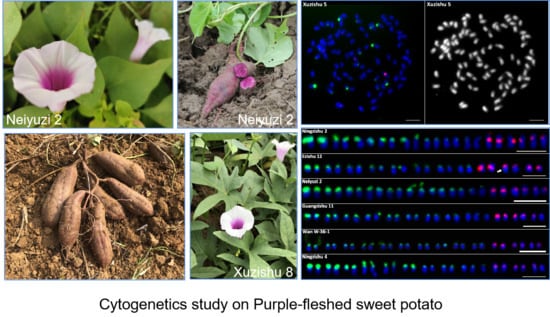
Comparative Chromosomal Localization of 45S and 5S rDNA Sites in 76 Purple-Fleshed Sweet Potato Cultivars
Abstract
:1. Introduction
2. Results
2.1. Chromosome Numbers
2.2. Number of 45S rDNA and 5S rDNA Sites
2.3. Distribution of 45S rDNA and 5S rDNA Sites
2.4. Colocalization of 45S and 5S rDNA Sites
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Chromosome Preparation
4.3. Probe Preparation
4.4. FISH and Signal Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistics Database. 2017. Available online: http://www.fao.org/faostat/ (accessed on 10 June 2020).
- Xie, F.L.; Burklew, C.E.; Yang, Y.F.; Liu, M.; Xiao, P.; Zhang, B.H.; Qiu, D.Y. De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome. Planta 2012, 236, 101–113. [Google Scholar] [CrossRef]
- Katayama, K.; Kitahara, K.; Sakai, T.; Kai, Y.; Yoshinaga, M. Resistant and digestible starch contents in sweet potato cultivars and lines. J. Appl. Glycosci. 2011, 58, 53–59. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Zeng, M.M.; Chen, J.; Jiao, Y.Z.; Niu, F.X.; Tao, G.J.; Zhang, S.; Qin, F.; He, Z.Y. Identification and quantitation of anthocyanins in purple-fleshed sweet potatoes cultivated in China by UPLC-PDA and UPLC-QTOF-MS/MS. J. Agricult. Food Chem. 2016, 64, 171–177. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Zhang, S.S.; Wang, F.B.; Zhao, N.; He, S.Z.; Liu, Q.C.; Zhai, H. Comparative transcriptome analysis of purple-fleshed sweet potato provides insights into the molecular mechanism of anthocyanin biosynthesis. Front. Agricult. Sci. Eng. 2018, 5, 214–225. [Google Scholar]
- Yoshimoto, M.; Okuno, S.; Yamaguchi, M.; Yamakawa, O. Antimutagenicity of deacylated anthocyanins in purple-fleshed sweetpotato. Biosci. Biotechnol. Biochem. 2001, 65, 1652–1655. [Google Scholar] [CrossRef]
- Chen, P.N.; Chu, S.C.; Chiou, H.L.; Chiang, C.L.; Yang, S.F.; Hsieh, Y.S. Cyanidin 3-glucoside and peonidin 3-glucoside inhibit tumor cell growth and induce apoptosis in vitro and suppress tumor growth in vivo. Nutr. Cancer 2005, 53, 232–243. [Google Scholar] [CrossRef]
- Luceri, C.; Giovannelli, L.; Pitozzi, V.; Toti, S.; Castagnini, C.; Routaboul, J.M.; Lepiniec, L.; Larrosa, M.; Dolara, P. Liver and colon DNA oxidative damage and gene expression profiles of rats fed Arabidopsis thaliana mutant seeds containing contrasted flavonoids. Food Chem. Toxicol. 2008, 46, 1213–1220. [Google Scholar] [CrossRef]
- Terahara, N.; Shimizu, T.; Kato, Y.; Nakamura, M.; Maitani, T.; Yamaguchi, M.; Goda, Y. Six diacylated anthocyanins from the storage roots of purple sweet potato, Ipomoea batatas. Biosci. Biotechnol. Biochem. 1999, 63, 1420–1424. [Google Scholar] [CrossRef] [Green Version]
- Terahara, N.; Konczak-Islam, I.; Nakatani, M.; Yamakawa, O.; Goda, Y.; Honda, T. Anthocyanins in callus induced from purple storage root of Ipomoea batatas L. Phytochemistry 2000, 54, 919–922. [Google Scholar] [CrossRef]
- Tian, Q.G.; Konczak, I.; Schwartz, S.J. Probing anthocyanin profiles in purple sweet potato cell line (Ipomoea batatas L. Cv. Ayamurasaki) by high-performance liquid chromatography and electrospray ionization tandem mass spectrometry. J. Agricult. Food Chem. 2005, 53, 6503–6509. [Google Scholar] [CrossRef]
- Magoon, M.L.; Krishnan, R.; Bai, K.V. Cytological evidence on the origin of sweet potato. Theor. Appl. Genet. 1970, 40, 360–366. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Yang, S.Q.; Li, Z.A.; Zhang, Y.X.; Wang, Y.Z.; Cheng, C.Y.; Li, J.; Chen, J.F.; Lou, Q.F. Comparative chromosomal localization of 45S and 5S rDNAs and implications for genome evolution in Cucumis. Genome 2016, 59, 449–457. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Webster, M.A.; Wright, J.; Cocker, J.M.; Smith, M.C.; Badakshi, F.; Harrison, P.H.; Gilmartin, P.M. Integration of genetic and physical maps of the Primula vulgaris S locus and localization by chromosome in situ hybridization. N. Phytol. 2015, 208, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.Y.; Zhang, Z.H.; Zong, X.; Huang, S.W.; Li, Z.Y.; Han, Y.H. A high-resolution cucumber cytogenetic map integrated with the genome assembly. BMC Genom. 2013, 14, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.H.; Zhang, Z.H.; Liu, C.X.; Liu, J.H.; Huang, S.W.; Jiang, J.M.; Jin, W.W. Centromere repositioning in cucurbit species: Implication of the genomic impact from centromere activation and inactivation. Proc. Natl. Acad. Sci. USA 2009, 106, 14937–14941. [Google Scholar] [CrossRef] [Green Version]
- Heslop-Harrison, J.S.; Schwarzacher, T. Organization of the plant genome in chromosomes. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef]
- Winterfeld, G.; Wolk, A.; Roser, M. Genome evolution in alpine oat-like grasses through homoploid hybridization and polyploidy. AoB Plants 2016, 8. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.Q.; Qin, X.D.; Cheng, C.Y.; Li, Z.; Lou, Q.F.; Li, J.; Chen, J.F. Organization and evolution of four differentially amplified tandem repeats in the Cucumis hystrix genome. Planta 2017, 246, 749–761. [Google Scholar] [CrossRef]
- He, L.; Braz, G.T.; Torres, G.A.; Jiang, J.M. Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 2018, 127, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.Y.; Zhang, T.; Wu, Y.F.; Zhang, W.L.; Zhang, P.D.; Xi, M.L.; Jiang, J.M. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 2020, 101, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galián, J.A.; Rosato, M.; Rosselló, J.A. Early evolutionary colocalization of the nuclear ribosomal 5S and 45S gene families in seed plants: Evidence from the living fossil gymnosperm Ginkgo biloba. Heredity 2012, 108, 640–646. [Google Scholar] [CrossRef] [Green Version]
- Garcia, S.; Garnatje, T.; Kovařík, A. Plant rDNA database: Ribosomal DNA loci information goes online. Chromosoma 2012, 121, 389–394. [Google Scholar] [CrossRef]
- Garcia, S.; Gálvez, F.; Gras, A.; Kovařík, A.; Garnatje, T. Plant rDNA database: Update and new features. Database 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapitan, N.L.V. Organization and evolution of higher plant nuclear genomes. Genome 1992, 35, 171–181. [Google Scholar] [CrossRef]
- Vitales, D.; Ambrosio, U.D.; Gálvez, F.; Kovařík, A.; Garcia, S. Third release of the plant rDNA database with updated content and information on telomere composition and sequenced plant genomes. Plant Syst. Evol. 2017, 303, 1115–1121. [Google Scholar] [CrossRef] [Green Version]
- Pontes, O.; Neves, N.; Silva, M.; Lewis, M.S.; Madlung, A.; Comai, L.; Viegas, W.; Pikaard, C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 2004, 101, 18240–18245. [Google Scholar] [CrossRef] [Green Version]
- Pedrosa-Harand, A.; de Almeida, C.C.S.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006, 112, 924–933. [Google Scholar] [CrossRef]
- Li, K.P.; Wu, Y.X.; Zhao, H.; Wang, Y.; Lu, X.M.; Wang, J.M.; Xu, Y.; Li, Z.Y.; Han, Y.H. Cytogenetic relationships among Citrullus species in comparison with some genera of the tribe Benincaseae (Cucurbitaceae) as inferred from rDNA distribution patterns. BMC Evol. Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [Green Version]
- Srisuwan, S.; Sihachakr, D.; Siljak-Yakovlev, S. The origin and evolution of sweet potato (Ipomoea batatas Lam.) and its wild relatives through the cytogenetic approaches. Plant Sci. 2006, 171, 424–433. [Google Scholar] [CrossRef]
- Choi, E.Y.; Seo, J.H.; Seo, B.B. Sequence polymorphism and chromosomal localization of 5S rDNA of three cultivated varieties of sweet potato (Ipomoea batatas (L.) Lam.). Genes Genom. 2009, 31, 325–332. [Google Scholar] [CrossRef]
- Tang, J.L.; Qi, D.S.; Zhang, Y.; Liu, H.J.; Sun, J.Y.; Cao, Q.H.; Ma, D.F.; Li, Z.Y. FISH analysis of chromosomes of sweet potato (Ipomoea batatas cv.Xushu No.18). Hereditas (Beijing) 2010, 32, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Y.; Yu, L.X.; Cai, Z.X.; Zhang, A.; Jin, W.W.; Han, Y.H.; Li, Z.Y. Comparative karyotype analysis among six Ipomoea species based on two newly identified repetitive sequences. Genome 2019, 62, 243–252. [Google Scholar] [CrossRef]
- Gao, R.F. Genetic Diversity Analysis of Purple-Fleshed Sweet Potato Bred in China Cultivars Revealed by Morphology and SSR Markers. Master’s Thesis, Chinese Academy of Agricultural Sciences, Xuzhou, China, 2019. [Google Scholar]
- Snowdon, R.J.; Friedt, W.; Köhler, A.; Köhler, W. Molecular cytogenetic localisation and characterisation of 5S and 25S rDNA loci for chromosome identification in oilseed rape (Brassica napus L.). Ann. Bot. 2000, 86, 201–204. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Lau, K.H.; Cao, Q.H.; Hamilton, J.P.; Sun, H.H.; Zhou, C.X.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.Y.; et al. Genome sequences of two diploid wild relatives of cultivated sweet potato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iovene, M.; Zhang, T.; Lou, Q.F.; Buell, C.R.; Jiang, J.M. Copy number variation in potato—An asexually propagated autotetraploid species. Plant J. 2013, 75, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Hardigan, M.A.; Crisovan, E.; Hamiltion, J.P.; Kim, J.; Laimbeer, P.; Leisner, C.P.; Manrique-Carpintero, N.C.; Newton, L.; Pham, G.M.; Vaillancourt, B.; et al. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 2016, 28, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Pham, G.M.; Newton, L.; Wiegert-Rininger, K.; Vaillancourt, B.; Douches, D.S.; Buell, C.R. Extensive genome heterogeneity leads to preferential allele expression and copy number-dependent expression in cultivated potato. Plant J. 2017, 92, 624–637. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.L.; Xu, T.L.; Wang, J.; Luo, L.; Yu, C.; Dong, G.M.; Pan, H.T.; Zhang, Q.X. Distribution of 45S rDNA in modern rose cultivars (Rosa hybrida), Rosa rugosa, and their interspecific hybrids revealed by fluorescence in situ hybridization. Cytogenet. Genome Res. 2016, 149, 226–235. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, R.; Nagpure, N.S.; Kushwaha, B.; Mani, I.; Chauhan, U.K.; Lakra, W.S. Population distribution of 45S and 5S rDNA in golden mahseer, Tor putitora: Population-specific FISH marker. J. Genet. 2009, 88, 315–320. [Google Scholar] [CrossRef]
- Liu, B.; Davis, T.M. Conservation and loss of ribosomal RNA gene sites in diploid and polyploid Fragaria (Rosaceae). BMC Plant Biol. 2011, 11, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sochorová, J.; Coriton, O.; Kuderová, A.; Lunerová, J.; Chèvre, A.; Kovařík, A. Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus. Ann. Bot. 2017, 119, 13–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Huang, M.; Li, H.; Zhang, L.; Gao, F.; Wang, P.; Hu, Y.; Yan, S.H.; Zhao, L.; Zhang, Q.; Tan, J.J.; et al. Plant 45S rDNA clusters are fragile sites and their instability is associated with epigenetic alterations. PLoS ONE 2012, 7, e35139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutrillaux, A.M.; Carton, B.; Cacheux, L.; Dutrillaux, B. Interstitial NORs, fragile sites, and chromosome evolution: A not so simple relationship-the example of Melolontha melolontha and genus Protaetia (Coleoptera: Scarabaeidae). Cytogenet. Genome Res. 2016, 149, 304–311. [Google Scholar] [CrossRef]
- Thomas, H.M.; Harper, J.A.; Meredith, M.R.; Morgan, W.G.; Thomas, I.D.; Timms, E.; King, I.P. Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res. 1996, 4, 486–490. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef]
- Mondin, M.; Aguiar-Perecin, M.L.R. Heterochromatin patterns and ribosomal DNA loci distribution in diploid and polyploid Crotalaria species (Leguminosae, Papilionoideae), and inferences on karyotype evolution. Genome 2011, 54, 718–726. [Google Scholar] [CrossRef]
- Rosato, M.; Kovařík, A.; Garilleti, R.; Rosselló, J.A. Conserved organisation of 45S rDNA sites and rDNA gene copy number among major clades of early land plants. PLoS ONE 2016, 11, e0162544. [Google Scholar] [CrossRef]
- Mantovani, M.; dos Douglas, L.; Santos, A.; Moreira-Filho, O. Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica 2005, 123, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Leitch, I.J.; Heslop-Harrison, J.S. Physical mapping of the 18S-5.8-26S rRNA genes in barley by in situ hybridization. Genome 1992, 35, 1013–1018. [Google Scholar] [CrossRef]
- Hanson, R.E.; Islam-Faridi, M.N.; Percival, E.A.; Crane, C.F.; Ji, Y.F.; McKnight, T.D.; Stelly, D.M.; Price, H.J. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 1996, 105, 55–61. [Google Scholar] [CrossRef]
- Martins, C.; Galetti, P.M., Jr. Conservative distribution of 5S rDNA loci in Schizodon (Pisces, Anostomidae) chromosomes. Chromosome Res. 2000, 8, 353–355. [Google Scholar] [CrossRef]
- Sun, J.Y. Relationships in Series Batatas Based on Cytogenetic Studies and Chloroplast Genome and Repetitive Sequence Analysis. Ph.D. Thesis, Jiangsu Normal University, Xuzhou, China, 2019. [Google Scholar]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [Green Version]
- Roa, F.; Guerra, M. Non-random distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet. Genome Res. 2015, 146, 243–249. [Google Scholar] [CrossRef]
- Taketa, S.; Harrison, G.E.; Heslop-Harrison, J.S. Comparative physical mapping of the 5S and 18S-25S rDNA in nine wild Hordeum species and cytotypes. Theor. Appl. Genet. 1999, 98, 1–9. [Google Scholar] [CrossRef]
- Kamisugi, Y.; Nakayama, S.; O’Neil, C.M.; Mathias, R.J.; Trick, M.; Fukui, K. Visualization of the Brassica self-incompatibility S-locus on identified oilseed rape chromosomes. Plant Mol. Biol. 1998, 38, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Zhang, Z.H.; Huang, S.W.; Jin, W.W. An integrated molecular cytogenetic map of Cucumis sativus L. chromosome 2. BMC Genet. 2011, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.M.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Genetics 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [Green Version]
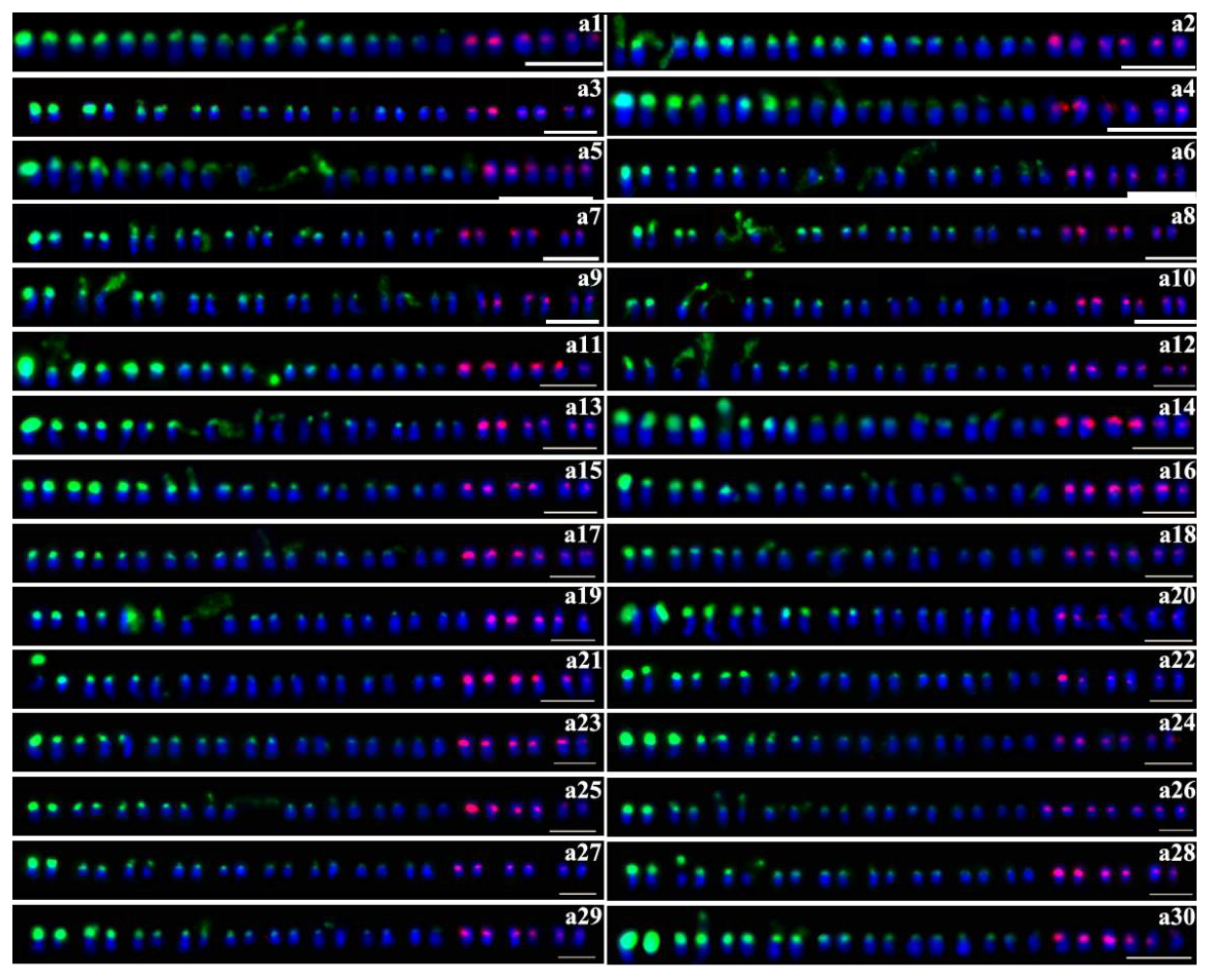
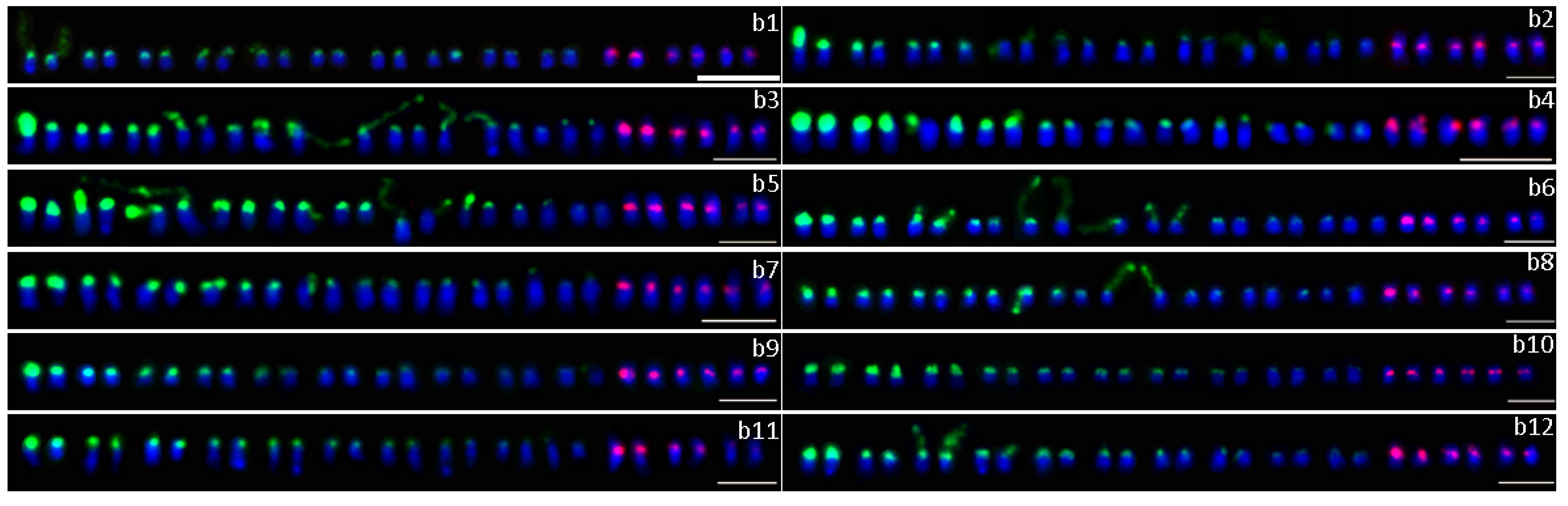

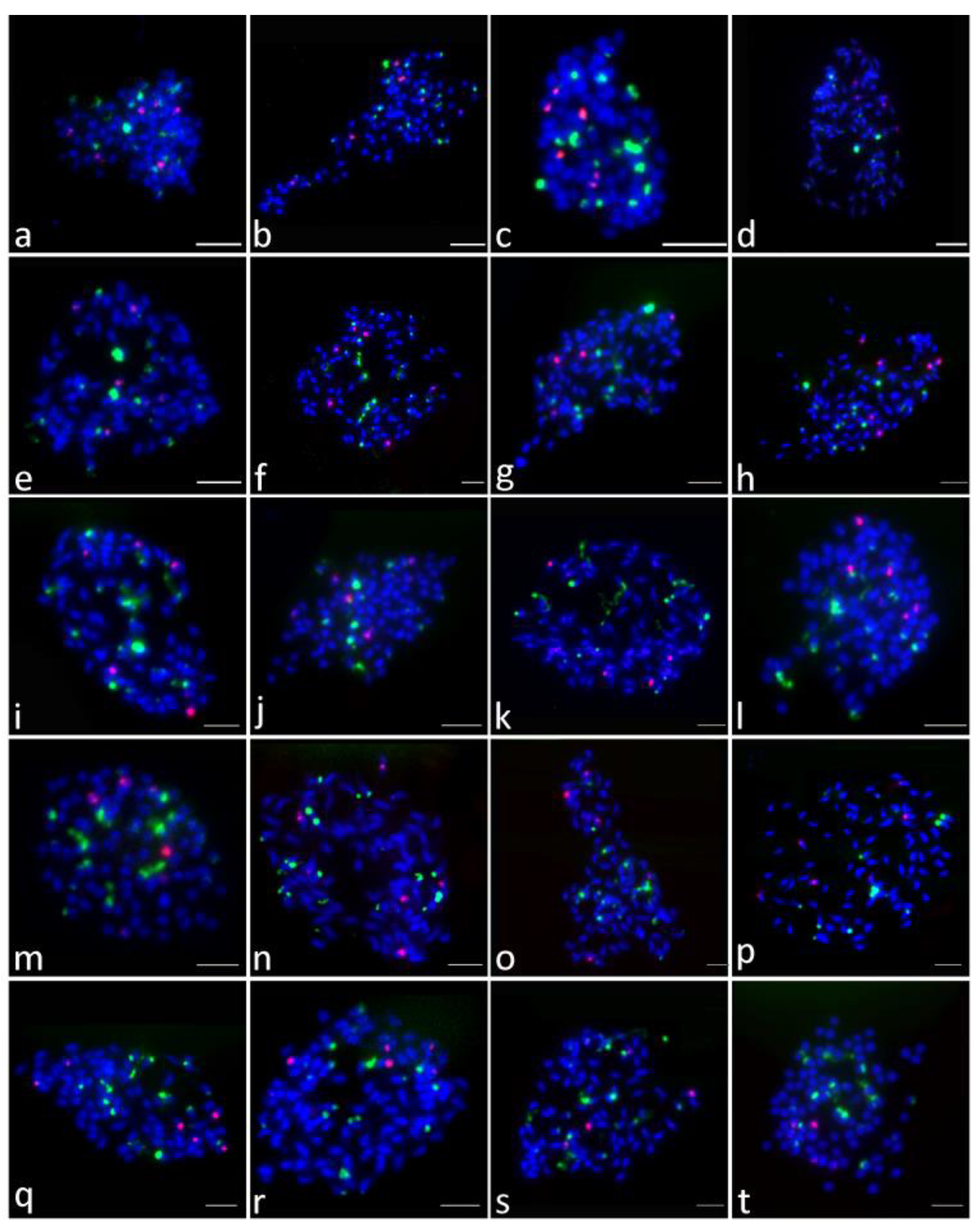
| No. | Cultivars | Chromosome Number (2n) | No. of 45S rDNA | No. of 5S rDNA | No. | Cultivars | Chromosome Number (2n) | No. of 45S rDNA | No. of 5S rDNA |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Quzishu 57 | 90 | 18(1S,12M,5W) | 6 | 39 | Chuanzishu 4 | 90 | 18(1S,11M,6W) | 6 |
| 2 | Neiyuzi 2 | 92 | 18(6S, 8M,4W) | 6 | 40 | Fushu 24 | 90 | 20(4S,8M,8W) | 6 |
| 3 | Qinzishu 2 | 90 | 18(3S,11M,4W) | 6 | 41 | Guijingshu 8 | 90 | 18(3S,11M,4W) | 6 |
| 4 | Longzishu 4 | 90 | 18(6S,10M,2W) | 6 | 42 | Chuanzishu 2 | 88 | 18(2S,12M,4W) | 6 |
| 5 | Longzishu 6 | 90 | 18(3S,13M,2W) | 6 | 43 | Guijingshu 9 | 90 | 18(4S,14M) | 6 |
| 6 | Zhezishu 1 | 90 | 18(4S,12M,2W) | 6 | 44 | Fangzishu 9 | 90 | 20(2S,16M,2W) | 6 |
| 7 | Qinzishu 3 | 90 | 18(3S,13M,2W) | 6 | 45 | Shangxuzi 1 | 90 | 20(3S,11M,6W) | 6 |
| 8 | Ningzishu 3 | 90 | 18(2S,6M,10W) | 6 | 46 | Jixuzi 2 | 90 | 16(4S,9M,3W) | 6 |
| 9 | Jizishu 2 | 90 | 18(1S,13M,4W) | 6 | 47 | Yuzixiang 10 | 90 | 18(4S,6M,8W) | 5 |
| 10 | Longjinshu 1 | 90 | 18(2S,9M,7W) | 6 | 48 | Xuzishu 3 | 90 | 18(2S,10M,6W) | 6 |
| 11 | Fuzishu 1 | 90 | 18(2S,13M,3W) | 6 | 49 | Yusuzi 43 | 90 | 16(1S,11M,4W) | 6 |
| 12 | Taizhong 11 | 90 | 18(1S,12M,5W) | 6 | 50 | Yuzishu 7 | 90 | 20(2S,14M,4W) | 6 |
| 13 | Yanzishu 3 | 89 | 18(1S,13M,4W) | 6 | 51 | Pengzishu 1 | 90 | 20(2S,11M,7W) | 6 |
| 14 | Jizishu 1 | 90 | 18(3S,12M,3W) | 6 | 52 | Yuzishu 3 | 90 | 18(2S,8M,8W) | 6 |
| 15 | Wan W36-1 | 90 | 20(16M,4W) | 6 | 53 | Yuzi 263 | 88 | 18(2S,12M,4W) | 6 |
| 16 | Longjinshu 3 | 90 | 18(2S,14M,2W) | 6 | 54 | Xuzishu 5 | 90 | 18(2S,14M,2W) | 6 |
| 17 | Zhanzishu 2 | 90 | 20(3S,14M,3W) | 6 | 55 | Guijingshu 6 | 90 | 16(1S,14M,1W) | 6 |
| 18 | Jizishu 2 | 90 | 18(2S,14M,2W) | 6 | 56 | Guijingshu 7 | 90 | 18(1S,10M,7W) | 6 |
| 19 | Yanzishu 2 | 90 | 19(2S,12M,5W) | 6 | 57 | Guijingshu 3 | 90 | 18(3S,7M,8W) | 6 |
| 20 | Ningzishu 4 | 90 | 21(4S,12M,5W) | 6 | 58 | Nanzishu 008 | 91 | 18(2S,10M,6W) | 6 |
| 21 | Ningzishu 1 | 88 | 18(4S,10M,4W) | 6 | 59 | Nanzishu 014 | 90 | 16(1S,13M,2W) | 6 |
| 22 | Xuzishu 2 | 88 | 18(1S,13M,4W) | 6 | 60 | Nanzishu 015 | 90 | 16(1S,14M,1W) | 6 |
| 23 | Ningzishu 2 | 90 | 16(2S,10M,4W) | 6 | 61 | Qianzishu 1 | 91 | 17(1S,13M,3W) | 6 |
| 24 | Ningzishu 5 | 89 | 18(2S,11M,5W) | 6 | 62 | Funingzi 3 | 90 | 21(1S,18M,2W) | 6 |
| 25 | Luozishu 1 | 90 | 18(1S,11M,6W) | 6 | 63 | Funingzi 4 | 90 | 20(4S,11M,5W) | 6 |
| 26 | Xuzishu 8 | 90 | 18(6S,8M,4W) | 6 | 64 | Quanzishu 96 | 90 | 18(2S,12M,4W) | 7 |
| 27 | Xuzishu 6 | 90 | 20(1S,12M,7W) | 6 | 65 | Puzishu 3 | 90 | 19(4S,8M,7W) | 6 |
| 28 | Yanzishu 4 | 90 | 18(4S,12M,2W) | 6 | 66 | Puzishu 18 | 89 | 18(2S,13M,3W) | 6 |
| 29 | Jizishu 1 | 90 | 18(3S,9M,6W) | 6 | 67 | Xushu33 | 91 | 18(1S,13M,4W) | 6 |
| 30 | Guizishu 1 | 90 | 16(3S,9M,4W) | 6 | 68 | Jizishu 3 | 90 | 18(2S,12M,4W) | 6 |
| 31 | Mianzishu 9 | 90 | 20(1S,16M,3W) | 6 | 69 | Guangzishu 9 | 90 | 20(2S,12M,6W) | 6 |
| 32 | Mianyuzi 11 | 90 | 20(4S,12M,4W) | 6 | 70 | Guangzishu 10 | 90 | 18(1S,14M,3W) | 6 |
| 33 | Wanzi 56 | 90 | 18(2S,13M,3W) | 6 | 71 | Guangzishu 11 | 89 | 19(2S,11M,6W) | 6 |
| 34 | Ezishu 13 | 90 | 20(6S,11M,3W) | 6 | 72 | Guangzishu 1 | 90 | 20(2S,15M,3W) | 6 |
| 35 | Ezishu 12 | 89 | 17(2S,12M,3W) | 6 | 73 | Guangzishu 2 | 90 | 20(2S,16M,2W) | 6 |
| 36 | Guizishu 3 | 90 | 20(2S,15M,3W) | 6 | 74 | Guangzishu 8 | 90 | 20(2S,13M,5W) | 6 |
| 37 | Guiziwei 1 | 90 | 18(5S,12M,1W) | 6 | 75 | Jizishu 18 | 90 | 21(1S,17M,3W) | 6 |
| 38 | Fushu 404 | 92 | 20(2S,12M,6W) | 6 | 76 | Ayamurasaki | 90 | 18(2S,14M,2W) | 6 |
| Probe Name | Sequence and Fluorochrome Label | Sequences Used to Develop Probes (GenBank Accession Number) |
|---|---|---|
| 5S-1 | TAMRA-5′GGATGCGATCATACCAGCACTAAT GCACCGGATCCCATCAGAACTCCGCAGTTAA GCGT3′ | 1–59 bases in coding region of 5S rRNA from Arabidopsis thaliana (GenBank AJ307346.2) |
| 5S-2 | TAMRA-5′GCTTGGGCGAGAGTAGTACTAGGA TGGGTGACCTCTCGGGAAATCCTCGTGTTGC ATC3′ | 60–118 bases in coding region of 5S rRNA from Arabidopsis thaliana (GenBank AJ307346.2) |
| 45S-1 | 6-FAM -5′AAAACGACTCTCGGCAACGGATAT CTCGGCTCTCGCATCGATGAAGAACGTAGCG AAAT3′ | Coding region of 5.8S rRNA from Arabidopsis thaliana (GenBank NR141643.1) |
| 45S-2 | 6-FAM -5′TACCTGGTTGATCCTGCCAGTAGTC ATATGCTTGTCTCAAAGATTAAGCCATGCAT GTG3′ | Coding region of 18S rRNA from Arabidopsis thaliana (GenBank NR141642.1) |
| 45S-3 | 6-FAM-5′CCCGCTGAGTTTAAGCATATCAATA AGCGGAGGAAAAGAAACTAACAAGGATTC CCTTA3′ | Coding region of 25S rRNA from Arabidopsis thaliana (GenBank X52320.1) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, D.; Chen, L.; Sun, J.; Zhang, L.; Gao, R.; Li, Q.; Han, Y.; Li, Z. Comparative Chromosomal Localization of 45S and 5S rDNA Sites in 76 Purple-Fleshed Sweet Potato Cultivars. Plants 2020, 9, 865. https://doi.org/10.3390/plants9070865
Su D, Chen L, Sun J, Zhang L, Gao R, Li Q, Han Y, Li Z. Comparative Chromosomal Localization of 45S and 5S rDNA Sites in 76 Purple-Fleshed Sweet Potato Cultivars. Plants. 2020; 9(7):865. https://doi.org/10.3390/plants9070865
Chicago/Turabian StyleSu, Dan, Lei Chen, Jianying Sun, Luyue Zhang, Runfei Gao, Qiang Li, Yonghua Han, and Zongyun Li. 2020. "Comparative Chromosomal Localization of 45S and 5S rDNA Sites in 76 Purple-Fleshed Sweet Potato Cultivars" Plants 9, no. 7: 865. https://doi.org/10.3390/plants9070865