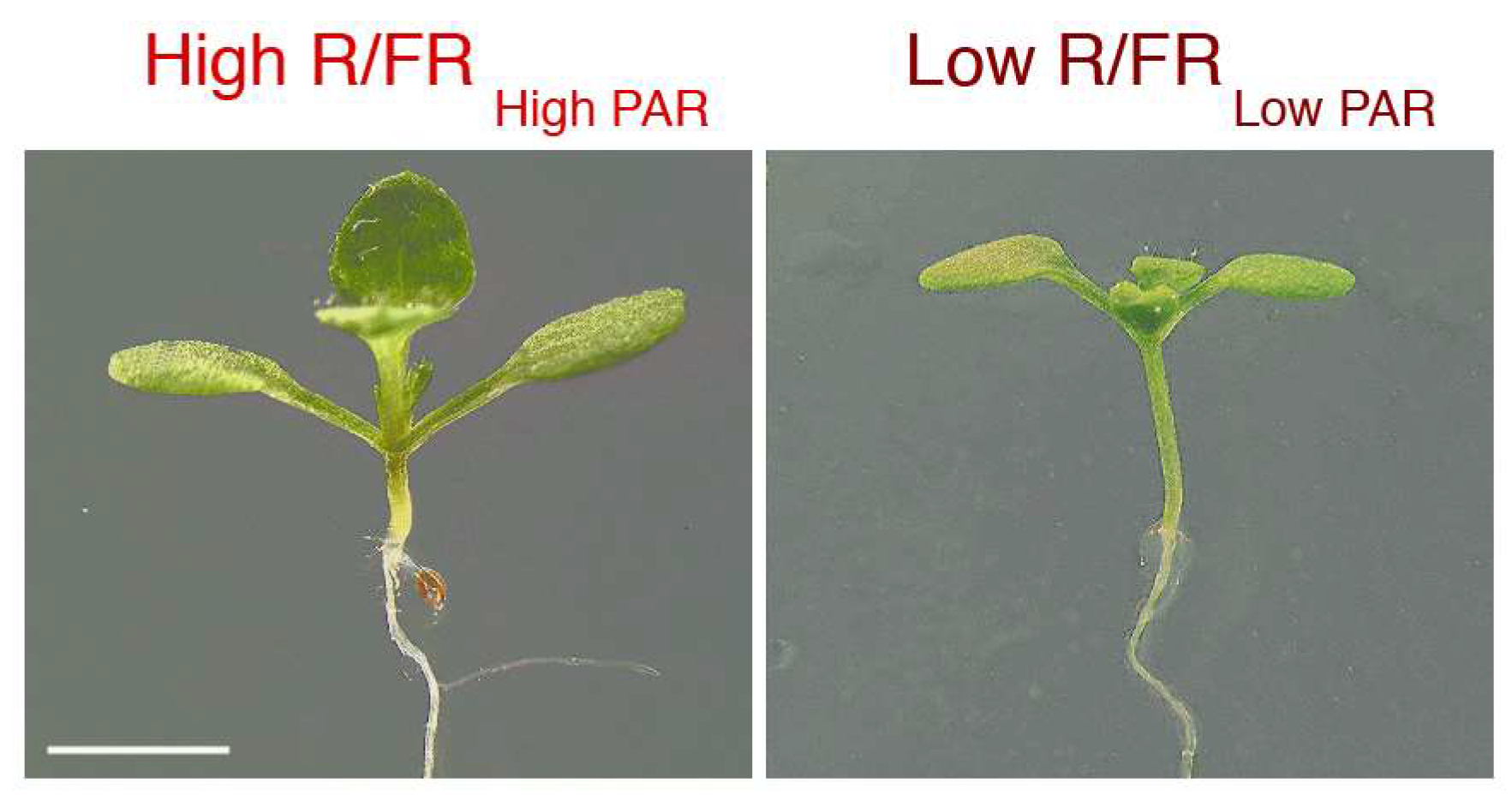
1. Introduction
2. Photoreceptors in the Control of Shade Avoidance
2.1. Phytochromes
2.2. Cryptochromes
2.3. UVR8
3. HD-Zip Transcription Factors in the Control of Shade Avoidance
3.1. HD-Zips II
3.2. HD-Zips III
4. Auxin as a Driver of the Shade Avoidance Response
4.1. Auxin Homeostasis
4.2. Auxin Transport
4.3. Auxin Signaling
5. From Arabidopsis to Crops
5.1. Maize
5.2. Tomato
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| BBX | B-BOX |
| DELLA proteins | Named after their conserved N-terminal |
| DIC | Differential Interference Contrast |
| GMO | Genetically Modified Organisms |
| GUS | β-glucuronidase |
References
- Gommers, C.M.M.; Visser, E.J.W.; St Onge, K.R.; Voesenek, L.A.C.J.; Pierik, R. Shade tolerance: When growing tall is not an option. Trends Plant Sci. 2013, 18, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.; Lopez-Garcia, M.; Phrathep, O.P.; Lawson, T.; Oulton, R.; Whitney, H.M. Photonic multilayer structure of Begonia chloroplasts enhances photosynthetic efficiency. Nat. Plants 2016, 2, 16162. [Google Scholar] [CrossRef] [PubMed]
- Gommers, C.M.; Keuskamp, D.H.; Buti, S.; van Veen, H.; Koevoets, I.T.; Reinen, E.; Voesenek, L.A.; Pierik, R. Molecular profiles of contrasting shade response strategies in wild plants: Differential control of immunity and shoot elongation. Plant Cell 2017, 29, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Pierik, R.; De Wit, M. Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J. Exp. Bot. 2014, 65, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.P.; Hayes, S.; Franklin, K.A. Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol. 2016, 33, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.L.; Pierik, R. The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, A.S.; Fankhauser, C. Plant Strategies for Enhancing Access to Sunlight. Curr. Biol. 2017, 27, R931–R940. [Google Scholar] [CrossRef] [PubMed]
- Ciolfi, A.; Sessa, G.; Sassi, M.; Possenti, M.; Salvucci, S.; Carabelli, M.; Morelli, G.; Ruberti, I. Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 2013, 163, 331–353. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Mathews, S.; Sharrock, R. Phytochrome gene diversity. Plant Cell Environ. 1997, 20, 666–671. [Google Scholar] [CrossRef]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.; Whitelam, G.C. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 1997, 20, 840–844. [Google Scholar] [CrossRef]
- Devlin, P.F.; Yanovsky, M.J.; Kay, S.A. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 2003, 133, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.; Bradley, J.M.; Harberd, N.P.; Whitelam, G.C. Photoresponses of light-grown phyA mutants of Arabidopsis: Phytochrome A is required for the perception of daylength extensions. Plant Physiol. 1994, 105, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Roig-Villanova, I.; Khan, S.; Shanahan, H.; Quail, P.H.; Martinez-Garcia, J.F.; Devlin, P.F. A novel high-throughput in vivo molecular screen for shade avoidance mutants identifies a novel phyA mutation. J. Exp. Bot. 2011, 62, 2973–2987. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- de Lucas, M.; Prat, S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014, 202, 1126–1141. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Choi, G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 2017, 35, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, H.; Park, J.; Kim, W.; Yoo, J.; Lee, N.; Yoon, T.Y.; Choi, G. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 2016, 28, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E.; Cohn, M.M.; Quail, P.H. Phytochrome signaling in green Arabidopsis seedlings: Impact assessment of a mutually negative phyB–PIF feedback loop. Mol. Plant 2012, 5, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ljung, K.; Breton, G.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; Ecker, J.R.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Hornitschek, P.; Kohnen, M.V.; Lorrain, S.; Rougemont, J.; Ljung, K.; López-Vidriero, I.; Franco-Zorrilla, J.M.; Solano, R.; Trevisan, M.; Pradervand, S.; et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012, 71, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Ruberti, I.; Sessa, G.; Ciolfi, A.; Possenti, M.; Carabelli, M.; Morelli, G. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnol. Adv. 2012, 30, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Steindler, C.; Matteucci, A.; Sessa, G.; Weimar, T.; Ohgishi, M.; Aoyama, T.; Morelli, G.; Ruberti, I. Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development 1999, 125, 4235–4245. [Google Scholar]
- Carabelli, M.; Turchi, L.; Ruzza, V.; Morelli, G.; Ruberti, I. Homeodomain-Leucine Zipper II family of transcription factors to the limelight: Central regulators of plant development. Plant Signal. Behav. 2013, 8, e25447. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Morelli, G.; Whitelam, G.; Ruberti, I. Twilight-zone and canopy shade induction of the ATHB-2 homeobox gene in green plants. Proc. Natl. Acad. Sci. USA 1996, 93, 3530–3535. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Praekelt, U.; Stoddart, W.M.; Billingham, O.E.; Halliday, K.J.; Whitelam, G.C. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 2003, 131, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Roig-Villanova, I.; Bou, J.; Sorin, C.; Devlin, P.F.; Martínez-García, J.F. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006, 141, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Tepperman, J.M.; Cohn, M.M.; Monte, E.; Al-Sady, B.; Erickson, E.; Quail, P.H. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 2012, 24, 1398–1419. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Carabelli, M.; Sassi, M.; Ciolfi, A.; Possenti, M.; Mittempergher, F.; Becker, J.; Morelli, G.; Ruberti, I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005, 19, 2811–2815. [Google Scholar] [CrossRef] [PubMed]
- Hornitschek, P.; Lorrain, S.; Zoete, V.; Michielin, O.; Fankhauser, C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009, 28, 3893–3902. [Google Scholar] [CrossRef] [PubMed]
- Ruzza, V.; Sessa, G.; Sassi, M.; Morelli, G.; Ruberti, I. Auxin coordinates shoot and root development during shade avoidance response. In Auxin and Its Role in Plant Development; Zažímalová, E., Petrasek, J., Benková, E., Eds.; Springer-Verlag: Wien, Austria, 2014; pp. 349–412. [Google Scholar]
- Galstyan, A.; Cifuentes-Esquivel, N.; Bou-Torrent, J.; Martinez-Garcia, J.F. The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 2011, 66, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Roig-Villanova, I.; Bou-Torrent, J.; Galstyan, A.; Carretero-Paulet, L.; Portolés, S.; Rodríguez-Concepción, M.; Martínez-García, J.F. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 2007, 26, 4756–4767. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Esquivel, N.; Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Sessa, G.; Salla Martret, M.; Roig-Villanova, I.; Ruberti, I.; Martínez-García, J.F. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013, 75, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, J.F.; Gallemí, M.; Molina-Contreras, M.J.; Llorente, B.; Bevilaqua, M.R.R.; Quail, P.H. The shade avoidance syndrome in Arabidopsis: The antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS ONE 2014, 9, e109275. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.J.; Hiltbrunner, A. Molecular mechanisms and ecological function of far-red light signalling. Plant Cell Environ. 2017, 40, 2509–2529. [Google Scholar] [CrossRef] [PubMed]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Xu, G.; Tang, W.; Jing, Y.; Ji, Q.; Fei, Z.; Lin, R. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 2013, 25, 1657–1673. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodríguez-Concepción, M.; Halliday, K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.J.; Menon, C.; zur Oven-Krockhaus, S.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.D.; Huq, E.; Hiltbrunner, A. Light-activated Phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by disrupting the COP1-SPA complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- McNellis, T.W.; von Arnim, A.G.; Araki, T.; Komeda, Y.; Miséra, S.; Deng, X.-W. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 1994, 6, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Rolauffs, S.; Fackendahl, P.; Sahm, J.; Fiene, G.; Hoecker, U. Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Phys. 2012, 160, 2015–2027. [Google Scholar] [CrossRef] [PubMed]
- Pacín, M.; Semmoloni, M.; Legris, M.; Finlayson, S.A.; Casal, J.J. Convergence Of Constitutive Photomorphogenesis 1 and Phytochrome Interacting Factor Signalling during Shade Avoidance. New Phytol. 2016, 211, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 2010, 64, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.J.; Maloof, J.N.; Wu, S.H. COP1 mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 2011, 156, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Dvornyk, V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS ONE 2015, 10, e0135940. [Google Scholar] [CrossRef] [PubMed]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.O.; van der Horst, G.T.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, B.; Su, J.; Liao, J.; Lin, C.; Oka, Y. Cryptochromes Orchestrate Transcription Regulation of Diverse Blue Light Responses in Plants. Photochem. Photobiol. 2017, 93, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.M.; Jaillais, Y.; Pedmale, U.V.; Moreno, J.E.; Chory, J.; Ballaré, C.L. Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 2011, 67, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Keuskamp, D.H.; Sasidharan, R.; Vos, I.; Peeters, A.J.; Voesenek, L.A.C.J.; Pierik, R. Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 2011, 67, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Huang, S.S.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.-A.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Shade Avoidance. Arabidopsis Book 2012, 10, e0157. [Google Scholar] [CrossRef] [PubMed]
- Rizzini, L.; Favory, J.-J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Ulm, R.; Jenkins, G. Q&A: How do plants sense and respond to UV-B radiation? BMC Biol. 2015, 13, 45. [Google Scholar]
- Oravecz, A.; Baumann, A.; Mate, Z.; Brzezinska, A.; Molinier, J.; Oakeley, E.J.; Adam, E.; Schäfer, E.; Nagy, F.; Ulm, R. Constitutively photomorphogenic1 is required for the UV-B response in Arabidopsis. Plant Cell 2006, 18, 1975–1990. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Jenkins, G.I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008, 146, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Favory, J.J.; Stec, A.; Gruber, H.; Rizzini, L.; Oravecz, A.; Funk, M.; Albert, A.; Cloix, C.; Jenkins, G.I.; Oakeley, E.J.; et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009, 28, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Velanis, C.N.; Jenkins, G.I.; Franklin, K.A. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. USA 2014, 111, 11894–11899. [Google Scholar] [CrossRef] [PubMed]
- Mazza, C.A.; Ballaré, C.L. Photoreceptors UVR8 and phytochrome B cooperate to optimize plant growth and defense in patchy canopies. New Phytol. 2015, 207, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Ruberti, I.; Sessa, G.; Lucchetti, S.; Morelli, G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 1991, 10, 1787–1791. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Morelli, G.; Ruberti, I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993, 12, 3507–3517. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Tron, A.E.; Bertoncini, C.W.; Palena, C.M.; Chan, R.L.; Gonzales, D.H. Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucl. Acids Res. 2001, 29, 4866–4872. [Google Scholar] [CrossRef] [PubMed]
- Brandt, R.; Salla-Martret, M.; Bou-Torrent, J.; Musielak, T.; Stahl, M.; Lanz, C.; Ott, F.; Schmid, M.; Greb, T.; Schwarz, M.; et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012, 72, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Turchi, L.; Baima, S.; Morelli, G.; Ruberti, I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 2015, 66, 5043–5053. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Carabelli, M.; Ruberti, I.; Baima, S.; Lucchetti, S.; Morelli, G. Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In Molecular-Genetic Analysis of Plant Development and Metabolism; Puigdomenech, P., Coruzzi, G., Eds.; NATO-ASI Series; Springer-Verlag: Berlin/Heidelberg, Germany, 1994; Volume 81, pp. 411–426. [Google Scholar]
- Sessa, G.; Steindler, C.; Morelli, G.; Ruberti, I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-Zip proteins. Plant Mol. Biol. 1998, 38, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Johansson, H.; Hanson, J.; Engstrom, P.; Soderman, E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Agalou, A.; Purwantomo, S.; Overnaes, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Ciarbelli, A.R.; Ciolfi, A.; Salvucci, S.; Ruzza, V.; Possenti, M.; Carabelli, M.; Fruscalzo, A.; Sessa, G.; Morelli, G.; Ruberti, I. The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol. Biol. 2008, 68, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Romani, F.; Reinheimer, R.; Florent, S.N.; Bowman, J.L.; Moreno, J.E. Evolutionary history of HOMEODOMAIN LEUCINE ZIPPER transcription factors during plant transition to land. New Phytol. 2018, 219, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Merelo, P.; Paredes, E.B.; Heisler, M.G.; Wenkel, S. The shady side of leaf development: The role of the REVOLUTA/KANADI1 module in leaf patterning and auxin-mediated growth promotion. Curr. Opin. Plant Biol. 2017, 35, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Turchi, L.; Carabelli, M.; Ruzza, V.; Possenti, M.; Sassi, M.; Peñalosa, A.; Sessa, G.; Salvi, S.; Forte, V.; Morelli, G.; Ruberti, I. Arabidopsis HD-Zip II transcription factors control embryo development and meristem function. Development 2013, 140, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Links, M.G.; Rozwadowski, K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010, 152, 1109–1134. [Google Scholar] [CrossRef] [PubMed]
- Ohgishi, M.; Oka, A.; Morelli, G.; Ruberti, I.; Aoyama, T. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J. 2001, 25, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Ohgishi, M.; Goda, H.; Higuchi, K.; Shimada, Y.; Yoshida, S.; Koshiba, T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002, 32, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Wenkel, S.; Emercy, J.; Hou, B.; Evans, M.M.S.; Barton, M.K. A feedback regulatory module formed by little zipper and HD-ZIPIII genes. Plant Cell 2007, 19, 3379–3390. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Kim, S.-G.; Lee, M.; Lee, I.; Park, H.Y.; Seo, P.J.; Jung, J.H.; Kwon, E.J.; Suh, S.W.; Paek, K.H.; Park, C.M. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 2008, 20, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Baima, S.; Forte, V.; Possenti, M.; Peñalosa, A.; Leoni, G.; Salvi, S.; Felici, B.; Ruberti, I.; Morelli, G. Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol. Plant 2014, 7, 1006–1025. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Salla-Martret, M.; Bou-Torrent, J.; Roig-Villanova, I.; Martínez-García, J.F. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009, 59, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Ueoka-Nakanishi, H.; Hori, N.; Ishida, K.; Ono, N.; Yamashino, T.; Nakamichi, N.; Mizuno, T. Characterization of shade avoidance responses in Lotus japonicus. Biosci. Biotechnol. Biochem. 2011, 75, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Kumar, R.; Ranjan, A.; Pelletier, J.M.; Townsley, B.T.; Ichihashi, Y.; Martinez, C.C.; Zumstein, K.; Harada, J.J.; Maloof, J.N.; et al. Light-induced indeterminacy alters shade-avoiding tomato leaf morphology. Plant Physiol. 2015, 169, 2030–2047. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, G.; Zhao, B.; Wang, B.; Lang, Z.; Zhang, C.; Wang, H. Regulatory modules controlling early shade avoidance response in maize seedlings. BMC Genom. 2016, 17, 269. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Possenti, M.; Sessa, G.; Ruzza, V.; Morelli, G.; Ruberti, I. Arabidopsis HD-Zip II proteins regulate the exit from proliferation during leaf development in canopy shade. J. Exp. Bot. 2018. [Google Scholar] [CrossRef] [PubMed]
- Trigg, S.A.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.C.; et al. CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 2017, 14, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, N.; Vanharen, H.; Inzé, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Bar, M.; Ori, N. Leaf development and morphogenesis. Development 2014, 141, 4219–4230. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Salla-Martret, M.; Brandt, R.; Musielak, T.; Palauquim, J.C.; Martinez-Garcia, J.F.; Wenkel, S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012, 7, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Reymond, M.C.; Brunoud, G.; Chauvet, A.; Martínez-Garcia, J.F.; Martin-Magniette, M.L.; Monéger, F.; Scutt, C.P. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 2012, 24, 2812–2825. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Mayo, V.M.; Marsch-Martínez, N.; de Folter, S. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J. 2012, 71, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ye, H.; Guo, H.; Johnson, A.; Zhang, M.; Lin, H.; Yin, Y. Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 2014, 77, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, Y.; Wang, H.; Ma, X.; Wang, B.; Wu, G.; Wang, H. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 2017, 8, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated squamosa promoter binding protein-like (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.R.; Long, J.A. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 2010, 464, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Baima, S.; Nobili, F.; Sessa, G.; Lucchetti, S.; Ruberti, I.; Morelli, G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 1995, 121, 4171–4182. [Google Scholar] [PubMed]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Kubo, M.; Demura, T.; Fukuda, H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 2005, 46, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.K.; Zalewski, C.S.; Bowman, J.L. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 2006, 173, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.K.; Bowman, J.L. Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 2006, 16, 1911–1917. [Google Scholar] [CrossRef] [PubMed]
- Donner, T.J.; Sherr, I.; Scarpella, E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 2009, 136, 3235–3246. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Reinhart, B.J.; Bartel, D.P.; Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Harrar, Y.; Lin, C.; Reinhart, B.; Newell, N.R.; Talavera-Rauh, F.; Hokin, S.A.; Barton, M.K.; Kerstetter, R.A. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 2014, 26, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Liu, T.; Newell, N.R.; Magnani, E.; Huang, T.; Kerstetter, R.; Michaels, S.; Barton, M.K. Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 2013, 25, 3228–3249. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Xie, Y.; Straub, D.; Eguen, T.; Brandt, R.; Stahl, M.; Martínez-García, J.F.; Wenkel, S. Meta-Analysis of Arabidopsis KANADI1 Direct Target Genes Identifies a Basic Growth-Promoting Module Acting Upstream of Hormonal Signaling Pathways. Plant Phys. 2015, 169, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- Merelo, P.; Ram, H.; Pia Caggiano, M.; Ohno, C.; Ott, F.; Straub, D.; Graeff, M.; Cho, S.K.; Yang, S.W.; Wenkel, S.; et al. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA 2016, 113, 11973–11978. [Google Scholar] [CrossRef] [PubMed]
- de Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, L. Hormonal Regulation in Shade Avoidance. Front. Plant Sci. 2017, 8, 1527. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Sellaro, R.; Zurbriggen, M.D.; Casal, J.J. Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 2018, 69, 213–228. [Google Scholar] [CrossRef]
- Nozue, K.; Harmer, S.L.; Maloof, J.N. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PIF5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011, 448, 358–361. [Google Scholar]
- Hersch, M.; Lorrain, S.; De Wit, M.; Trevisan, M.; Ljung, K.; Bergmann, S.; Fankhauser, C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 6515–6520. [Google Scholar] [CrossRef] [PubMed]
- Pucciariello, O.; Legris, M.; Costigliolo Rojas, C.; Iglesias, M.J.; Hernando, C.E.; Dezar, C.; Vazquez, M.; Yanovsky, M.J.; Finlayson, S.A.; Prat, S.; et al. Rewiring of auxin signaling under persistent shade. Proc. Natl. Acad. Sci. USA. 2018, 115, 5612–5617. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Procko, C.; Crenshaw, C.M.; Ljung, K.; Noel, J.P.; Chory, J. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 2014, 165, 1285–1301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [PubMed]
- Kohnen, M.V.; Schmid-Siegert, E.; Trevisan, M.; Petrolati, L.A.; Senechal, F.; Muller-Moule, P.; Maloof, J.; Xenarios, I.; Fankhauser, C. Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 2016, 28, 2889–2904. [Google Scholar] [CrossRef] [PubMed]
- Muller-Moule, P.; Nozue, K.; Pytlak, M.L.; Palmer, C.M.; Covington, M.F.; Wallace, A.D.; Harmer, S.L.; Maloof, J.N. YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. Peer J. 2016, 4, e2574. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Possenti, M.; Sessa, G.; Ciolfi, A.; Sassi, M.; Morelli, G.; Ruberti, I. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007, 21, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guo, Y.; Novak, O.; Chen, W.; Ljung, K.; Noel, J.P.; Chory, J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2016, 2, 16025. [Google Scholar] [CrossRef] [PubMed]
- Michaud, O.; Fiorucci, A.S.; Xenarios, I.; Fankhauser, C. Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 7444–7449. [Google Scholar] [CrossRef] [PubMed]
- Pantazopoulou, C.K.; Bongers, F.J.; Küpers, J.J.; Reinen, E.; Das, D.; Evers, J.B.; Anten, N.P.R.; Pierik, R. Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, 7450–7455. [Google Scholar] [CrossRef] [PubMed]
- Keuskamp, D.H.; Pollmann, S.; Voesenek, L.A.; Peeters, A.J.; Pierik, R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 2010, 107, 22740–22744. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Wang, J.; Ruberti, I.; Vernoux, T.; Xu, J. Shedding light on auxin movement: Light-regulation of polar auxin transport in the photocontrol of plant development. Plant Signal. Behav. 2013, 8, e23355. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Yan, F.; Zourelidou, M.; Wang, M.; Ljung, K.; Fastner, A.; Hammes, U.Z.; Di Donato, M.; Geisler, M.; Schwechheimer, C.; et al. SHADE AVOIDANCE 4 is required for proper auxin distribution in the hypocotyl. Plant Physiol. 2017, 173, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Christie, J.M. Plant phototropic growth. Curr. Biol. 2015, 25, R384–R389. [Google Scholar] [CrossRef] [PubMed]
- Morelli, G.; Ruberti, I. Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol. 2000, 122, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Morelli, G.; Ruberti, I. Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci. 2002, 7, 399–404. [Google Scholar] [CrossRef]
- Sassi, M.; Lu, Y.; Zhang, Y.; Wang, J.; Dhonukshe, P.; Blilou, I.; Dai, M.; Li, J.; Gong, X.; Jaillais, Y.; et al. COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 2012, 139, 3402–3412. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, Q.; Gao, X.; Jiang, C.; Harberd, N.P.; Fu, X. Shoot-to-Root Mobile Transcription Factor HY5 Coordinates Plant Carbon and Nitrogen Acquisition. Curr. Biol. 2016, 26, 640–646. [Google Scholar] [CrossRef] [PubMed]
- van Gelderen, K.; Kang, C.; Paalman, R.; Keuskamp, D.; Hayes, S.; Pierik, R. Far-Red Light Detection in the Shoot Regulates Lateral Root Development through the HY5 Transcription Factor. Plant Cell 2018, 30, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Péret, B.; Middleton, A.M.; French, A.P.; Larrieu, A.; Bishopp, A.; Njo, M.; Wells, D.M.; Porco, S.; Mellor, N.; Band, L.R.; et al. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol. Syst. Biol. 2013, 9, 699. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Redman, J.C.; Haas, B.J.; Tanimoto, G.; Town, C.D. Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 2004, 38, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 11, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Possenti, M.; Sessa, G.; Ciolfi, A.; Sassi, M.; Morelli, G.; Ruberti, I. A novel regulatory circuit underlying plant response to canopy shade. Plant Signal. Behav. 2008, 3, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Sellaro, R.; Yanovsky, M.J.; Casal, J.J. Repression of shade avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 2011, 68, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three Auxin Response Factors Promote Hypocotyl Elongation. Plant Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Maddonni, G.A.; Otegui, M.E.; Andrieu, B.; Chelle, M.; Casal, J.J. Maize leaves turn away from neighbors. Plant Physiol. 2002, 130, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fellner, M.; Horton, L.A.; Cocke, A.E.; Stephens, N.R.; Ford, E.D.; Van Volkenburgh, E. Light interacts with auxin during leaf elongation and leaf angle development in young corn seedlings. Planta 2003, 216, 366–376. [Google Scholar] [PubMed]
- Dubois, P.G.; Olsefski, G.T.; Flint-Garcia, S.; Setter, T.L.; Hoekenga, O.A.; Brutnell, T.P. Physiological and genetic characterization of end-of-day far-red light response in maize seedlings. Plant Physiol. 2010, 154, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, C.C.; Trupkin, S.A.; Ghiglione, H.; Slafer, G.; Casal, J.J. Low red/far-red ratios delay spike and stem growth in wheat. J. Exp. Bot. 2010, 61, 3151–3162. [Google Scholar] [CrossRef] [PubMed]
- Carriedo, L.G.; Maloof, J.N.; Brady, S.M. Molecular control of crop shade avoidance. Curr. Opin. Plant Biol. 2016, 30, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Canopy light signals and crop yield in sickness and in health. ISRN Agron. 2013, 2013, 650439. [Google Scholar] [CrossRef]
- Küpers, J.J.; van Gelderen, K.; Pierik, R. Location Matters: Canopy Light Responses over Spatial Scales. Trends Plant Sci. 2018, 23, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, M.J.; Farmer, P.R.; Brutnell, T.P. Structure and expression of maize phytochrome family homeologs. Genetics 2004, 167, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, M.J.; Kennedy, L.M.; Costich, D.E.; Brutnell, T.P. Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. Plant J. 2007, 49, 338–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, C.; Li, L. Shade-induced stem elongation in rice seedlings: Implication of tissue-specific phytohormone regulation. J. Integr. Plant Biol. 2016, 58, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Kurepin, L.V.; Emery, R.J.N.; Pharis, R.P.; Reid, D.M. Uncoupling light quality from light irradiance effects in Helianthus annuus shoots: Putative roles for plant hormones in leaf and internode growth. J. Exp. Bot. 2007, 58, 2145–2157. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, A.C.; Strasser, B.; Cerdán, P.D.; Amasino, R.M. Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 2008, 148, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Cagnola, J.I.; Ploschuk, E.; Benech-Arnold, T.; Finlayson, S.A.; Casal, J.J. Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol. 2012, 160, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhang, Y.; Xiao, Y.; Liu, F.; Wang, M.; Zhu, X.; Liu, P.; Sun, Q.; Wang, W.; Peng, M.; et al. Transcriptome response of cassava leaves under natural shade. Sci. Rep. 2016, 6, 31673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mayba, O.; Pfeiffer, A.; Shi, H.; Tepperman, J.M.; Speed, T.P.; Quail, P.H. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013, 9, e1003244. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Zhang, H.; Song, X.; Jiang, Y.; Liang, R.; Li, G. Functional Characterization of the Maize Phytochrome-Interacting Factors PIF4 and PIF5. Front. Plant Sci. 2017, 8, 2273. [Google Scholar] [CrossRef] [PubMed]
- Bush, S.M.; Carriedo, L.; Fulop; Ichihashi, Y.; Covington, M.F.; Kumar, R.; Ranjan, A.; Chitwood, D.H.; Headland, L.; Filiault, D.L.; et al. Auxin signaling is a common factor underlying natural variation in tomato shade avoidance. bioRxiv 2015, 031088. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Headland, L.R.; Kumar, R.; Peng, J.; Maloof, J.N.; Sinha, N.R. The developmental trajectory of leaflet morphology in wild tomato species. Plant Physiol. 2012, 158, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Ranjan, A.; Kumar, R.; Ichihashi, Y.; Zumstein, K.; Headland, L.R.; Ostria-Gallardo, E.; Aguilar-Martínez, J.A.; Bush, S.; Carriedo, L.; et al. Resolving distinct genetic regulators of tomato leaf shape within a heteroblastic and ontogenetic context. Plant Cell 2014, 26, 3616–3629. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Headland, L.R.; Filiault, D.L.; Kumar, R.; Jiménez-Gómez, J.M.; Schrager, A.V.; Park, D.S.; Peng, J.; Sinha, N.R.; Maloof, J.N. Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 2012, 7, e29570. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Galstyan, A.; Gallemí, M.; Cifuentes-Esquivel, N.; Molina-Contreras, M.J.; Salla-Martret, M.; Jikumaru, Y.; Yamaguchi, S.; Kamiya, Y.; Martínez-García, J.F. Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J. Exp. Bot. 2014, 65, 2937–2947. [Google Scholar] [CrossRef] [PubMed]
- de Wit, M.; Ljung, K.; Fankhauser, C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 2015, 208, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Mochizuki, N.; Deguchi, A.; Nagano, A.J.; Suzuki, T.; Nagatani, A. Auxin Contributes to the Intraorgan Regulation of Gene Expression in Response to Shade. Plant Physiol. 2018, 177, 847–862. [Google Scholar] [CrossRef] [PubMed]
- Iannacone, R.; Mittempergher, F.; Morelli, G.; Panio, G.; Perito, A.; Ruberti, I.; Sessa, G.; Cellini, F. Influence of an Arabidopsis dominant negative athb2 mutant on tomato plant development. Acta Hortic. 2008, 789, 263–276. [Google Scholar] [CrossRef]
- Tan, W.; Zhang, D.; Zhou, H.; Zheng, T.; Yin, Y.; Lin, H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018, 14, e1007336. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.A.; Jawdy, S.S.; Gunter, L.E.; Allen, S.M.; Yang, X.; Tuskan, G.A.; Wullschleger, S.D.; Weston, D.J. New Phytol. 2012, 196, 726–737. [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple Pathways in the Control of the Shade Avoidance Response. Plants 2018, 7, 102. https://doi.org/10.3390/plants7040102
Sessa G, Carabelli M, Possenti M, Morelli G, Ruberti I. Multiple Pathways in the Control of the Shade Avoidance Response. Plants. 2018; 7(4):102. https://doi.org/10.3390/plants7040102
Chicago/Turabian StyleSessa, Giovanna, Monica Carabelli, Marco Possenti, Giorgio Morelli, and Ida Ruberti. 2018. "Multiple Pathways in the Control of the Shade Avoidance Response" Plants 7, no. 4: 102. https://doi.org/10.3390/plants7040102
APA StyleSessa, G., Carabelli, M., Possenti, M., Morelli, G., & Ruberti, I. (2018). Multiple Pathways in the Control of the Shade Avoidance Response. Plants, 7(4), 102. https://doi.org/10.3390/plants7040102




