Green Extraction of Alkaloids and Polyphenols from Peumus boldus Leaves with Natural Deep Eutectic Solvents and Profiling by HPLC-PDA-IT-MS/MS and HPLC-QTOF-MS/MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phytochemical Profiling of Peumus boldus Methanol Extract
2.1.1. Identification of P. boldus Phenolic Compounds
2.1.2. Identification of P. boldus Alkaloids.
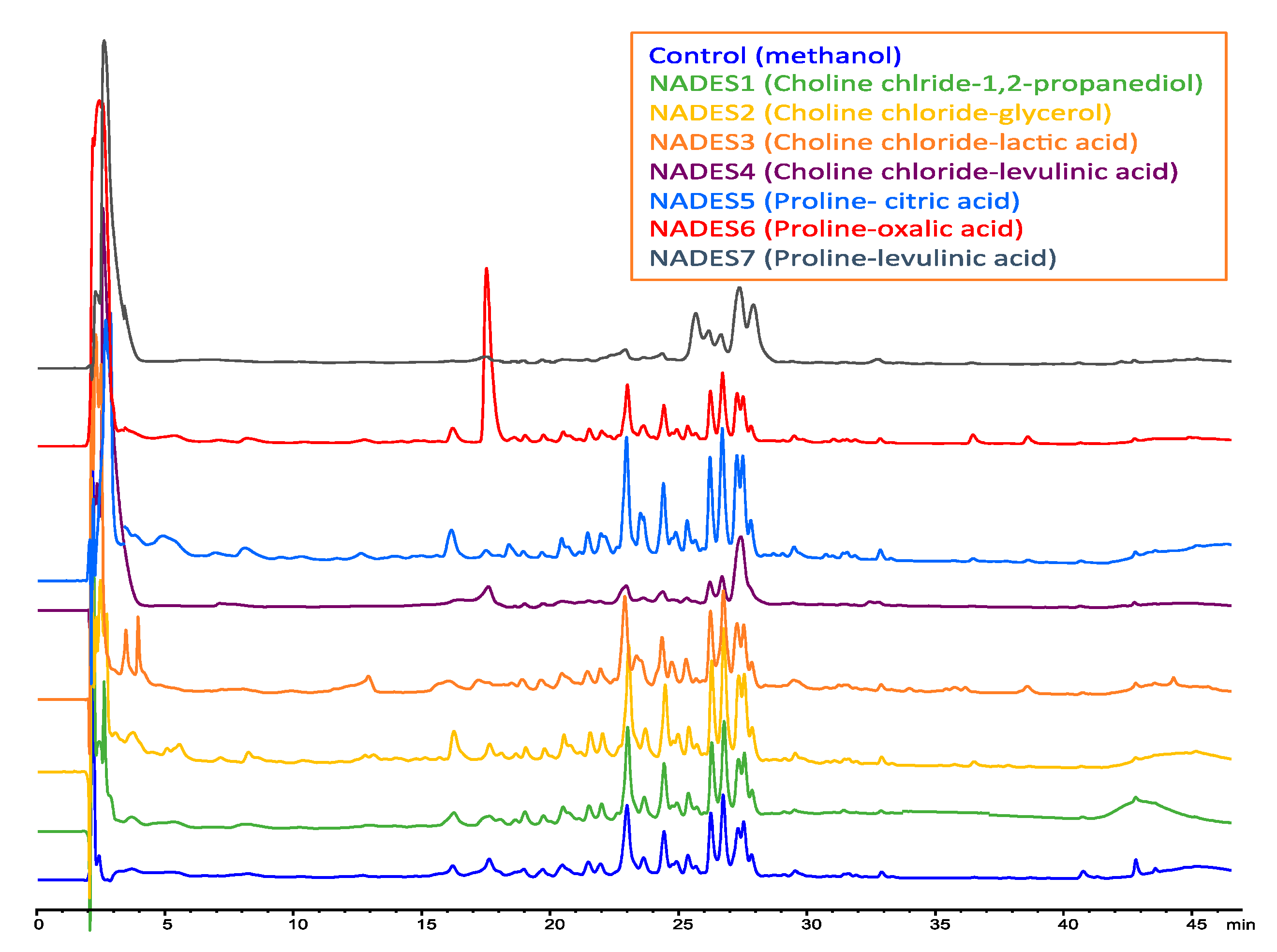
2.2. Extractability of Boldine from Peumus boldus Leaves with Diverse NADES
2.3. Extraction Yields of Total Polyphenols from Peumus boldus Leaves
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Preparation of NADES
3.3. Plant Material and Extraction
3.4. Analysis of Extracts
3.4.1. Total Polyphenol Content
3.4.2. Qualitative and Quantitative HPLC-PDA-IT-MS/MS Analysis
3.4.3. Q-ToF High-Resolution Mass Spectrometry Measurements
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernández, J.; Lagos, P.; Rivera, P.; Zamorano-Ponce, E. Effect of boldo (Peumus boldus Molina) infusion on lipoperoxidation induced by cisplatin in mice liver. Phytother. Res. 2009, 3, 1024–1027. [Google Scholar] [CrossRef] [PubMed]
- Looser, G. ¿Cuál es el verdadero nombre botánico del peumo y del boldo? Rev. Chil. Hist. Nat. 1935, 39, 203–211. [Google Scholar]
- Speisky, H.; Squella, J.A.; Núñez-Vergara, L.J. Activity of boldine on rat ileum. Planta Med. 1991, 57, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Speisky, H.; Cassels, B. Boldo and boldine: An emerging case of natural drug development. Pharmacol. Res. 1994, 29, 1–12. [Google Scholar] [CrossRef]
- O’Brien, P.; Carrasco-Pozo, C.; Speisky, H. Boldine and its antioxidant or health-promoting properties. Chem. Biol. Interact. 2006, 159, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Simirgiotis, M.J.; Schmeda-Hirschmann, G. Direct identification of phenolic constituents in boldo Folium (Peumus boldus Mol.) infusions by high-performance liquid chromatography with diode array detection and electrospray ionization tandem mass spectrometry. J. Chromatogr. A 2010, 1217, 443–449. [Google Scholar] [CrossRef]
- Fuentes-Barros, G.; Castro-Saavedra, S.; Liberona, L.; Acevedo-Fuentes, W.; Tirapegui, C.; Mattar, C.; Cassels, B. Variation of the alkaloid content of Peumus boldus (boldo). Fitoterapia 2018, 127, 179–185. [Google Scholar] [CrossRef]
- Bradley, P.R. British Herbal Compendium: A Handbook of Scientific Information on Widely Used Plant Drugs; British Herbal Medicine Association: Bournemouth, UK, 2006; p. 2. [Google Scholar]
- EMA-HMPC. Community Herbal Monograph on Peumus boldus Molina, Folium. Available online: https://www.ema.europa.eu/en/documents/herbal-opinion/opinion-hmpc-european-union-herbal-monograph-peumus-boldus-molina-folium_en.pdf (accessed on 1 February 2020).
- Schmeda-Hirschmann, G.; Rodríguez, J.A.; Theoduloz, C.; Astudillo, S.L.; Feresin, G.E.; Tapia, A. Free-radical Scavengers and Antioxidants from Peumus boldus Mol. (“Boldo”). Free Radic. Res. 2003, 37, 447–452. [Google Scholar] [CrossRef]
- Quezada, M.; Asencio, M.; Valle, J.M.; Aguilera, J.M.; Gomez, B. Antioxidant activity of crude extract, alkaloid fraction and flavonoid fraction from boldo (Peumus boldus Molina) leaves. J. Food Sci. 2004, 69, 371–376. [Google Scholar] [CrossRef]
- Cermanova, J.; Kadova, Z.; Zagorova, M.; Hroch, M.; Tomsik, P.; Nachtigal, P.; Kudlackova, Z.; Pavek, P.; Dubecka, M.; Ceckova, M.; et al. Boldine enhances bile production in rats via osmotic and Farnesoid X receptor dependent mechanisms. Toxicol. Appl. Pharmacol. 2015, 285, 12–22. [Google Scholar] [CrossRef]
- Gomez, G.; Velarde, V. Boldine Improves Kidney Damage in the Goldblatt 2K1C Model Avoiding the Increase in TGF-β. Int. J. Mol. Sci. 2018, 19, 1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konrath, E.; Santin, K.; Nassif, M.; Latini, A.; Henriques, A.; Salbego, C. Antioxidant and pro-oxidant properties of boldine on hippocampal slices exposed to oxygen-glucose deprivation in vitro. Neurotoxicology 2008, 29, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Schirckel, S.; Bittner, M. La salud en nuestras manos: Plantas medicinales en Chile, Riqueza Natural y Científica, 2nd ed.; Lamas y Cía Ltd.: Concepción, Chile, 2010; p. 238. [Google Scholar]
- Soto, C.; Caballero, E.; Pérez, E.; Zúniga, M. Effect of extraction conditions on total phenolic content and antioxidant capacity of pretreated wild Peumus boldus leaves from Chile. Food Bioprod. Process. 2013, 92, 328–333. [Google Scholar] [CrossRef]
- Lu, P.; Sun, H.; Zhang, L.; Hu, H.; Zhang, L.; Zhao, F.; Ge, C.; Yao, M.; Wang, T.; Li, J. Isocorydine targets the drug-resistant cellular side population through PDCD4-related apoptosis in hepatocellular carcinoma. Mol. Med. 2012, 18, 1136–1146. [Google Scholar] [CrossRef]
- Pastene, E.; Parada, V.; Avello, M.; Ruiz, A.; García, A. Catechin-based Procyanidins from Peumus boldus Mol. Aqueous Extract Inhibit Helicobacter pylori Urease and Adherence to Adenocarcinoma Gastric Cells. Phytother. Res. 2014, 28, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Abbott, A.P.; Boothby, D.; Capper, G.; Davies, D.L.; Rasheed, R.K. Deep Eutectic Solvents Formed between Choline Chloride and Carboxylic Acids: Versatile Alternatives to Ionic Liquids. J. Am. Chem. Soc. 2004, 126, 9142–9147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; De Oliveira Vigier, K.; Royer, S.; Jerome, F. Deep eutectic solvents: Syntheses, properties and applications. Chem. Soc. Rev. 2012, 41, 7108–7146. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef] [Green Version]
- Abidin, Z.; Hamdi, M.; Maan, H.; Adeeb, H.; Subramanian, J.N. New horizons in the extraction of bioactive compounds using deep eutectic solvents: A review. Anal. Chim. Acta 2017, 979, 1–23. [Google Scholar] [CrossRef]
- Choi, Y.H.; van Spronsen, J.; Dai, Y.; Verberne, M.; Hollmann, F.; Arends, I.W.C.E.; Witkamp, G.J.; Verpoorte, R. Are Natural Deep Eutectic Solvents the Missing Link in Understanding Cellular Metabolism and Physiology? Plant Physiol. 2011, 156, 1701–1705. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Friesen, J.B.; McAlpine, J.B.; Lankin, D.C.; Chen, S.N.; Pauli, G.F. Natural Deep Eutectic Solvents: Properties, Applications, and Perspectives. J. Nat. Prod. 2018, 81, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; van Spronsen, J.; Witkamp, G.J.; Verpoorte, R.; Choi, Y.H. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta 2013, 766, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.M.; Wang, L.W.; Gao, Z.; Zhuang, B.; Yin, Q.; Liu, E.H. Green and efficient extraction of different types of bioactive alkaloids using deep eutectic solvents. Microchem. J. 2019, 145, 345–353. [Google Scholar] [CrossRef]
- Takla, S.S.; Shawky, E.; Hammoda, H.; Darwish, F. Green techniques in comparison to conventional ones in the extraction of Amaryllidaceae alkaloids: Best solvents selection and parameters optimization. J. Chromatogr. A 2018, 1567, 99–110. [Google Scholar] [CrossRef]
- Shawky, E.; Takla, S.S.; Hammoda, H.; Darwish, F. Evaluation of the influence of green extraction solvents on the cytotoxic T activities of Crinum (Amaryllidaceae) alkaloid extracts using in-vitro-in-silico approach. J. Ethnopharmacol. 2018, 227, 139–149. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cheng, P.; Wang, W.; Yan, S.; Tang, Q.; Liu, D.; Xie, H. Rapid Investigation and Screening of Bioactive Components in Simo Decoction via LC-Q-TOF-MS and UF-HPLC-MD Methods. Molecules 2018, 23, 1792. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.; Yadav, A.; Dabur, R. Interactions of a medicinal climber Tinospora cordifolia with supportive interspecific plants trigger the modulation in its secondary metabolic profiles. Sci. Rep. 2019, 9, 14327. [Google Scholar] [CrossRef]
- Bakiri, A.; Hubert, J.; Reynaud, R.; Lanthony, S.; Harakat, D.; Renault, J.H.; Nuzillard, J.M. Computer-Aided 13C NMR Chemical Profiling of Crude Natural Extracts without Fractionation. J. Nat. Prod. 2017, 80, 1387–1396. [Google Scholar] [CrossRef]
- Soares, E.R.; da Silva, F.M.; de Almeida, R.A.; de Lima, B.R.; da Silva Filho, F.A.; Barison, A.; Koolen, H.H.; Pinheiro, M.L.B.; de Souza, A.D. Direct infusion ESI-IT-MSn alkaloid profile and isolation of tetrahydroharman and other alkaloids from Bocageopsis pleiosperma maas (Annonaceae). Phytochem. Anal. 2015, 26, 339–345. [Google Scholar] [CrossRef]
- Nikolic, D.; Gödecke, T.; Chen, S.N.; White, J.; Lankin, D.; Pauli, G.F.; Van Breemen, R. Mass spectrometric dereplication of nitrogen-containing constituents of black cohosh (Cimicifuga racemosa L.). Fitoterapia 2012, 83, 441–460. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Bajpai, V.; Kumar, S.; Singh, A.K.; Kumar, B. Analysis of isoquinoline alkaloids from Mahonia leschenaultia and Mahonia napaulensis roots using UHPLC-Orbitrap-MSn and UHPLC-QqQLIT-MS/MS. J. Pharm. Anal. 2017, 7, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Del Mar-Contreras, M.; Noureddine, B.; Gómez-Caravaca, A.; Gálvez, J.; Segura-Carretero, A. Alkaloids Profiling of Fumaria capreolata by Analytical Platforms Based on the Hyphenation of Gas Chromatography and Liquid Chromatography with Quadrupole-Time-of-Flight Mass Spectrometry. Int. J. Anal. Chem. 2017, 2017, 5178729. [Google Scholar]
- De Lima, B.R.; da Silva, F.M.A.; Soares, E.R.; de Almeida, R.A.; da Silva-Filho, F.A.; Barison, A.; Costa, E.V.; Koolen, H.H.F.; de Souza, A.D.L.; Pinheiro, M.L.B. Integrative Approach Based on Leaf Spray Mass Spectrometry, HPLC-DAD-MS/MS, and NMR for Comprehensive Characterization of Isoquinoline-Derived Alkaloids in Leaves of Onychopetalum amazonicum R. E. Fr. J. Braz. Chem. Soc. 2020, 31, 79–89. [Google Scholar] [CrossRef]
- Duan, L.; Dou, L.L.; Guo, L.; Li, P.; Liu, E.H. Comprehensive evaluation of deep eutectic solvents in extraction of bioactive natural products. ACS Sustain. Chem. Eng. 2016, 4, 2405–2411. [Google Scholar] [CrossRef]
- Bajkacz, S.; Adamek, J. Evaluation of new natural deep eutectic solvents for the extraction of isoflavones from soy products. Talanta 2017, 168, 329–335. [Google Scholar] [CrossRef]
- Katsampa, P.; Valsamedo, E.; Grigorakis, S.; Makris, D. A green ultrasound-assisted extraction process for the recovery of antioxidant polyphenols and pigments from onion solid wastes using Box–Behnken experimental design and kinetics. Ind. Crops Prod. 2015, 77, 535–543. [Google Scholar] [CrossRef]
- Ruesgas-Ramón, M.; Figueroa-Espinoza, M.C.; Durand, E. Application of Deep Eutectic Solvents (DES) for Phenolic Compounds Extraction: Overview, Challenges, and Opportunities. J. Agric. Food Chem. 2017, 65, 3591–3601. [Google Scholar] [CrossRef]
- González, C.; Mustafa, N.; Wilson, E.; Verpoorte, R.; Choi, Y. Application of natural deep eutectic solvents for the “green” extraction of vanillin from vanilla pods. Flavour Fragr. J. 2018, 33, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Savi, L.; Carpiné, D.; Waszczynskyj, N.; Ribani, R.; Haminiuk, C. Influence of temperature, water content and type of organic acid on the formation, stability and properties of functional natural deep eutectic solvents. Fluid Phase Equilibria 2019, 488, 40–47. [Google Scholar] [CrossRef]
- Mulia, K.; Krisanti, E.; Terahadi, F.; Putri, S. Selected Natural Deep Eutectic Solvents for the Extraction of α-Mangostin from Mangosteen (Garcinia mangostana L.) Pericarp. Int. J. Technol. 2015, 6, 1211. [Google Scholar] [CrossRef] [Green Version]
- Rogalinski, T.; del Valle, J.M.; Zetzl, C.; Brunner, G. Extraction of boldo (Peumus boldus Mol.) leaves with hot pressurized water and supercritical CO2. Food Res. Int. 2003, 38, 203–213. [Google Scholar]
- Schwanz, M. Desenvolvimento e validacão de método analiítico para quantificacão da boldina em Peumus boldus Mol. (Monimiaceae) e avaliacão preliminar de sua estabilidade. Ph.D. Thesis, University of Rio Grande do Sul, Porto Alegre, Brazil, April 2006. [Google Scholar]
- Espic, M. Evaluación de la producción de biomasa aérea y del rendimiento en aceite esencial y boldina, en boldo (Peumus boldus Mol.) en la comuna de Papudo V región. Bachelor’s Thesis, Faculty of Forestry Sciences, University of Chile, Santiago, Chile, 2007. [Google Scholar]
- Camara, C.I.; Bornancini, C.A.; Cabrera, J.L.; Ortega, M.G.; Yudi, L.M. Quantitative analysis of boldine alkaloid in natural extracts by cyclic voltammetry at a liquid–liquid interface and validation of the method by comparison with high performance liquid chromatography. Talanta 2010, 83, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Petigny, L.; Périno-Issartier, S.; Wajsman, J.; Chemat, F. Batch and Continuous Ultrasound Assisted Extraction of Boldo Leaves (Peumus boldus Mol.). Int. J. Mol. Sci. 2013, 14, 5750–5764. [Google Scholar] [CrossRef] [PubMed]
- Petigny, L.; Périno, S.; Minuti, M.; Visinoni, F.; Wajsman, J.; Chemat, F. Simultaneous Microwave Extraction and Separation of Volatile and Non-Volatile Organic Compounds of Boldo Leaves. From Lab to Industrial Scale. Int. J. Mol. Sci. 2014, 15, 7183–7198. [Google Scholar] [CrossRef] [Green Version]
- Gómez, A.V.; Tadini, C.C.; Biswas, A.; Buttrum, M.; Kim, S.; Boddu, V.M.; Cheng, H.N. Microwave-assisted extraction of soluble sugars from banana puree with natural deep eutectic solvents (NADES). LWT 2019, 107, 79–88. [Google Scholar] [CrossRef]
- Si, Y.Y.; Sun, S.W.; Liu, K.; Liu, Y.; Shi, H.L.; Zhao, K.; Wang, J.; Wang, W. Novel Deep Eutectic Solvent Based on Levulinic Acid and 1,4-Butanediol as an Extraction Media for Bioactive Alkaloid Rutaecarpine. Processes 2019, 7, 171. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Garzon, J.; Friesen, J.B.; Zhang, Y.; McAlpine, J.B.; Lankin, D.C.; Chen, S.-N.; Pauli, G.F. Countercurrent assisted quantitative recovery of metabolites from plant-associated natural deep eutectic solvents. Fitoterapia 2016, 112, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Smink, D.; Kersten, S.R.A.; Schuur, B. Recovery of lignin from deep eutectic solvents by liquid-liquid extraction. Sep. Purif. Technol. 2020, 235, 116127. [Google Scholar] [CrossRef]
- Kim, K.H.; Dutta, T.; Sun, J.; Simmons, N.; Singh, S. Biomass pretreatment using deep eutectic solvents from lignin derived phenols. Green Chem. 2018, 20, 809–815. [Google Scholar] [CrossRef]
- Liang, X.; Fu, Y.; Chang, J. Effective separation, recovery and recycling of deep eutectic solvent after biomass fractionation with membrane-based methodology. Sep. Purif. Technol. 2019, 210, 409–416. [Google Scholar] [CrossRef]
- Sanap, A.K.; Shankarling, G.S. Eco-Friendly and recyclable media for rapid synthesis of tricyanovinylated aromatics using biocatalyst and deep eutectic solvent. Catal. Commun. 2014, 49, 58–62. [Google Scholar] [CrossRef]
- Phadtare, S.B.; Shankarling, G.S. Halogenation reactions in biodegradable solvent: Efficient bromination of substituted 1-aminoanthra-9, 10-quinone in deep eutectic solvent (choline chloride: Urea). Green Chem. 2010, 12, 458–462. [Google Scholar] [CrossRef]
- Singh, B.S.; Lobo, H.R.; Pinjari, D.V.; Jarag, K.J.; Pandit, A.B.; Shankarling, G.S. Ultrasound and deep eutectic solvent (DES): A novel blend of techniques for rapid and energy efficient synthesis of oxazoles. Ultrason. Sonochem. 2013, 20, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolibdic phosphotungstic acid reagent. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Bordiga, M.; Gómez-Alonso, S.; Locatelli, M.; Travaglia, F.; Coïsson, J.D.; Hermosín-Gutiérrez, I.; Arlorio, M. Phenolics characterization and antioxidant activity of six different pigmented Oryza sativa L. cultivars grown in Piedmont (Italy). Food Res. Int. 2014, 65, 282–290. [Google Scholar] [CrossRef]
- Directive 2002/657/EC. Commission Decision of 12 August 2002 Implementing Council Directive 96/23/EC Concerning the Performance of Analytical Methods and the Interpretation of Results (Notified under Document Number C(2002) 3044); L221/8-36; Official Journal of the European Communities: Brussels, Belgium, 2002.






| Peak * | tR (min) | λ Max (nm) | Identified Compound | [M + H]+ m/z | MS/MS Fragments | Ref. |
|---|---|---|---|---|---|---|
| 6 | 15.2 | 281, 302 | coclaurine | 286.1 | 269.0, 237.0, 209.0, 175.0, 137.0 | [29,30] |
| 7 | 15.8 | 283, 303 | N-methylcoclaurine | 300.1 | 269.1, 237.1, 209.0, 175.0, 137.0 | [29,30,31] |
| 8 | 16.3 | 282, 302 | laurolitsine | 314.0 | 297.1, 265.0 | [32] |
| 9 | 17.3 | 281, 302 | isoboldine | 328.1 | 297.1, 265.0, 237.1, 165.0 | [31,33] |
| 10 | 17.8 | 280, 303 | boldine | 328.1 | 297.1, 265.0, 237.0, 205.0 | [29,34] |
| 15 | 21.0 | 265, 282 | reticuline | 330.0 | 192.1, 175.1, 137.0 | [31,32,33,34,35,36] |
| 21 | 24.6 | 266, 303 | isocorydine | 342.1 | 296.1, 279.1, 264.0, 248.1, | [37] |
| 27 | 26.8 | 283, 302, | laurotetanine | 328.1 | 311.1, 279.1, 248.1, 219.1, 191.1 | [29,32] |
| 28 | 27.5 | 282, 303 | N-methyllaurotetanine | 342.2 | 311.1, 296.1, 280.1, 265.1, 253.1, 237.1 | [29,32] |
| Peak * | tR (min) | Identified Compound | Formula | Mass Experimental | Mass Calculated | Error ppm | [M + H]+ m/z | MS-MS Fragments |
|---|---|---|---|---|---|---|---|---|
| 6 | 25.64 | Coclaurine | C17H19NO3 | 285.13653 | 285.13649 | 0.14 | 286.14390 | 269.11697, 237.09093, 209.09577, 175.07523 |
| 7 | 26.12 | N-methylcoclaurine | C18H21NO3 | 299.15228 | 299.15214 | 0.47 | 300.15937 | 269.11659, 237.09041, 209.09544, 175.07485 |
| 8 | 26.51 | Laurolitsine | C18H19NO4 | 313.13133 | 313.13141 | 0.26 | 314.13859 | 297.11174, 265.08608, 209.09629, 165.06924 |
| 9 | 27.50 | Isoboldine | C19H21NO4 | 327.14718 | 327.14706 | 0.38 | 328.15444 | 297.11182, 265.08594, 237.09035, 165.06932 |
| 10 | 27.55 | Boldine | C19H21NO4 | 327.14716 | 327.14706 | 0.31 | 328.15445 | 297.11248, 265.08617, 237.09124, 205.06469 |
| 15 | 27.41 | Reticuline | C19H23NO4 | 329.16323 | 329.16271 | 1.6 | 330.17059 | 192.10222, 239.10637 175.07548, 137.05999 |
| 21 | 30.02 | Isocorydine | C20H23NO4 | 341.16282 | 341.16271 | 0.32 | 342.17014 | 296.10428, 279.10209, 264.07821, 248.08325 |
| 27 | 30.08 | Laurotetanine | C19H21NO4 | 327.14735 | 327.14706 | 0.9 | 328.13587 | 311.12739, 279.10199, 248.08321, 191.08531 |
| 28 | 32.55 | N-Methyl-laurotetanine | C20H23NO4 | 341.16313 | 341.16271 | 1.23 | 342.17048 | 311.12849, 296.10546, 280.11149, 265.08649 |
| Code | Component 1 (HBD) | Component 2 (HDA) | Molar Ratio | Water (%) | Ref. |
|---|---|---|---|---|---|
| NADES1 | Choline chloride | 1,2-propanediol | 1:3 | 20% | [25,37,38,39,40,41,42,43] |
| NADES2 | Choline chloride | Glycerol | 1:2 | 20% | [25,28,38] |
| NADES3 | Choline chloride | Lactic acid | 1:2 | 20% | [25,26,27,28] |
| NADES4 | Choline chloride | Levulinic acid | 1:1 | 20% | [25,26,27,28] |
| NADES5 | l-Proline | Citric acid | 1:2 | 20% | [25,26,27,28] |
| NADES6 | l-Proline | Oxalic acid | 1:1 | 20% | [25,26,27,28] |
| NADES7 | l-Proline | Levulinic acid | 1:1 | 20% | [25,26,27,28] |
| Extraction Method, Analysis | Boldine Yields | Reference |
|---|---|---|
| Ethanolic extract, HPLC | 0.14% | [10] |
| European Pharmacopoeia, HPLC-UV | 0.016 to 0.059% | [45] |
| European Pharmacopoeia, HPLC-UV | 0.01 to 0.05% | [46] |
| European Pharmacopoeia, HPLC-UV | 0.06% | [47] |
| European Pharmacopoeia, UHPLC-MS/MS | 0.01 to 0.018% | [7] |
| UAE (water) 23 W/cm2, 36 °C, 40 min | 0.148 % | [48] |
| MAE (water) 200 W, 7.5% S/L, 56 min | 0.122% | [49] |
| NADES6: Proline-oxalic acid, 340 rpm, 50 °C, 50 min, HPLC | 0.24% | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Vega, J.; Gómez-Alonso, S.; Pérez-Navarro, J.; Pastene-Navarrete, E. Green Extraction of Alkaloids and Polyphenols from Peumus boldus Leaves with Natural Deep Eutectic Solvents and Profiling by HPLC-PDA-IT-MS/MS and HPLC-QTOF-MS/MS. Plants 2020, 9, 242. https://doi.org/10.3390/plants9020242
Torres-Vega J, Gómez-Alonso S, Pérez-Navarro J, Pastene-Navarrete E. Green Extraction of Alkaloids and Polyphenols from Peumus boldus Leaves with Natural Deep Eutectic Solvents and Profiling by HPLC-PDA-IT-MS/MS and HPLC-QTOF-MS/MS. Plants. 2020; 9(2):242. https://doi.org/10.3390/plants9020242
Chicago/Turabian StyleTorres-Vega, Jeniffer, Sergio Gómez-Alonso, José Pérez-Navarro, and Edgar Pastene-Navarrete. 2020. "Green Extraction of Alkaloids and Polyphenols from Peumus boldus Leaves with Natural Deep Eutectic Solvents and Profiling by HPLC-PDA-IT-MS/MS and HPLC-QTOF-MS/MS" Plants 9, no. 2: 242. https://doi.org/10.3390/plants9020242





