Induction of Jasmonoyl-Isoleucine (JA-Ile)-Dependent JASMONATE ZIM-DOMAIN (JAZ) Genes in NaCl-Treated Arabidopsis thaliana Roots Can Occur at Very Low JA-Ile Levels and in the Absence of the JA/JA-Ile Transporter JAT1/AtABCG16
Abstract
:1. Introduction
2. Results
3. Discussion
3.1. The JA Pathway Can Be Activated Without a Strong JA-Ile Increase
3.2. Potential Artefacts of Low JA-Ile Measurements in Salt-Treated Roots
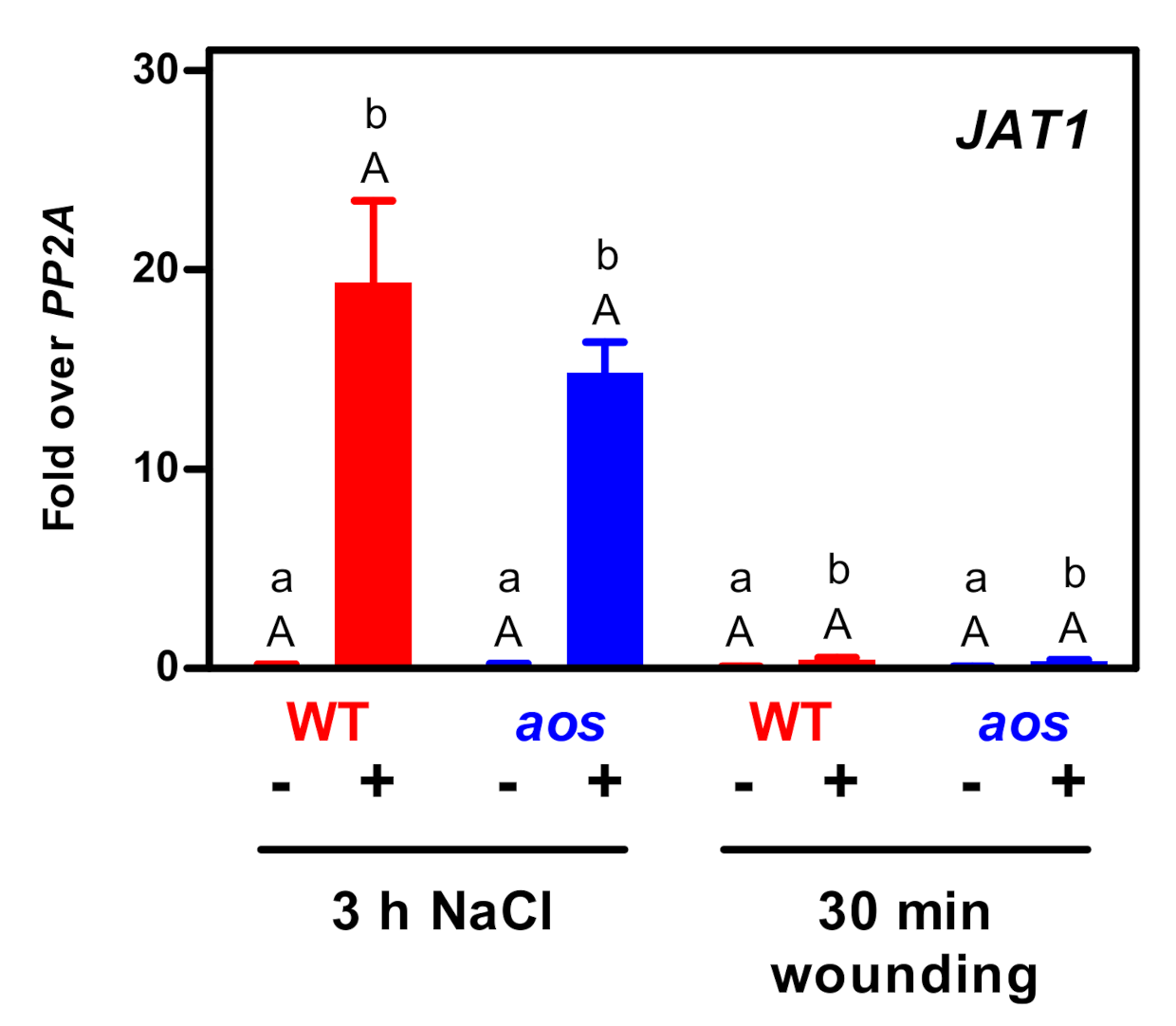
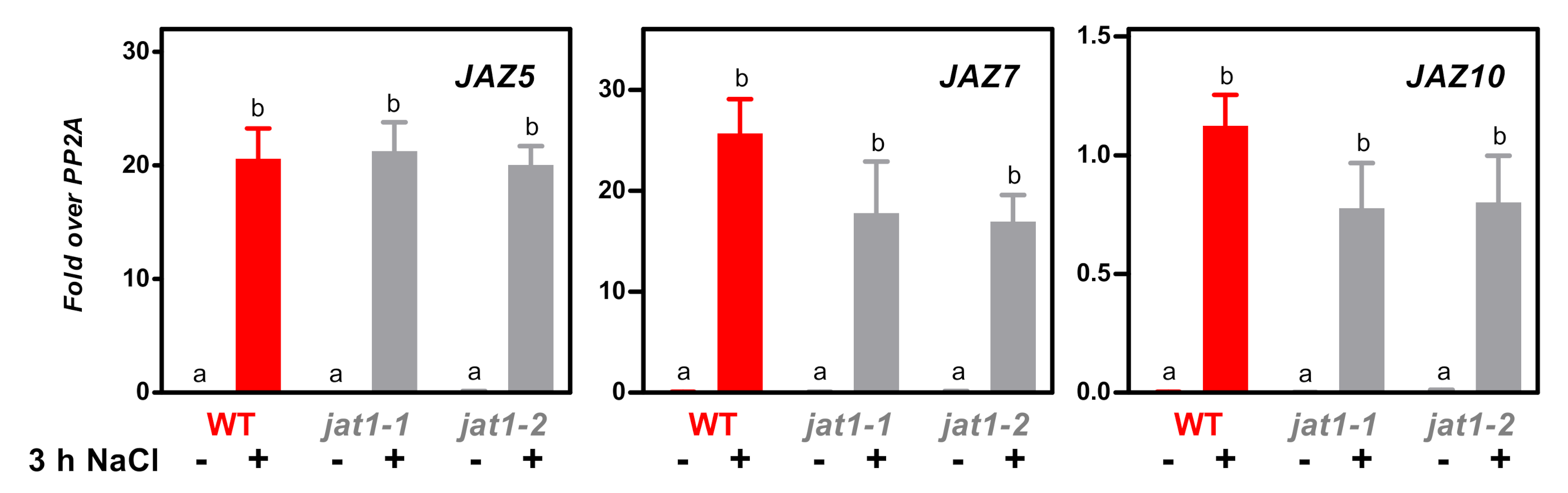
3.3. The JA/JA-Ile Transporter JAT1 Is Not Required for Salt-Induced Expression of JAZ Genes
3.4. Possible Activation Mechanisms of the JA-Ile Signaling Cascade at Low JA-Ile Levels
4. Materials and Methods
4.1. Plant Material, Growth Conditions and Treatment
4.2. Quantitative Reverse Transcription (qRT)-PCR
4.3. Phytohormone Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Feussner, I. The oxylipin pathways: Biochemistry and function. Ann. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Zheng, J.; Li, S.; Huang, G.; Skilling, S.J.; Wang, L.; Li, L.; Li, M.; Yuan, L.; Liu, P. Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential for jasmonate signaling. Mol. Plant 2017, 10, 695–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, 8930–8939. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.; Tham, C.; Duan, L.; Dinneny, J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef] [Green Version]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Gong, Q.; Bohnert, H.J. Dissecting salt stress pathways. J. Exp. Bot. 2006, 57, 1097–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grebner, W.; Stingl, N.E.; Oenel, A.; Mueller, M.J.; Berger, S. Lipoxygenase6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013, 161, 2159–2170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koster, J.; Thurow, C.; Kruse, K.; Meier, A.; Iven, T.; Feussner, I.; Gatz, C. Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 2012, 159, 391–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, D.; Mithofer, A.; Glawischnig, E.; Georgii, E.; Ghirardo, A.; Kanawati, B.; Schmitt-Kopplin, P.; Schnitzler, J.P.; Durner, J.; Gaupels, F. Short-term exposure to nitrogen dioxide provides basal pathogen resistance. Plant Physiol. 2018, 178, 468–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbona, V.; Argamasilla, R.; Gomez-Cadenas, A. Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. J. Plant Physiol. 2010, 167, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- de Ollas, C.; Arbona, V.; Gomez-Cadenas, A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015, 38, 2157–2170. [Google Scholar] [CrossRef] [Green Version]
- Seltmann, M.A.; Stingl, N.E.; Lautenschlaeger, J.K.; Krischke, M.; Mueller, M.J.; Berger, S. Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 2010, 152, 1940–1950. [Google Scholar] [CrossRef] [Green Version]
- Glombitza, S.; Dubuis, P.H.; Thulke, O.; Welzl, G.; Bovet, L.; Gotz, M.; Affenzeller, M.; Geist, B.; Hehn, A.; Asnaghi, C.; et al. Crosstalk and differential response to abiotic and biotic stressors reflected at the transcriptional level of effector genes from secondary metabolism. Plant Mol. Biol. 2004, 54, 817–835. [Google Scholar] [CrossRef] [Green Version]
- Koo, A.J.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubert, Y.; Widemann, E.; Miesch, L.; Pinot, F.; Heitz, T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 2015, 66, 3879–3892. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, C.E.; Acevedo-Acevedo, O.; Miranda, G.S.; Vergara-Barros, P.; Holuigue, L.; Figueroa, C.R.; Figueroa, P.M. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4209–4220. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.C.; Liao, P.M.; Kuo, W.W.; Lin, T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Piqueras, R.; Sanchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jia, H.; Wang, J. cGMP and ethylene are involved in maintaining ion homeostasis under salt stress in Arabidopsis roots. Plant Cell Rep. 2014, 33, 447–459. [Google Scholar] [CrossRef]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Saldierna Guzman, J.P.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A.; et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar]
- Tischer, S.V.; Wunschel, C.; Papacek, M.; Kleigrewe, K.; Hofmann, T.; Christmann, A.; Grill, E. Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 10280–10285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu-Mitao, Y.; Kakimoto, T. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol. 2014, 55, 1450–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruschhaupt, M.; Mergner, J.; Mucha, S.; Papacek, M.; Doch, I.; Tischer, S.V.; Hemmler, D.; Chiasson, D.; Edel, K.H.; Kudla, J.; et al. Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO J. 2019, 38, e101859. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Zhang, T.; Koo, A.J.; Stacey, G.; Tanaka, K. Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 2018, 176, 511–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant signaling networks involving Ca(2+) and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 2015, 6, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terrile, M.C.; Paris, R.; Calderon-Villalobos, L.I.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongue, C.A. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [Green Version]
- Heitz, T.; Smirnova, E.; Marquis, V.; Poirier, L. Metabolic control within the jasmonate biochemical pathway. Plant Cell Physiol. 2019, 60, 2621–2628. [Google Scholar] [CrossRef]
- Ellis, C.; Turner, J.G. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 2002, 215, 549–556. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Stingl, N.; Krischke, M.; Fekete, A.; Mueller, M.J. Analysis of defense signals in Arabidopsis thaliana leaves by ultra-performance liquid chromatography/tandem mass spectrometry: Jasmonates, salicylic acid, abscisic acid. Methods Mol. Biol. 2013, 1009, 103–113. [Google Scholar] [PubMed]
- de Vries, S.; de Vries, J.; von Dahlen, J.K.; Gould, S.B.; Archibald, J.M.; Rose, L.E.; Slamovits, C.H. On plant defense signaling networks and early land plant evolution. Commun. Integr. Biol. 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thurow, C.; Krischke, M.; Mueller, M.J.; Gatz, C. Induction of Jasmonoyl-Isoleucine (JA-Ile)-Dependent JASMONATE ZIM-DOMAIN (JAZ) Genes in NaCl-Treated Arabidopsis thaliana Roots Can Occur at Very Low JA-Ile Levels and in the Absence of the JA/JA-Ile Transporter JAT1/AtABCG16. Plants 2020, 9, 1635. https://doi.org/10.3390/plants9121635
Thurow C, Krischke M, Mueller MJ, Gatz C. Induction of Jasmonoyl-Isoleucine (JA-Ile)-Dependent JASMONATE ZIM-DOMAIN (JAZ) Genes in NaCl-Treated Arabidopsis thaliana Roots Can Occur at Very Low JA-Ile Levels and in the Absence of the JA/JA-Ile Transporter JAT1/AtABCG16. Plants. 2020; 9(12):1635. https://doi.org/10.3390/plants9121635
Chicago/Turabian StyleThurow, Corinna, Markus Krischke, Martin J. Mueller, and Christiane Gatz. 2020. "Induction of Jasmonoyl-Isoleucine (JA-Ile)-Dependent JASMONATE ZIM-DOMAIN (JAZ) Genes in NaCl-Treated Arabidopsis thaliana Roots Can Occur at Very Low JA-Ile Levels and in the Absence of the JA/JA-Ile Transporter JAT1/AtABCG16" Plants 9, no. 12: 1635. https://doi.org/10.3390/plants9121635



