Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Data Collection and Meta-Analysis
2.3. Gene Ontology (GO) Enrichment Analysis
2.4. TF, TR, and PK Identification and MapMan Analysis
2.5. Sequence Alignment and Gene Comparison
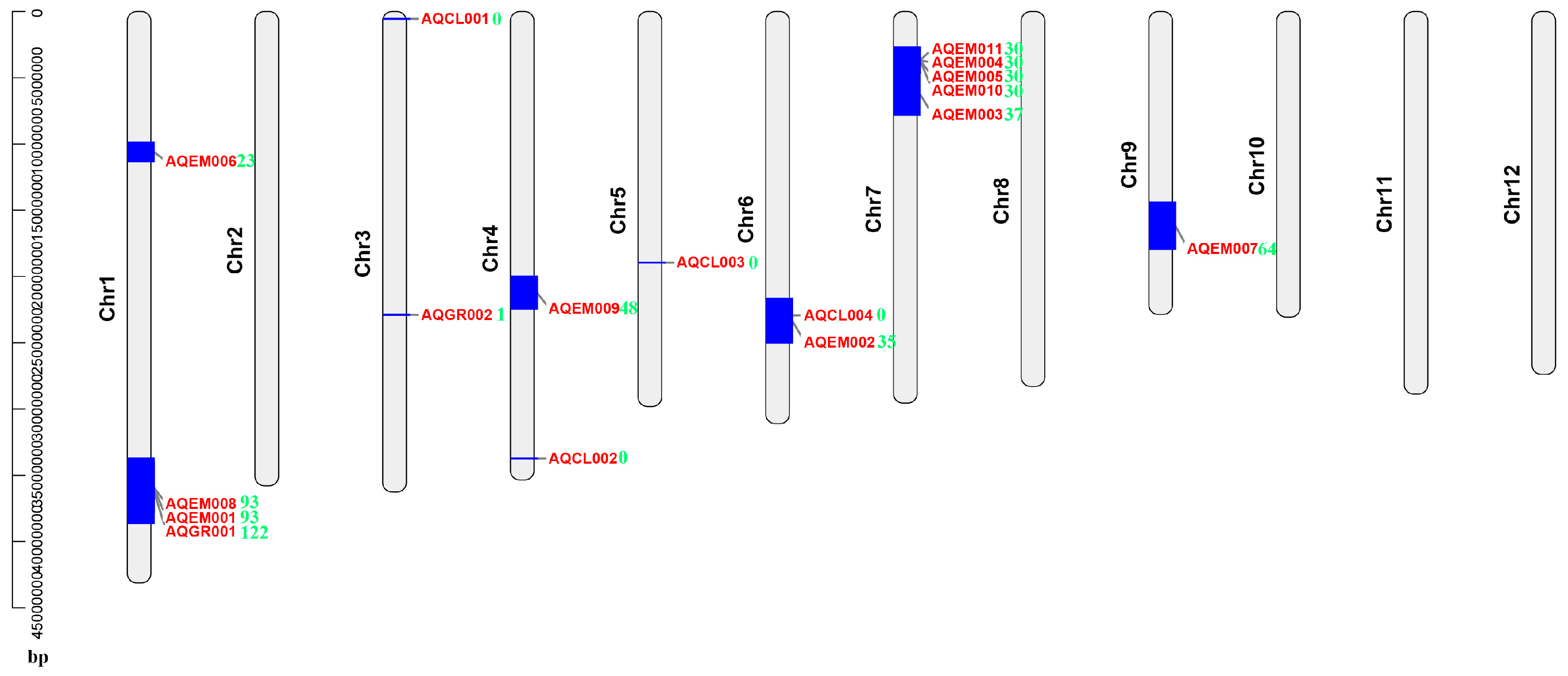
2.6. DEG Mapping on the Previously Identified Salt-Stress-Related QTL Intervals
2.7. Quantitative Real-Time PCR (qRT-PCR) Validation of DEGs
3. Result
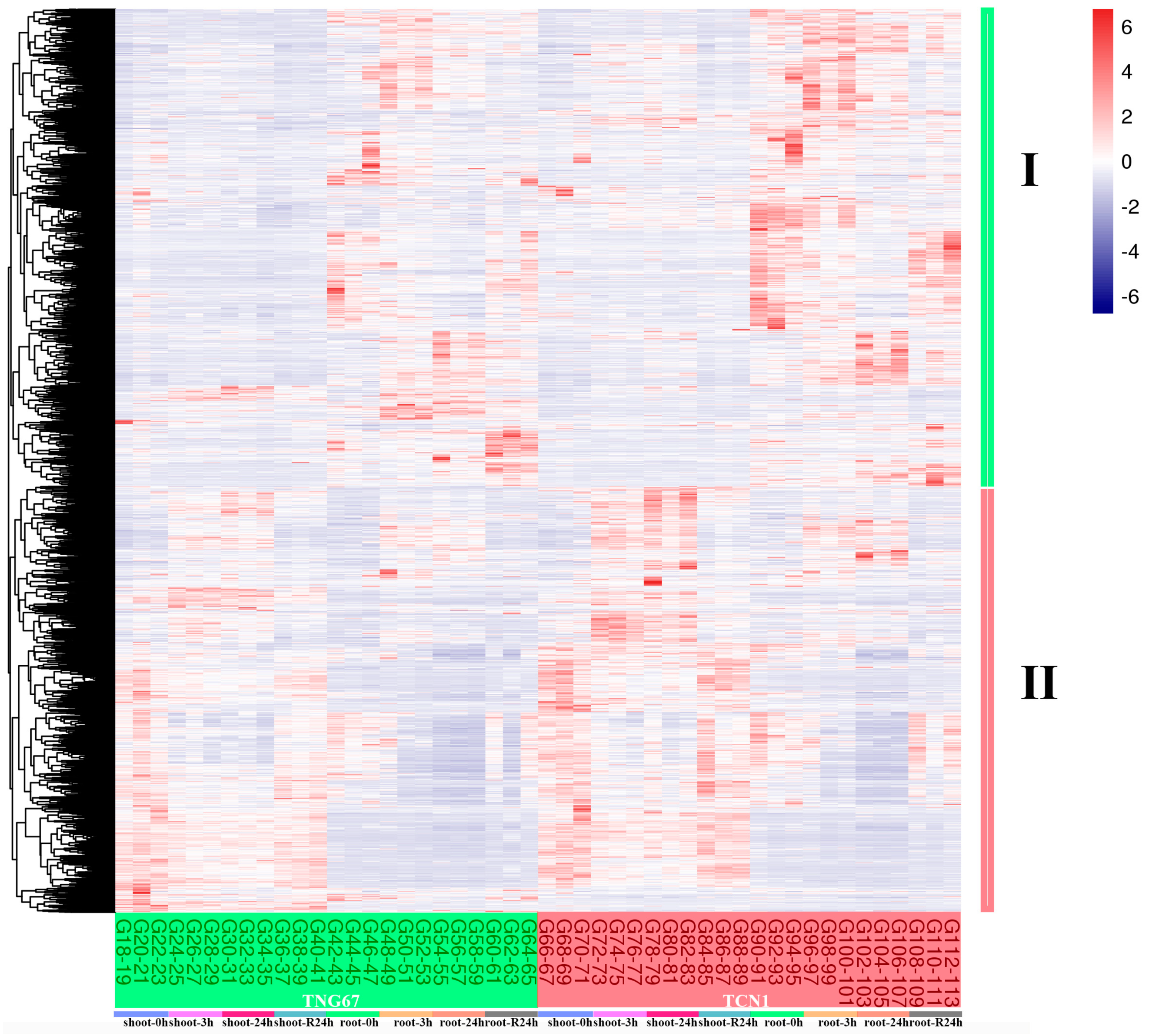
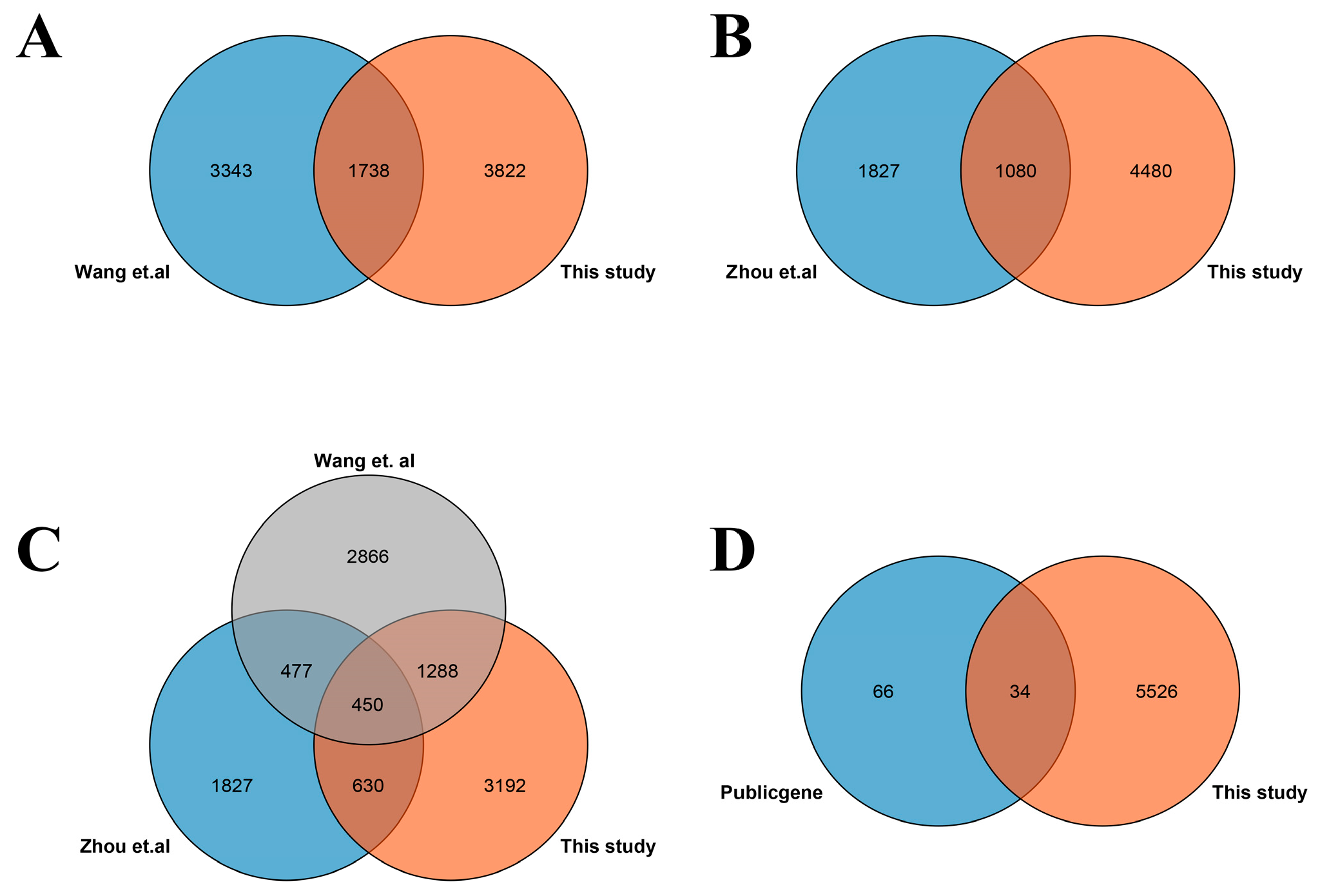
3.1. Identification of DEGs Involved in Salt Stress
3.2. GO Enrichment Analysis of Core Salt-Responsive Genes
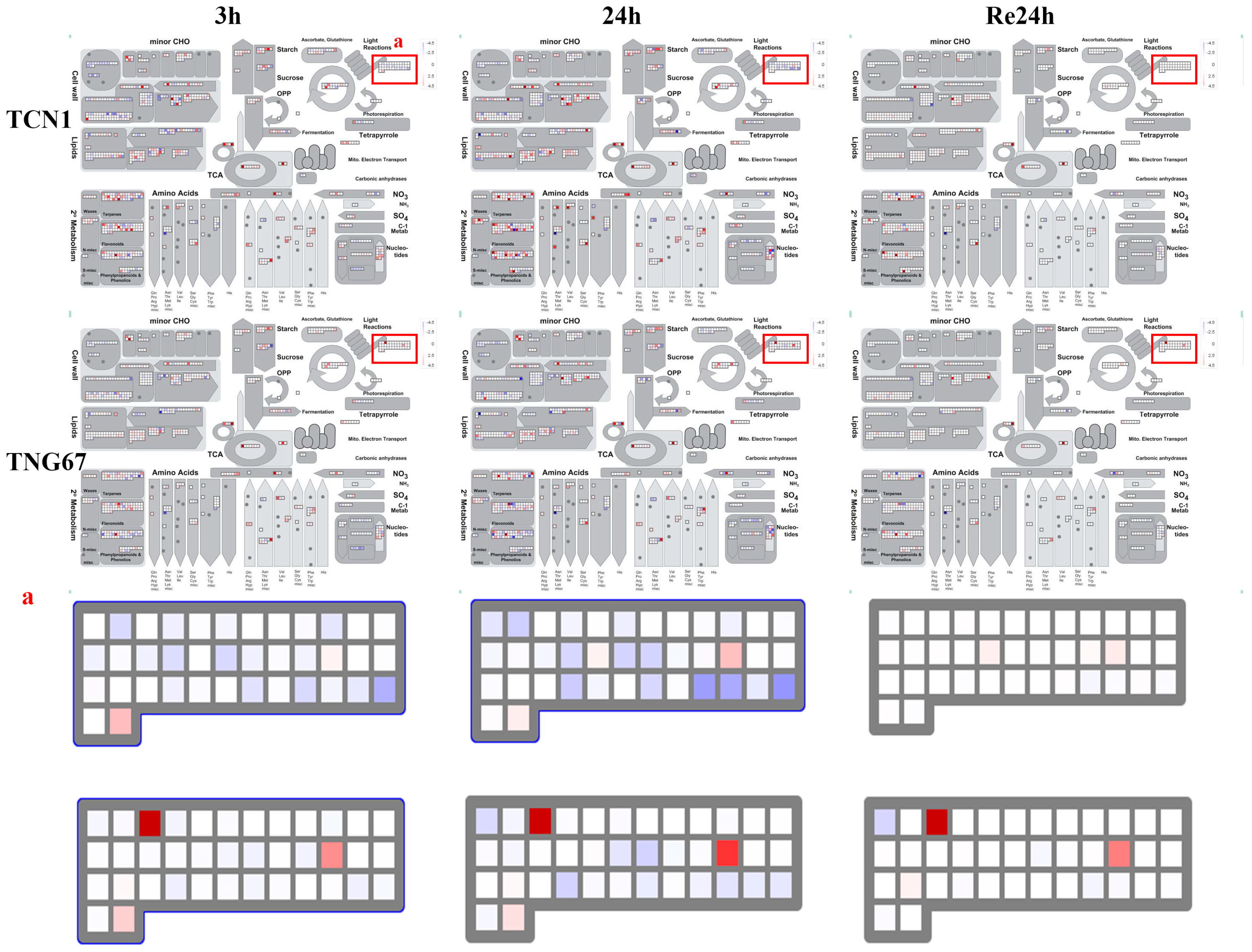
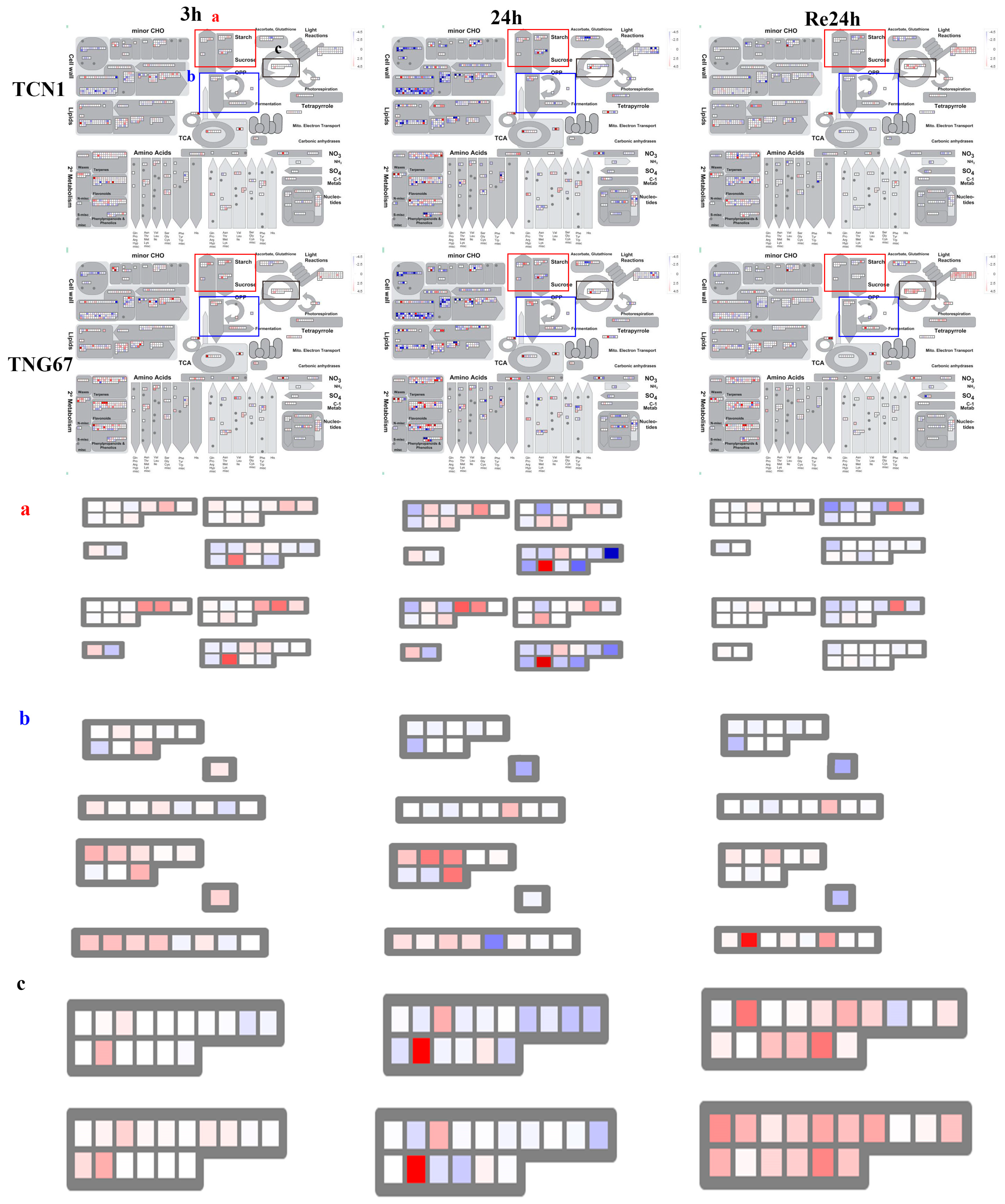
3.3. MapMan Analysis of All Core Salt-Reponsive Genes of Shoots and Roots
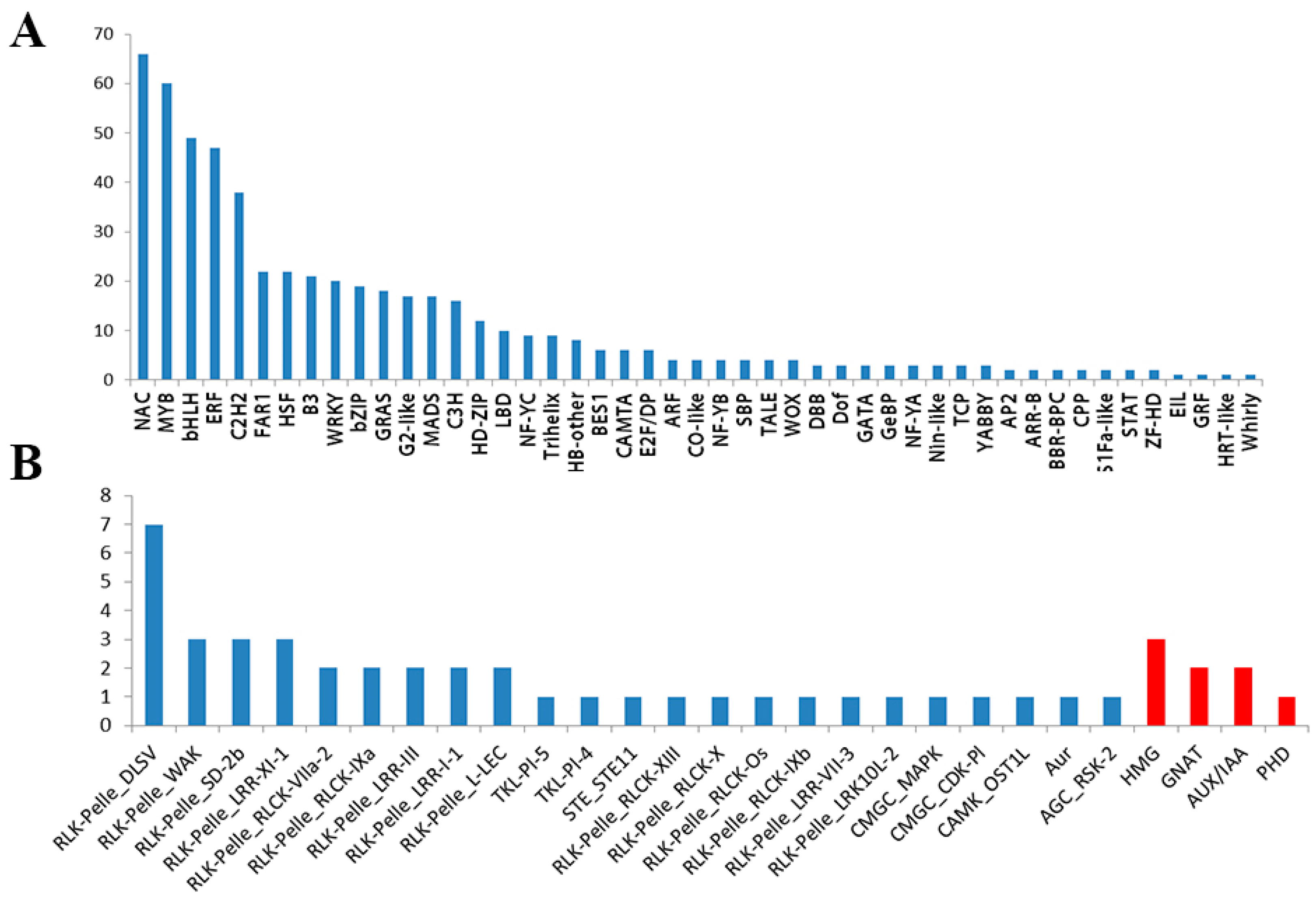
3.4. Identification of TFs, TRs, and PKs Involved in Salt Stress
3.5. Key Genes Involved in the Response to Salt Stress
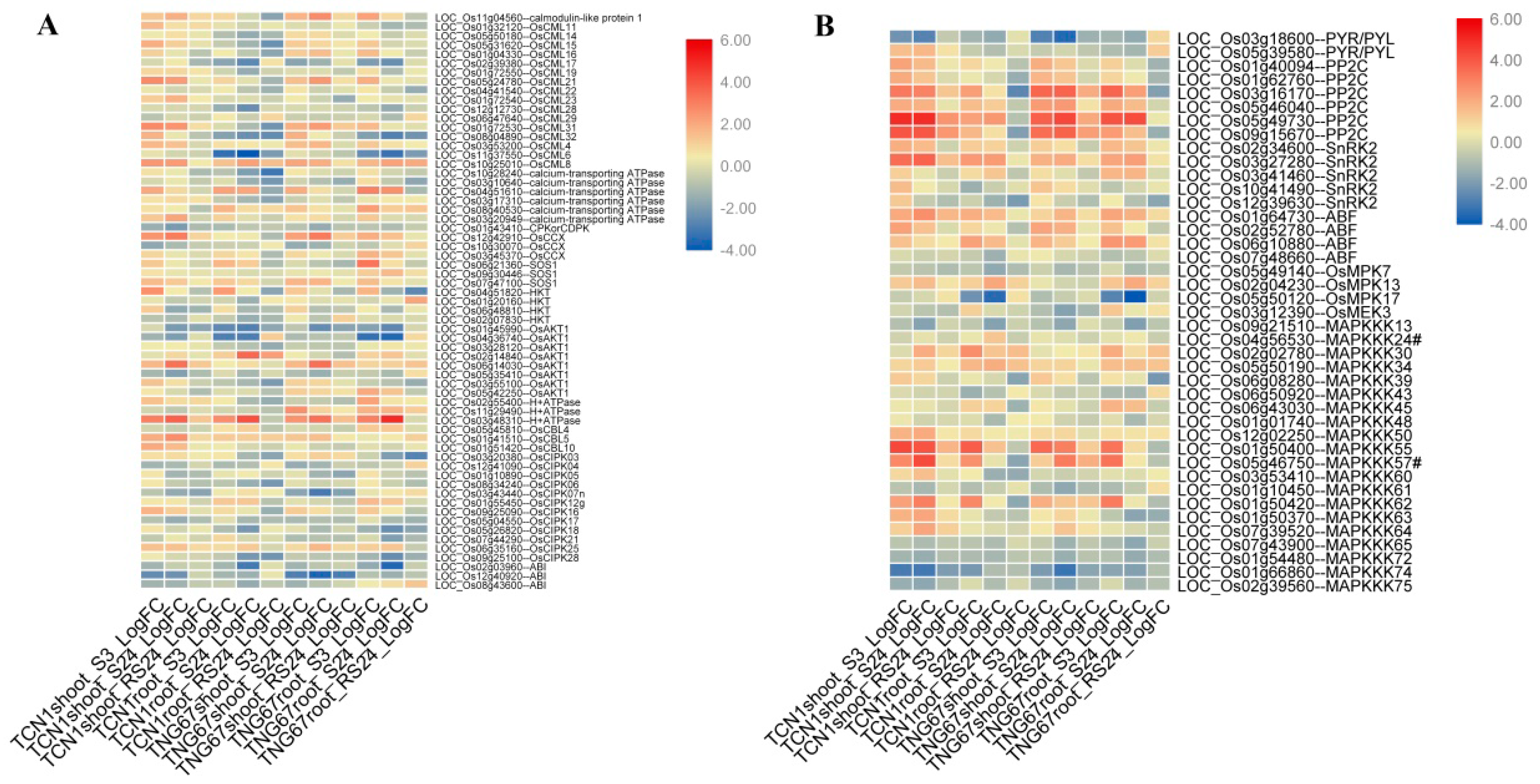
3.5.1. DEGs Involved in the Ca2+ Signal Transduction Pathway
3.5.2. DEGs Involved in the ABA Signal Transduction Pathway
3.5.3. DEGs Involved in the MAPK Cascade Pathway
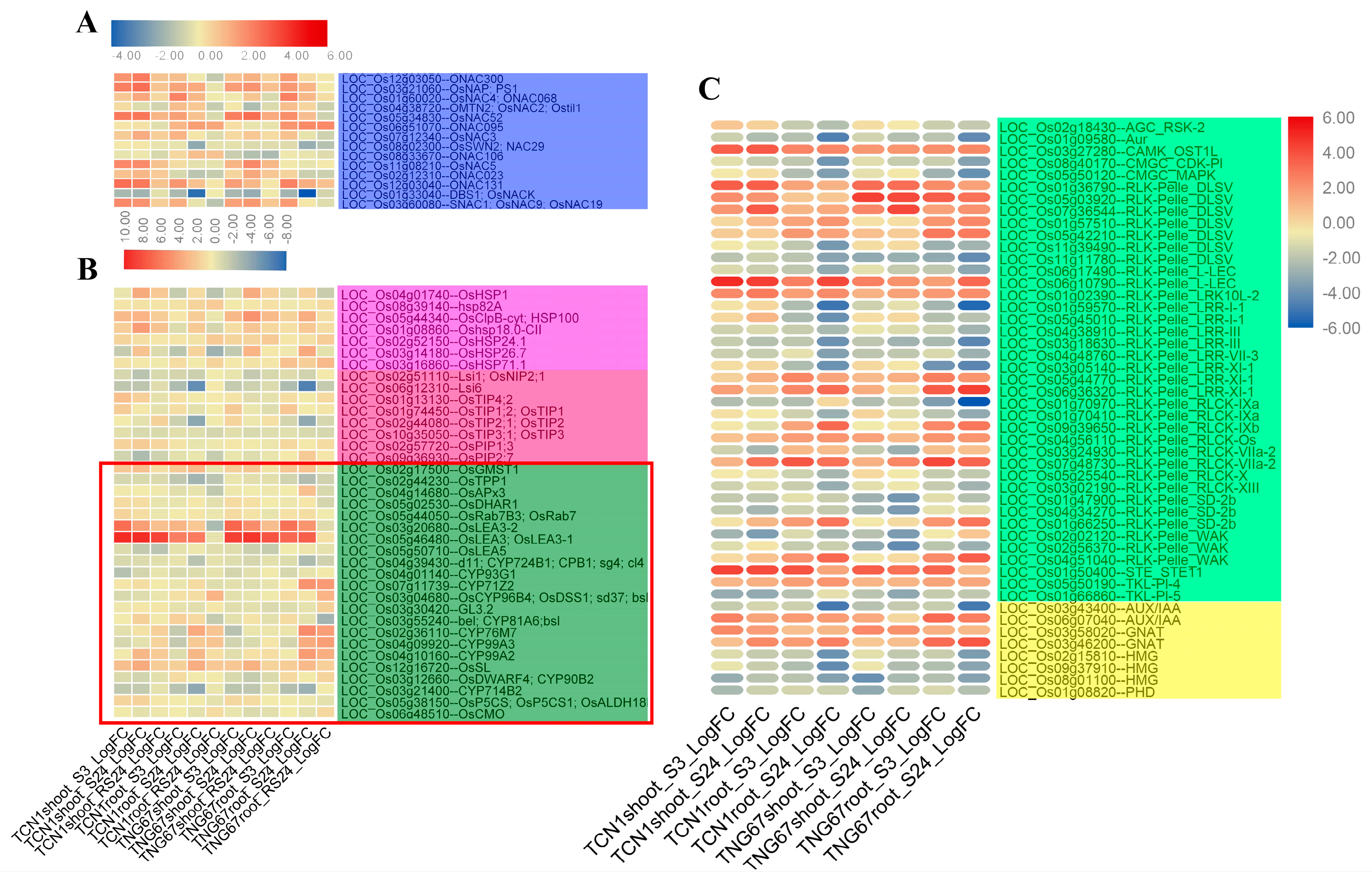
3.5.4. Key TFs and Functional Proteins Related to Salt Stress
3.5.5. TRs and PKs Involved in Salt Stress
3.6. Key Genes Associated with Higher Salt Stress Tolerance in TNG67
4. Discussion
4.1. The Ca2+ Signaling Pathway, ABA Signaling Transduction Pathway, and MAPK Cascade Pathway Play Important Roles under Salt Stress Conditions
4.2. Many TFs and PKs May Be Good Candidate Genes for Plant Salt Stress Tolerance Breeding
4.3. Common DEGs between This Study and Previous Studies Contain Important Salt-Responsive Genes
4.4. Multiple DEGs Mapped to the Previously Identified Salt-Stress-Related QTL Intervals and Could Be Good Candidate Genes for Rice Salt Stress Tolerance Breeding
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef] [PubMed]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef] [PubMed]
- Nouri, M.Z.; Moumeni, A.; Komatsu, S. Abiotic stresses: Insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 2015, 16, 20392–20416. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Na+ transport in plants. FEBS Lett. 2007, 581, 2247–2254. [Google Scholar] [CrossRef]
- Fatehi, F.; Alizadeh, H.; Brimavandi, T.; Struik, P.C. The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Mol. Biol. Rep. 2012, 39, 6387–6397. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Zhang, Y.; Fan, F.; Li, W.; Wang, F.; Zhong, W.; Wang, C.; Yang, J. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 2018, 8, 2085. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA-A Publ. RNA Soc. 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Karan, R.; DeLeon, T.; Biradar, H.; Subudhi, P.K. Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef]
- Sellmann, D.; Becker, T.; Knoch, F. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar]
- Lin, H.X.; Zhu, M.Z.; Yano, M.; Gao, J.P.; Liang, Z.W.; Su, W.A.; Hu, X.H.; Ren, Z.H.; Chao, D.Y. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445. [Google Scholar] [CrossRef] [PubMed]
- Rama, S.; Annapurna, B.; Mukesh, J. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 23719. [Google Scholar] [Green Version]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Yang, Y.W.; Chen, H.C.; Jen, W.F.; Liu, L.Y.; Chang, M.C. Comparative transcriptome analysis of shoots and roots of TNG67 and TCN1 rice seedlings under cold stress and following subsequent recovery: Insights into metabolic pathways, phytohormones, and transcription factors. PLoS ONE 2015, 10, e0131391. [Google Scholar] [CrossRef]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef]
- Chao, D.Y.; Luo, Y.H.; Shi, M.; Luo, D.; Lin, H.X. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005, 15, 796–810. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, X.; Jiao, Y.; Qin, Y.; Liu, X.; He, K.; Chen, C.; Ma, L.; Jian, W.; Xiong, L. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007, 63, 591–608. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, H.; Kawahara, Y.; Sakai, H.; Kanamori, H.; Wakimoto, H.; Yamagata, H.; Oono, Y.; Wu, J.; Ikawa, H.; Itoh, T. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genom. 2010, 11, 683. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.). PLos ONE 2016, 11, e0146242. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Wei, B.; Wang, L.; Du, C.; Hu, G.Z.; Wang, L.; Jin, Y.; Kong, D.L. Identification of differentially expressed genes regulated by transcription factors in glioblastomas by bioinformatics analysis. Mol. Med. Rep. 2015, 11, 2548–2554. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, S.; Li, L.; Zhang, L.; Xian, S.; Wang, Y.; Cheng, Y. Identification of differentially expressed genes and signaling pathways in ovarian cancer by integrated bioinformatics analysis. Oncotargets Ther. 2018, 11, 1457–1474. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Yi, Z.; Chen, J.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar]
- Goffard, N.; Weiller, G. Extending MapMan: Application to legume genome arrays. Bioinformatics 2006, 22, 2958–2959. [Google Scholar] [CrossRef]
- Molla, K.A.; Debnath, A.B.; Ganie, S.A.; Mondal, T.K. Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice ( Oryza sativa L.). BMC Plant Biol. 2015, 15, 122. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica. Plants 2019, 8, 30. [Google Scholar] [CrossRef]
- Wilkins, K.A.; Matthus, E.; Swarbreck, S.M.; Davies, J.M. Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 2016, 7, 1296. [Google Scholar] [CrossRef]
- Mo, C.; Wan, S.; Xia, Y.; Ren, N.; Zhou, Y.; Jiang, X. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 2018, 9, 269. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Redillas, M.C.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Y.; Feng, Q.; Gong, H.; Li, W.; Zhan, Q.; Cheng, B.; Xia, J. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015, 6, 6258. [Google Scholar] [CrossRef] [Green Version]
- García-Morales, S.; Gómez-Merino, F.C.; Trejo-Téllez, L.I. NAC transcription factor expression, amino acid concentration and growth of elite rice cultivars upon salt stress. Acta Physiol. Plant. 2014, 36, 1927–1936. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Wan, S.; Wang, W.; Zhou, T.; Zhang, Y.; Chen, J.; Xiao, B.; Yang, Y.; Yu, Y. Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 2018, 84, 481–492. [Google Scholar] [CrossRef]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- De, Z.A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar]
- Yang, Z.; Lu, R.; Dai, Z.; Yan, A.; Tang, Q.; Cheng, C.; Xu, Y.; Yang, W.; Su, J. Salt-Stress response mechanisms using de Novo transcriptome sequencing of salt-tolerant and sensitive Corchorus spp. genotypes. Genes 2017, 8, 226. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Zhang, S.; Haider, I.; Kohlen, W.; Jiang, L.; Bouwmeester, H.; Meijer, A.H.; Schluepmann, H.; Liu, C.M.; Ouwerkerk, P.B.F. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 2012, 80, 571–585. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Gao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 86–101. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Xiang, J.; Ran, J.; Zou, J.; Zhou, X.; Liu, A.; Zhang, X.; Peng, Y.; Tang, N.; Luo, G.; Chen, X. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Vaid, N.; Pandey, P.; Srivastava, V.K.; Tuteja, N. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol. Biol. 2015, 88, 193–206. [Google Scholar] [CrossRef]
- Li, C.H.; Sun, Y. The Receptor-Like Kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 2014, 26, 2538. [Google Scholar] [CrossRef] [PubMed]
- Seung Yeob, L.; Jeong Ho, A.; Young Soon, C.; Doh Won, Y.; Myung Chul, L.; Jong Cheol, K.; Kyu Seong, L.; Moo Young, E. Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol. Cells 2006, 21, 192. [Google Scholar]


| Gene_Ontology_term | Cluter_frequency | p-Value |
|---|---|---|
| heat acclimation (GO:0010286); | 5.70% | 0 |
| response to cadmium ion (GO:0046686); | 14.25% | 4.16E-10 |
| response to water deprivation (GO:0009414); | 14.81% | 1.59E-09 |
| response to cold (GO:0009409); | 13.11% | 2.01E-09 |
| response to salt stress (GO:0009651); | 14.53% | 8.65E-08 |
| positive regulation of transcription, DNA-templated (GO:0045893); | 10.83% | 9.72E-08 |
| response to heat (GO:0009408); | 7.12% | 1.66E-07 |
| hyperosmotic salinity response (GO:0042538); | 7.41% | 3.62E-07 |
| response to high light intensity (GO:0009644); | 6.27% | 4.43E-07 |
| response to abscisic acid (GO:0009737); | 11.68% | 3.43E-06 |
| negative regulation of abscisic acid-activated signaling pathway (GO:0009788); | 3.70% | 5.57E-06 |
| response to hydrogen peroxide (GO:0042542); | 5.70% | 0.0000161 |
| negative regulation of seed dormancy process (GO:1902039); | 1.14% | 0.0001145 |
| leaf senescence (GO:0010150); | 4.84% | 0.0001683 |
| negative regulation of protein kinase activity (GO:0006469); | 1.42% | 0.0002851 |
| toxin catabolic process (GO:0009407); | 4.56% | 0.0005736 |
| cell proliferation (GO:0008283); | 4.00% | 0.0010226 |
| negative regulation of transcription, DNA-templated (GO:0045892); | 4.84% | 0.0018958 |
| response to chitin (GO:0010200); | 8.55% | 0.0022302 |
| response to hypoxia (GO:0001666); | 2.85% | 0.0038614 |
| response to organic substance (GO:0010033); | 6.27% | 0.0043247 |
| response to wounding (GO:0009611); | 8.26% | 0.0045121 |
| protein folding (GO:0006457); | 4.84% | 0.0050309 |
| release of seed from dormancy (GO:0048838); | 1.14% | 0.005182 |
| hydrogen peroxide catabolic process (GO:0042744); | 3.13% | 0.0056712 |
| glucose catabolic process (GO:0006007); | 3.42% | 0.0062763 |
| ethylene biosynthetic process (GO:0009693); | 3.13% | 0.007078 |
| ethylene-activated signaling pathway (GO:0009873); | 3.99% | 0.0072325 |
| PSII associated light-harvesting complex II catabolic process (GO:0010304); | 2.00% | 0.0081537 |
| photoinhibition (GO:0010205); | 1.42% | 0.0083607 |
| Gene_Ontology_term | Cluter_frequency | p-Value |
|---|---|---|
| hyperosmotic salinity response (GO:0042538); | 8.39% | 0 |
| response to cold (GO:0009409); | 13.12% | 0 |
| response to cadmium ion (GO:0046686); | 13.12% | 0 |
| response to salt stress (GO:0009651); | 15.05% | 0 |
| response to oxidative stress (GO:0006979); | 7.74% | 0 |
| response to nitrate (GO:0010167); | 7.10% | 2.164E-10 |
| defense response to fungus (GO:0050832); | 10.97% | 5.793E-10 |
| response to desiccation (GO:0009269); | 4.30% | 8.921E-10 |
| nitrate transport (GO:0015706); | 7.10% | 1.275E-09 |
| salicylic acid mediated signaling pathway (GO:0009863); | 5.16% | 2.674E-08 |
| response to water deprivation (GO:0009414); | 10.54% | 3.62E-08 |
| root hair elongation (GO:0048767); | 6.88% | 4.066E-08 |
| response to abscisic acid (GO:0009737); | 10.98% | 4.342E-07 |
| lateral root morphogenesis (GO:0010102); | 2.80% | 7.002E-07 |
| oxidation-reduction process (GO:0055114); | 12.69% | 2.418E-06 |
| response to auxin (GO:0009733); | 7.31% | 5.595E-06 |
| defense response to nematode (GO:0002215); | 1.94% | 6.042E-06 |
| oligopeptide transport (GO:0006857); | 3.87% | 2.173E-05 |
| response to cyclopentenone (GO:0010583); | 3.87% | 2.421E-05 |
| response to wounding (GO:0009611); | 8.60% | 2.649E-05 |
| lipid transport (GO:0006869); | 2.79% | 0.0002419 |
| plant-type cell wall organization (GO:0009664); | 5.38% | 0.0002581 |
| transition metal ion transport (GO:0000041); | 3.44% | 0.0003009 |
| selenate transport (GO:0080160); | 1.075% | 0.0005686 |
| lignin biosynthetic process (GO:0009809); | 3.23% | 0.000737 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage. Plants 2019, 8, 64. https://doi.org/10.3390/plants8030064
Kong W, Zhong H, Gong Z, Fang X, Sun T, Deng X, Li Y. Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage. Plants. 2019; 8(3):64. https://doi.org/10.3390/plants8030064
Chicago/Turabian StyleKong, Weilong, Hua Zhong, Ziyun Gong, Xinyi Fang, Tong Sun, Xiaoxiao Deng, and Yangsheng Li. 2019. "Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage" Plants 8, no. 3: 64. https://doi.org/10.3390/plants8030064
APA StyleKong, W., Zhong, H., Gong, Z., Fang, X., Sun, T., Deng, X., & Li, Y. (2019). Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage. Plants, 8(3), 64. https://doi.org/10.3390/plants8030064







