Soybean Resistance to Soybean Mosaic Virus
Abstract
:1. Introduction
2. Biological Properties and Transmission of SMV
2.1. SMV Genome and Gene Function
2.2. Biological and Molecular Properties of SMV Infection and Transmission
3. Resistance Genes (R-Genes): Soybean Response to SMV Infection
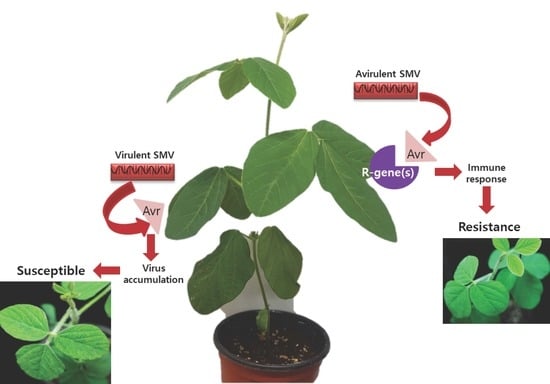
3.1. NLR Gene Family-Mediated Resistance to SMV
3.2. Rsv Genes
3.3. Rsc Genes
4. Independent Host Factors Involved in Soybean-SMV Interaction
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the World 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. J. Food Secur. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Pagano, M.C.; Miransari, M. The importance of Soybean Production Worldwide; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–26. [Google Scholar]
- Hill, J.H.; Whitham, S.A. Control of virus diseases in soybeans. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar] [PubMed]
- Malapi-Nelson, M.; Wen, R.-H.; Ownley, B.; Hajimorad, M. Co-infection of soybean with Soybean mosaic virus and Alfalfa mosaic virus results in disease synergism and alteration in accumulation level of both viruses. Plant Dis. 2009, 93, 1259–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, G.L.; Hill, C.B. 13 Diseases of Soybean and Their Management; CABI: USA, 2010; p. 276. [Google Scholar]
- Hajimorad, M.; Domier, L.L.; Tolin, S.; Whitham, S.; Saghai Maroof, M. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvez, L.C.; Banerjee, J.; Pinar, H.; Mitra, A. Engineered plant virus resistance. Plant Sci. 2014, 228, 11–25. [Google Scholar] [CrossRef]
- Choi, B.K.; Koo, J.M.; Ahn, H.J.; Yum, H.J.; Choi, C.W.; Ryu, K.H.; Chen, P.; Tolin, S. Emergence of Rsv resistance breaking Soybean mosaic virus isolates from Korean soybean cultivars. Virus Res. 2005, 112, 42–51. [Google Scholar] [CrossRef]
- Koo, J.; Choi, B.; Ahn, H.; Yum, H.; Choi, C. First report of an Rsv resistance-breaking isolate of Soybean mosaic virus in Korea. Plant Pathol. 2005, 54, 573. [Google Scholar] [CrossRef]
- Kang, B.-C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef] [Green Version]
- Calvo, M.; Martínez-Turiño, S.; García, J.A. Resistance to Plum pox virus Strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant-Microbe Interact. 2014, 27, 1291–1301. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [Green Version]
- Baebler, S.; Witek, K.; Petek, M.; Stare, K.; Tusek-Znidaric, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Zyta, D.; Marczewski, W.; et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J. Exp. Bot. 2014, 65, 1095–1109. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Loss and gain of elicitor function of soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajimorad, M.; Hill, J. Rsv1-mediated resistance against Soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 2001, 14, 587–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1–mediated extreme resistance to Soybean mosaic virus. Mol. Plant Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Alazem, M.; Tseng, K.-C.; Chang, W.-C.; Seo, J.-K.; Kim, K.-H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 581. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; Widyasari, K.; Kim, K.H. An Avirulent Strain of Soybean Mosaic Virus Reverses the Defensive Effect of Abscisic Acid in a Susceptible Soybean Cultivar. Viruses 2019, 11, 879. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; He, M.H.; Moffett, P.; Lin, N.S. Abscisic Acid Induces Resistance against Bamboo Mosaic Virus through Argonaute2 and 3. Plant Physiol. 2017, 174, 339–355. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.; Yu, K.; Wang, A. Pathogenesis of Soybean mosaic virus in soybean carrying Rsv1 gene is associated with miRNA and siRNA pathways, and breakdown of AGO1 homeostasis. Virology 2015, 476, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, H.; Niu, H.; Luo, J.; Zhi, H. Soybean Cytochrome b5 Is a Restriction Factor for Soybean Mosaic Virus. Viruses 2019, 11, 546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019, 99, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Maroof, S.; Jeong, S.C.; Gunduz, I.; Tucker, D.; Buss, G.; Tolin, S. Pyramiding of soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci. 2008, 48, 517–526. [Google Scholar] [CrossRef]
- Shakiba, E.; Chen, P.; Shi, A.; Li, D.; Dong, D.; Brye, K. Two novel alleles at the Rsv 3 locus for resistance to Soybean mosaic virus in PI 399091 and PI 61947 soybeans. Crop Sci. 2012, 52, 2587–2594. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genomics 2017, 292, 811–822. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J.; et al. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H.; et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Liu, N.; Yang, Z.; Zheng, G.; Zhi, H. Fine mapping and identification of the soybean RSC4 resistance candidate gene to soybean mosaic virus. Plant Breed. 2011, 130, 653–659. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of R SC8 resistance candidate genes to soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Wang, D.-G.; Lin, Z.; Kai, L.; Ying, M.; Wang, L.-Q.; Yang, Y.-Q.; Yang, Y.-H.; Zhi, H.-J. Marker-assisted pyramiding of soybean resistance genes RSC4, RSC8, and RSC14Q to soybean mosaic virus. J. Integr. Agric. 2017, 16, 2413–2420. [Google Scholar] [CrossRef]
- Cho, E.-K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.; Zhi, H.; Gai, J. Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Tian, Z.; Li, K.; Li, H.; Huang, Z.; Hu, G.; Zhang, L.; Zhi, H. Identification and variation analysis of soybean mosaic virus strains in Shandong, Henan and Anhui provinces of China. Soybean Science 2013, 32, 806–809. [Google Scholar]
- Riechmann, J.L.; Lain, S.; García, J.A. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 1992, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Olspert, A.; Chung, B.Y.W.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [Green Version]
- Verchot, J.; Carrington, J.C. Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 1995, 69, 3668–3674. [Google Scholar] [CrossRef] [Green Version]
- Kapust, R.B.; Tözsér, J.; Copeland, T.D.; Waugh, D.S. The P1′ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002, 294, 949–955. [Google Scholar] [CrossRef] [Green Version]
- Maliogka, V.I.; Salvador, B.; Carbonell, A.; Saenz, P.; León, D.S.; Oliveros, J.C.; Delgadillo, M.O.; García, J.A.; Simón-Mateo, C. Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Mol. Plant Pathol. 2012, 13, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Salvador, B.; Saenz, P.; Yangüez, E.; Quiot, J.B.; Quiot, L.; Delgadillo, M.O.; Garcia, J.A.; Simón-Mateo, C. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 2008, 9, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Carrington, J.C. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 2001, 285, 71–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Kadoury, D.; Gal-On, A.; Huet, H.; Wang, Y.; Raccah, B. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: Effects on aphid transmission and binding to purified virions. J. Gen. Virol. 1998, 79, 897–904. [Google Scholar] [CrossRef]
- Seo, J.K.; Kang, S.H.; Seo, B.Y.; Jung, J.K.; Kim, K.H. Mutational analysis of interaction between coat protein and helper component-proteinase of Soybean mosaic virus involved in aphid transmission. Mol. Plant Pathol. 2010, 11, 265–276. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Carrington, J.C. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 1998, 95, 461–470. [Google Scholar] [CrossRef] [Green Version]
- Wen, R.-H.; Khatabi, B.; Ashfield, T.; Maroof, M.S.; Hajimorad, M. The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 2013, 26, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Eggenberger, A.; Hajimorad, M.; Hill, J. Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef] [Green Version]
- Luan, H.; Shine, M.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Strain-specific P3 of Soybean mosaic virus elicits Rsv1-mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1-genotype soybean. Virology 2006, 345, 156–166. [Google Scholar] [CrossRef] [Green Version]
- Chowda-Reddy, R.; Sun, H.; Chen, H.; Poysa, V.; Ling, H.; Gijzen, M.; Wang, A. Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Mol. Plant Microbe Interact. 2011, 24, 37–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, R.-H.; Hajimorad, M. Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 2010, 400, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, X.-Y.; Chen, J.; Shi, Y.-H.; Chen, J.-P. The ‘6K1’protein of a strain of Soybean mosaic virus localizes to the cell periphery. Arch. Virol. 2007, 152, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Jensen, P.E.; Schaad, M.C. Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 1998, 14, 393–400. [Google Scholar] [CrossRef]
- Seo, J.-K.; Lee, S.-H.; Kim, K.-H. Strain-specific cylindrical inclusion protein of Soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant Microbe Interact. 2009, 22, 1151–1159. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Hajimorad, M.; Eggenberger, A.L.; Tsang, S.; Whitham, S.A.; Hill, J.H. Cytoplasmic inclusion cistron of Soybean mosaic virus serves as a virulence determinant on Rsv3-genotype soybean and a symptom determinant. Virology 2009, 391, 240–248. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Wang, A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI-and COPII-dependent manner. J. Virol. 2008, 82, 12252–12264. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Huang, T.-S.; McNeil, J.; Laliberté, J.-F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Michon, T.; Estevez, Y.; Walter, J.; German-Retana, S.; Le Gall, O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 2006, 273, 1312–1322. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF (iso) 4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef] [Green Version]
- Carrington, J.C.; Freed, D.D.; Leinicke, A.J. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell 1991, 3, 953–962. [Google Scholar] [PubMed] [Green Version]
- Adams, M.J.; Antoniw, J.F.; Beaudoin, F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005, 6, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charon, J.; Theil, S.; Nicaise, V.; Michon, T. Protein intrinsic disorder within the Potyvirus genus: From proteome-wide analysis to functional annotation. Mol. Biosyst. 2016, 12, 634–652. [Google Scholar] [CrossRef]
- Hong, Y.; Hunt, A.G. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology 1996, 226, 146–151. [Google Scholar] [CrossRef]
- Kang, S.-H.; Lim, W.-S.; Hwang, S.-H.; Park, J.-W.; Choi, H.-S.; Kim, K.-H. Importance of the C-terminal domain of soybean mosaic virus coat protein for subunit interactions. J. Gen. Virol. 2006, 87, 225–229. [Google Scholar] [CrossRef]
- Steinlage, T.A.; Hill, J.; Nutter Jr, F. Temporal and spatial spread of Soybean mosaic virus (SMV) in soybeans transformed with the coat protein gene of SMV. Phytopathology 2002, 92, 478–486. [Google Scholar] [CrossRef]
- Helm, M.; Qi, M.; Sarkar, S.; Yu, H.; Whitham, S.A.; Innes, R.W. Engineering a decoy substrate in soybean to enable recognition of the Soybean Mosaic Virus NIa protease. Mol. Plant Microbe Interact. 2019, 32, 760–769. [Google Scholar] [CrossRef]
- Reagan, B.C.; Burch-Smith, T.M. Viruses Reveal the Secrets of Plasmodesmal Cell Biology. Mol. Plant Microbe Interact. 2019, 33, 26–39. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B. Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef] [Green Version]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genomics 2014, 15, 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Nachappa, P.; Culkin, C.T.; Saya, P.M.; Han, J.; Nalam, V.J. Water stress modulates soybean aphid performance, feeding behavior, and virus transmission in soybean. Front. Plant Sci. 2016, 7, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jossey, S.; Hobbs, H.A.; Domier, L.L. Role of Soybean mosaic virus–encoded proteins in seed and aphid transmission in soybean. Phytopathology 2013, 103, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Noman, A.; Aqeel, M.; Lou, Y. PRRs and NB-LRRs: From signal perception to activation of plant innate immunity. Int. J. Mol. Sci. 2019, 20, 1882. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [Green Version]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef] [Green Version]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyers, B.C.; Morgante, M.; Michelmore, R.W. TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002, 32, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-Z.; Fang, Y.; Pang, H. The current status of the soybean-soybean mosaic virus (SMV) pathosystem. Front. Microbiol. 2016, 7, 1906. [Google Scholar] [CrossRef] [PubMed]
- Khatabi, B.; Fajolu, O.; Wen, R.H.; Hajimorad, M. Evaluation of N orth A merican isolates of S oybean mosaic virus for gain of virulence on Rsv-genotype soybeans with special emphasis on resistance-breaking determinants on Rsv4. Mol. Plant Pathol. 2012, 13, 1077–1088. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Chen, S.; Shu, L.; Palmer, R.G.; Xing, G.; Li, Y.; Yang, S.; Yu, D.; Zhao, T. Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome. J. Integr. Plant Biol. 2014, 56, 1009–1019. [Google Scholar] [CrossRef]
- Zhan, Y.; Zhi, H.; Yu, D.; Gai, J. Identification and distribution of SMV strains in huang-huai valleys. Agric. Sci. China 2006, 39, 2009–2015. [Google Scholar]
- Hayes, A.; Jeong, S.; Gore, M.; Yu, Y.; Buss, G.; Tolin, S.; Maroof, M.S. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Shi, A.; Chen, P.; Li, D.; Zheng, C.; Zhang, B.; Hou, A. Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol. Breed. 2009, 23, 113. [Google Scholar] [CrossRef]
- Wang, J.; Shine, M.; Gao, Q.-M.; Navarre, D.; Jiang, W.; Liu, C.; Chen, Q.; Hu, G.; Kachroo, A. Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 2014, 165, 1269–1284. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.P.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I.E. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, C.; Hwang, S.H.; Fang, I.R.; Kwon, S.I.; Park, S.R.; Ahn, I.; Kim, J.B.; Hwang, D.J. Molecular characterization of Oryza sativa WRKY 6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015, 208, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.J.; Bowman, B.C.; Jeong, N.; Yang, K.; Kastl, C.; Tolin, S.A.; Maroof, M.; Jeong, S.-C. The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 2011, 4, 55–64. [Google Scholar] [CrossRef]
- Tran, P.-T.; Widyasari, K.; Seo, J.-K.; Kim, K.-H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 513, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Gunduz, I. Genetic Analysis of Soybean Mosaic Virus Resistance in Soybean; Virginia Tech: Blacksburg, VA, USA, 2000. [Google Scholar]
- Jeong, S.; Kristipati, S.; Hayes, A.; Maughan, P.; Noffsinger, S.; Gunduz, I.; Buss, G.; Maroof, M. Genetic and sequence analysis of markers tightly linked to the soybean mosaic virus resistance gene, Rsv 3. Crop Sci. 2002, 42, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.-K.; Kwon, S.-J.; Cho, W.K.; Choi, H.-S.; Kim, K.-H. Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 2014, 4, 5905. [Google Scholar] [CrossRef] [Green Version]
- Hayes, A.J.; Ma, G.; Buss, G.R.; Maroof, M. Molecular marker mapping of Rsv 4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci. 2000, 40, 1434–1437. [Google Scholar] [CrossRef]
- Maroof, M.; Tucker, D.M.; Skoneczka, J.A.; Bowman, B.C.; Tripathy, S.; Tolin, S.A. Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 2010, 3, 14–22. [Google Scholar] [CrossRef]
- Gunduz, I.; Buss, G.R.; Chen, P.; Tolin, S.A. Genetic and phenotypic analysis of Soybean mosaic virus resistance in PI 88788 soybean. Phytopathology 2004, 94, 687–692. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Khatabi, B.; Hajimorad, M. Amino acid substitution in P 3 of S oybean mosaic virus to convert avirulence to virulence on R sv4-genotype soybean is influenced by the genetic composition of P 3. Mol. Plant Pathol. 2015, 16, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Klepadlo, M.; Chen, P.; Shi, A.; Mason, R.E.; Korth, K.L.; Srivastava, V.; Wu, C. Two tightly linked genes for Soybean mosaic virus resistance in soybean. Crop Sci. 2017, 57, 1844–1853. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, P.; Gergerich, R. Characterization of resistance to Soybean mosaic virus in diverse soybean germplasm. Crop Sci. 2005, 45, 2503–2509. [Google Scholar] [CrossRef]
- Shi, A.; Chen, P.; Zheng, C.; Hou, A.; Zhang, B. A PCR-based marker for the Rsv1 locus conferring resistance to soybean mosaic virus. Crop Sci. 2008, 48, 262–268. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, P.; Li, D.; Gergerich, R. New sources of resistance to Soybean mosaic virus in soybean. Can. J. Plant Pathol. 2008, 30, 595–603. [Google Scholar] [CrossRef]
- Buss, G.; Ma, G.; Chen, P.; Tolin, S. Registration of V94-5152 soybean germplasm resistant to soybean mosaic potyvirus. Crop Sci. 1997, 37, 1987–1988. [Google Scholar] [CrossRef]
- Ma, G.; Chen, P.; Buss, G.; Tolin, S. Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor. Appl. Genet. 1995, 91, 907–914. [Google Scholar] [CrossRef]
- Ilut, D.C.; Lipka, A.E.; Jeong, N.; Bae, D.N.; Kim, D.H.; Kim, J.H.; Redekar, N.; Yang, K.; Park, W.; Kang, S.-T.; et al. Identification of haplotypes at the Rsv4 genomic region in soybean associated with durable resistance to soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 453–468. [Google Scholar] [CrossRef]
- Fu, S.; Zhan, Y.; Zhi, H.; Gai, J.; Yu, D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 2006, 128, 63–69. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Cheng, H.; Hu, Z.; Chu, S.; Zhang, G.; Yu, D. Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J. Integr. Plant Biol. 2015, 57, 722–729. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, D.; Zhang, H.; Shen, Y.; Yang, Y.; Li, K.; Wang, L.; Yang, Y.; Zhi, H. Fine mapping of the RSC8 locus and expression analysis of candidate SMV resistance genes in soybean. Plant Breed. 2016, 135, 701–706. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Wang, D.; Liu, N.; Zhi, H. Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legume Genomics Genet. 2010, 1. [Google Scholar] [CrossRef]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.; Ren, R.; Yang, Y.; Zhi, H. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Zheng, G.-J.; Yang, Y.-Q.; Ying, M.; Yang, X.-F.; Chen, S.-Y.; Rui, R.; Wang, D.-G.; Yang, Z.-L.; Zhi, H.-J. Fine mapping and candidate gene analysis of resistance gene RSC3Q to soybean mosaic virus in Qihuang 1. J. Integr. Agric. 2014, 13, 2608–2615. [Google Scholar] [CrossRef] [Green Version]
- Li, H.C.; Zhi, H.J.; Gai, J.Y.; Guo, D.Q.; Wang, Y.W.; Li, K.; Bai, L.; Yang, H. Inheritance and gene mapping of resistance to soybean mosaic virus strain SC14 in soybean. J. Integr. Plant Biol. 2006, 48, 1466–1472. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, D.-G.; Li, H.-C.; Zheng, G.-J.; Yang, Y.-Q.; Li, H.-W.; Zhi, H.-J. Fine mapping of the R SC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 2011, 181, 127–135. [Google Scholar] [CrossRef]
- Li, K.; Ren, R.; Adhimoolam, K.; Gao, L.; Yuan, Y.; Liu, Z.; Zhong, Y.; Zhi, H. Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed. 2015, 134, 684–695. [Google Scholar] [CrossRef]
- Li, N.; Yin, J.L.; Li, C.; Wang, D.G.; Yang, Y.Q.; Karthikeyan, A.; Luan, H.X.; Zhi, H.J. NB-LRR gene family required for Rsc4-mediated resistance to Soybean mosaic virus. Crop Pasture Sci. 2016, 67, 541–552. [Google Scholar] [CrossRef]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Liu, J.-Z.; Horstman, H.D.; Braun, E.; Graham, M.A.; Zhang, C.; Navarre, D.; Qiu, W.-L.; Lee, Y.; Nettleton, D.; Hill, J.H.; et al. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011, 157, 1363–1378. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-Z.; Braun, E.; Qiu, W.-L.; Shi, Y.-F.; Marcelino-Guimarães, F.C.; Navarre, D.; Hill, J.H.; Whitham, S.A. Positive and negative roles for soybean MPK6 in regulating defense responses. Mol. Plant-Microbe Interact. 2014, 27, 824–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyán, J.; et al. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, N.; Sahu, P.P.; Puranik, S.; Prasad, M. Recent advances in plant–virus interaction with emphasis on small interfering RNAs (siRNAs). Mol. Biotechnol. 2013, 55, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Weng, K.F.; Shih, S.R.; Brewer, G. The evolving world of small RNAs from RNA viruses. Wiley Interdiscip. Rev. RNA 2016, 7, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Alazem, M.; Kim, K.H.; Lin, N.S. Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2538. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Shin, C. The role of plant small RNAs in NB-LRR regulation. Brief. Funct. Genomics 2015, 14, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Hinze, A.; Tucker, M.R.; Bouché, N.; Gasciolli, V.; Elmayan, T.; Lauressergues, D.; Jauvion, V.; Vaucheret, H.; Laux, T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009, 5, e1000646. [Google Scholar] [CrossRef] [Green Version]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [Green Version]
- Sarma, B.K.; Singh, H.B.; Fernando, D.; Silva, R.N.; Gupta, V.K. Enhancing plant disease resistance without R genes. Trends Biotechnol. 2016, 34, 523–525. [Google Scholar] [CrossRef]
- Gao, L.; Luo, J.; Ding, X.; Wang, T.; Hu, T.; Song, P.; Zhai, R.; Zhang, H.; Zhang, K.; Li, K. Soybean RNA interference lines silenced for eIF4E show broad potyvirus resistance. Mol. Plant Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Robaglia, C.; Gallois, J.-L. eIF4E resistance: Natural variation should guide gene editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef]
- Zhou, L.; He, H.; Liu, R.; Han, Q.; Shou, H.; Liu, B. Overexpression of GmAKT2 potassium channel enhances resistance to soybean mosaic virus. BMC Plant Biol. 2014, 14, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Niu, L.; Zhang, W.; Yang, J.; Xing, G.; He, H.; Guo, D.; Du, Q.; Qian, X.; Yao, Y. RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean. Plant Cell Rep. 2018, 37, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Furutani, N.; Hidaka, S.; Kosaka, Y.; Shizukawa, Y.; Kanematsu, S. Coat protein gene-mediated resistance to soybean mosaic virus in transgenic soybean. Breed. Sci. 2006, 56, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Jung, H.W.; Choi, H.K.; Lee, Y.-H.; Baek, I.-Y.; Ko, J.-M.; Jeong, S.-C.; Pack, I.S.; et al. Characterization of SMV resistance of soybean produced by genetic transformation of SMV-CP gene in RNAi. Plant Biotechnol. Rep. 2013, 7, 425–433. [Google Scholar] [CrossRef]
- Gao, L.; Ding, X.; Li, K.; Liao, W.; Zhong, Y.; Ren, R.; Liu, Z.; Adhimoolam, K.; Zhi, H. Characterization of Soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 2015, 128, 1489–1505. [Google Scholar] [CrossRef]
- Redekar, N.; Clevinger, E.; Laskar, M.; Biyashev, R.; Ashfield, T.; Jensen, R.V.; Jeong, S.-C.; Tolin, S.; Maroof, S. Candidate gene sequence analyses toward identifying Rsv3-type resistance to Soybean mosaic virus. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.-H.; Lee, J.-H.; Kim, D.-H.; Choi, H.K.; Jung, H.W.; et al. RNAi-Mediated Soybean Mosaic Virus (SMV) Resistance of a Korean Soybean Cultivar; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]


| Protein | Function for Virus | Function for Plant |
|---|---|---|
| P1 | Protease [41,42], Viral host range [43,44] | |
| HC-Pro | Long-distance movement [45], a ‘bridge’ between virion particles and aphid stylets in aphid transmission [46,47], suppression of host defense (RNA silencing) [48] | Virulence determinant [49,50] |
| P3 | Targets host elongation factors 1A (eEF1A) to facilitate SMV replication [51] | Effector of Rsv1 [50,52], Effector of Rsv4 [53] |
| PIPO | Movement [54] | |
| 6K1 | Cell-to-cell movement [55] | |
| CI | Required for genome replication and movement (cell-to-cell or long-distance movement) [56] | Effector of Rsv3 [57,58] |
| 6K2 | Formation of the virus replication complex [59,60] | |
| VPg | Binds specifically to eIF4E to initiate polyprotein translation [61,62] | |
| NIa-Pro | Proteinase [63,64] | |
| Nib | The catalytic subunit of RdRp [65,66,67] | |
| CP | A ‘bridge’ between virion particles and aphid stylets in aphid transmission [47], cell-to-cell movement, virus assembly [68,69] |
| R Gene | SMV Strain | Cultivar | Location | Effector | Type of R Gene |
|---|---|---|---|---|---|
| Rsv1 | G1–G6 [90] | Kwanggyo Marshall Odgen PI96983 PI507389 Raiden Suweon97 Kosuzu Susumaru PI39887 Jitsuka Clifford Tousan65 Corcisa PI61944 PI61947 [107,108] | Chromosome 13 | P3 [14,52] HC-pro [49] | NB-LRR-type of R-genes [89] |
| Rsv3 | G5,G6,G7 [98,99] | Columbia Hardee Tosan140 PI 339870 PI399091 L29 [90,108] | Chromosome 14 | CI [57,58] | CC-NB-LRR type or R-gene [96] |
| Rsv4 | G1–G7 [103] | PI486355 V94-5152 P188788 Haman Ilpumgeomjeong KAERI-GNT-220-7 PI 398593 PI438307 Rhosa Beeson [86,108,109,110,111] | Chromosome 2 | P3 [53,86,104] | Non-NLR genes (RNase-H family protein) [65] |
| Rsv5 | G1 [106] | York Dorman Arksoy Riple Calhoun Musen [106] | Chromosome 13 | Possibly P3 | unknown |
| R gene | SMV Strain | Cultivar | Location | Candidate Genes |
|---|---|---|---|---|
| Rsc7 | SC7 | Kefeng No.1 [112,113] | Chromosome 2 Linked markers (distance): Satt266 (43.7 cM) Satt634 (18.1 cM) Satt558 (26.6 cM) Satt157 (36.4 cM) Satt698 (37.9 cM) [112] Flanking markers: BARCSOYSSR_02_0621 BARCSOYSSR_02_0632 [113] | 15 candidate genes with one NBS-LRR type gene, one HSP40 gene and one serine carboxypeptidase-type gene [113]. |
| Rsc8 | SC8 | Kefeng No.1 [32] | Chromosome 2 Flanking markers: BARCSOYSSR_02_0610 BARCSOYSSR_02_0616 [32] Other markers: ZL-42 and ZL-52 | Glyma02g13310, Glyma02g13320, Glyma02g13400, Glyma02g13460, Glyma02g13470 [32] Glyma02g121500 and Glyma02g121600 (encoding MADS-box proteins) [114] |
| Rsc5 | SC5 | Kefeng No1 [28] | Chromosome 2 Flanking markers: Bin 352 Bin353 [28] | 11 candidate genes with Glyma02g13495 as the most plausible candidate [28] |
| Rsc20 | SC20 | Qihuang-1 [29] | Chromosome 13 Flanking markers: BARCSOYSSR_13_1099 BARCSOYSSR_13_1185 [29] | TIR-NBS-LRR type R genes: Glyma13g194700 and Glyma13g195100 [29]. |
| Rsc12 | SC12 | Qihuang-22 [115] | Chromosome 13 Flanking marker: Satt334 Sct_033 [115] | |
| Rsc3 | SC3 | Qihuang-1 [116] | Chromosome 13 [116] | Glyma13g25920, Glyma13g25950, Glyma13g25970, and Glyma13g26000 [116]. |
| Rsc3Q | SC3 | Qihuang-1 [117] | Chromosome 13 Flanking markers: BARCSOYSSR_13_1114 BARCSOYSSR_13_1136 [117] | Glyma13g25730, Glyma13g25750, Glyma13g25950, Glyma13g25970, and Glyma13g26000 [117]. |
| Rsc14Q | SC14 | Qihuang-1 [118,119] | Chromosome 13 Flanking markers: Sat_234 Sct_033 [118] Other markers: Satt334 MY750 [119] | |
| Rsc18 | SC18 | Kefeng No.1 [120] Qihuang-22 [120] | Chromosome 2 Flanking marker: BARCSOYSSR_02_0667 BARCSOYSSR_02_0670 [120] Chromosome 13 Flanking marker: SOYHSP176 Satt334 [120] | Glyma02g127800, Glyma02g128200 and Glyma02g128300 [120] |
| Rsc4 | SC4 | Dabaima [121] | Chromosome 14 Flanking markers: BARCSOYSSR_14_1413 BARCSOYSSR_14_1416 [31] | NB-LRR genes: Glyma14g38510 and Glyma14g38560 P450 family gene: Glyma14g38580 [31] |
| Host Factors | Roles in SMV Resistance | Reference |
|---|---|---|
| eEF1A | Targeted by P3, promotes SMV replication | [51] |
| GmEDR1, GmEDS1 GmPAD4 | Induce accumulation of SA, mediated resistance against SMV | [17] |
| GmHSP90 | Reduced the replication and movement of SMV-G2 (Rsv1-mediated resistance) | [17] |
| WRKY6 WRKY30 | Rsv1-mediated resistance against SMV-G2 | [17] |
| GmPP2C3a | Induces callose accumulation, restricts SMV movement | [100] |
| GmPEX14 | Induces burst of H2O2, (Rsc15-mediated resistance) | [30] |
| GmMPK4 | Negatively regulates SA accumulation and defense response | [123] |
| GmMPK6 | Repressor and activator in defense response | [124] |
| GmKR3 | Stimulates ABA accumulation | [25] |
| GmCYB5 | Targets the P3 protein to inhibit SMV accumulation | [24] |
| Tolerance Cultivar | Reference | |
|---|---|---|
| Transgenic GmAKT2 | Alter the level of potassium, reduce the spread of SMV | [136] |
| RNAi-mediated silencing of SMV P3 transgenic soybean | Exhibited stable and enhanced resistance to SMV SC3 and other potyviruses. | [137] |
| Transgenic GmKR3 | Enhances resistance against multiple viruses, including SMV-SC3, via ABA signaling | [25] |
| Attenuated SMV-Coat-protein mediated-resistance transgenic soybean | Highly resistant to SMV strain D and A (in Japan) | [138] |
| SMV-CP-RNAi transgenic soybean | Induces a functional gene silencing system and resulted in a viral-resistant phenotype. | [139] |
| Inverted repeat-SMV-HC-pro transgenic soybean | Induced RNA-mediated resistance via RNAi by targeting SMV-HC-pro | [140] |
| Soybean RNA interfere lines, silenced for eIF4E | Interferes viral replication cycles, increases broad-spectrum resistance against SMV-SC3, SC7,SC-15,SC18, and SMV-R | [134] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Widyasari, K.; Alazem, M.; Kim, K.-H. Soybean Resistance to Soybean Mosaic Virus. Plants 2020, 9, 219. https://doi.org/10.3390/plants9020219
Widyasari K, Alazem M, Kim K-H. Soybean Resistance to Soybean Mosaic Virus. Plants. 2020; 9(2):219. https://doi.org/10.3390/plants9020219
Chicago/Turabian StyleWidyasari, Kristin, Mazen Alazem, and Kook-Hyung Kim. 2020. "Soybean Resistance to Soybean Mosaic Virus" Plants 9, no. 2: 219. https://doi.org/10.3390/plants9020219