Impact of Solar Radiation on Gene Expression in Bacteria
Abstract
:1. Introduction
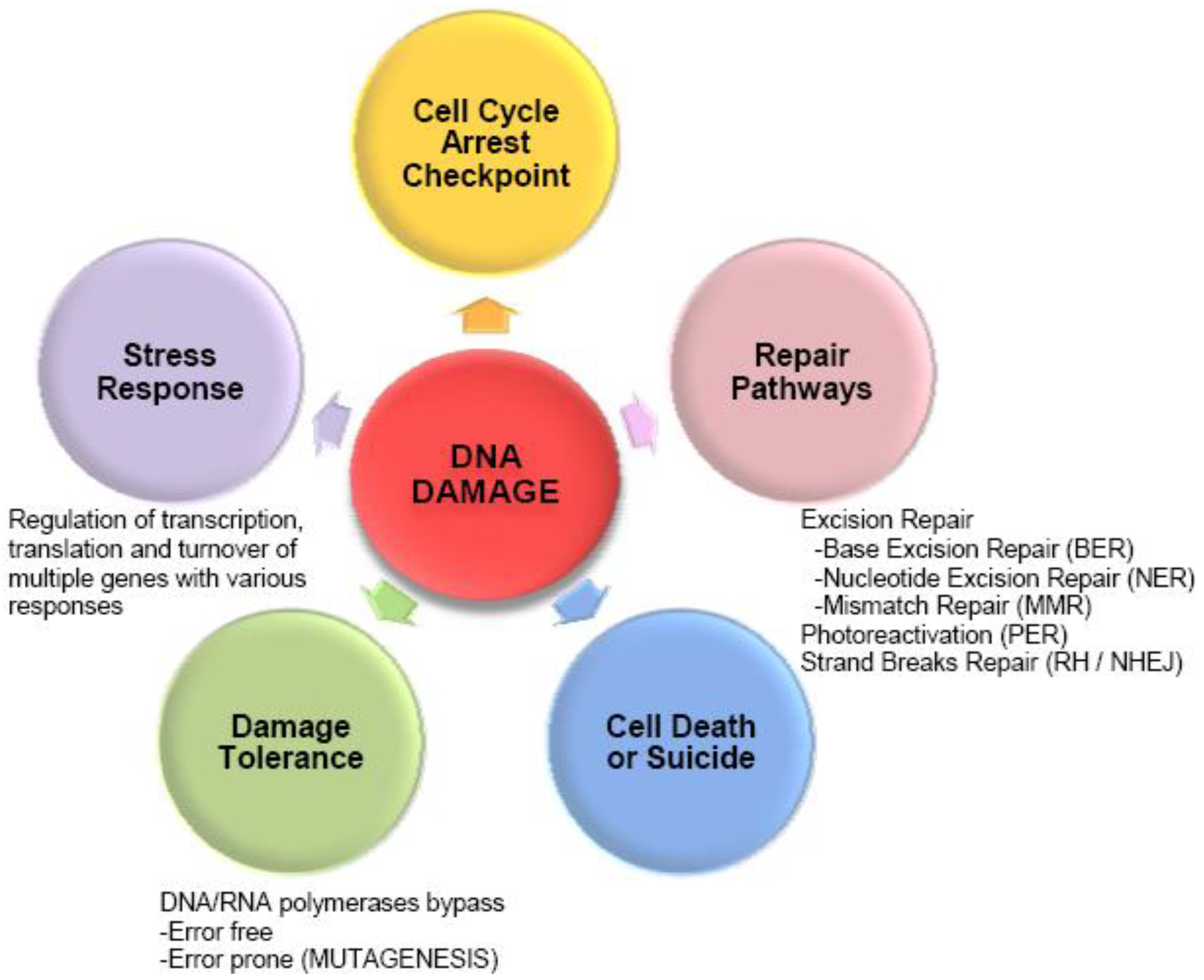
| Microorganism | Radiation | Gene Regulation | Application areas | References |
|---|---|---|---|---|
| Deinococcus gobiensis | Gamma radiation + UVC radiation | Transcriptome | Resistance analysis | [17] |
| Deinococcus radiodurans | Ionizing radiation | Transcriptome | Resistance analysis | [28,29] |
| Proteome | [30] | |||
| Proteome | Proteogenomics/ Resistance analysis | [31] | ||
| Nostoc commune | UVB radiation | Proteome | Resistance analysis | [32] |
| Deinococcus deserti | UV radiation, Ionizing radiation | Proteome | Proteogenomics | [33] |
| Ruegeria pomeroyi DSS-3 | Ionizing radiation | Proteome | Proteogenomics | [34] |
| Shewanella oneidensis MR-1 | UVA, UVB, UVC | Transcriptome + Proteome | Proteogenomics | [35] |
| Transcriptome | Sensitivity analysis | [36] | ||
| Ionizing radiation | Transcriptome | Sensitivity analysis | [37] | |
| Escherichia coli | UVA | Transcriptome | Bacterial Disinfection | [38] |
| Bacillus pumilus | “UV-Mars” “UV-Space” | Proteome | Astrobiology | [39] |
| Rhodospirillum rubrum | Ionizing radiation | Transcriptome/ Proteome | Astrobiology | [40] |
| Prochlorococcus marinus MED4 | High light intensity | Transcriptome | Microbial ecology | [41] |
| Proteome | [42] | |||
| Synechocystis sp. strain PCC 6803 | High light intensity + UVB | Transcriptome | Microbial ecology | [43,44] |
| Photobacterium angustum S14 | UVB | Proteome | Microbial ecology | [45] |
| Sphingopyxis alaskensis RB2256 | UVB, UVA, Visible light | Proteome | Microbial ecology | [46] |
2. Understanding the Extreme Radiation-Resistance of Key Bacterial Model
3. Sensitive vs. Resistant Bacteria
4. Bacterial Disinfection for Water Treatment and Astrobiology Applications
5. Response of Marine Bacteria to Solar Radiation: A Case Study in Microbial Ecology
6. UV-Induced Protein Damage
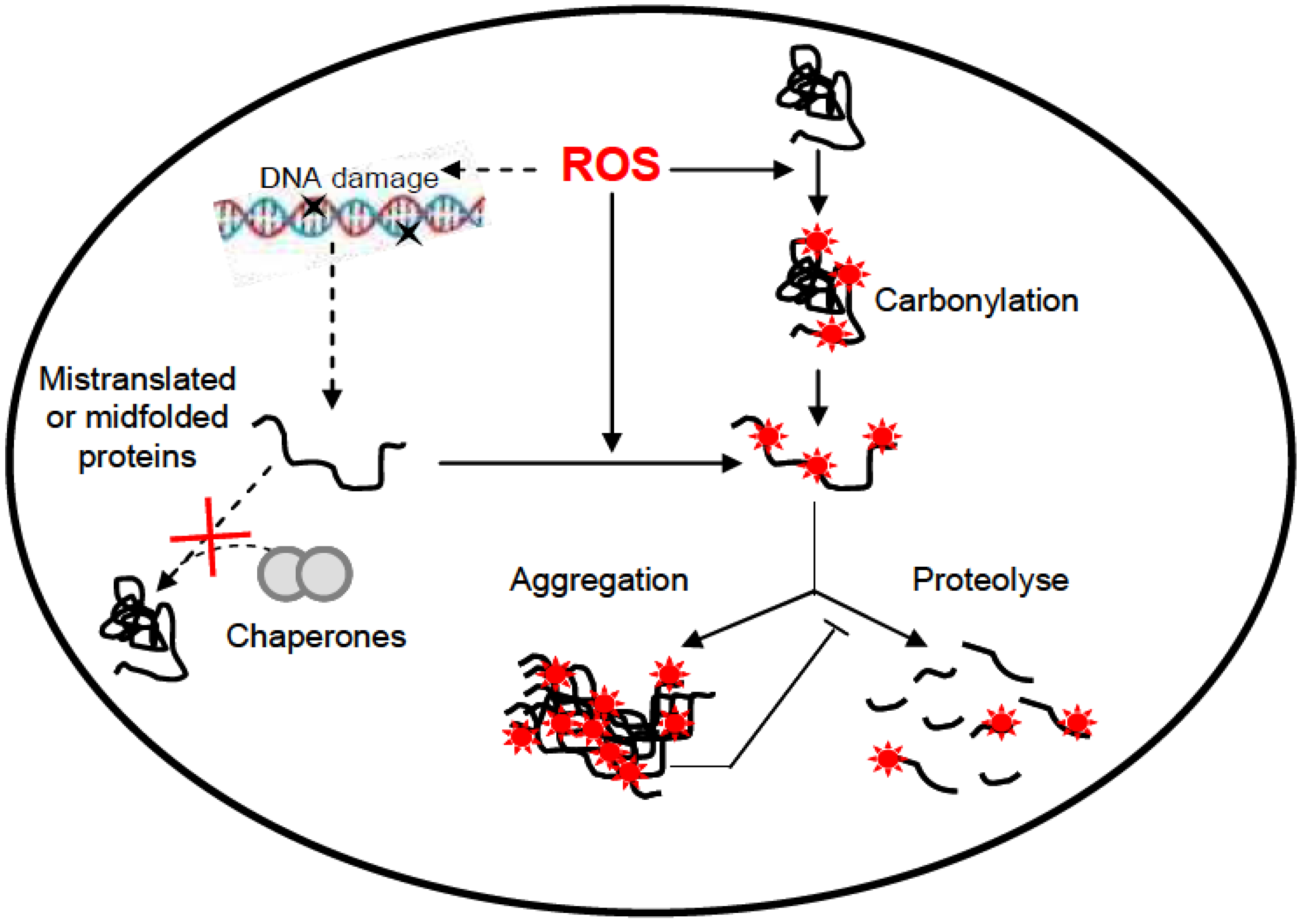
7. Conclusion
Acknowledgements
Conflicts of Interest
References
- Sinha, R.P.; Häder, D.P. UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef]
- Friedberg, E.C. DNA damage and repair. Nature 2003, 421, 436–440. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Meador, J.A.; Joux, F.; Douki, T. Effect of the GC content of DNA on the distribution of UVB-induced bipyrimidine photoproducts. Photochem. Photobiol. Sci. 2008, 7, 794–801. [Google Scholar] [CrossRef]
- Wurtmann, E.J.; Wolin, S.L. RNA under attack: Cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 34–49. [Google Scholar] [CrossRef]
- Fimognari, C.; Sestili, P.; Lenzi, M.; Bucchini, A.; Cantelli-Forti, G.; Hrelia, P. RNA as a new target for toxic and protective agents. Mutat. Res. 2008, 648, 15–22. [Google Scholar] [CrossRef]
- Cadet, J.; Sage, E.; Douki, T. Ultraviolet radiation-mediated damage to cellular DNA. Mut. Res. 2005, 571, 3–17. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Mechanisms of Free Radical Damage to DNA. In DNA & Free Radicals: Techniques, Mechanisms & Applications; Aruoma, O.I., Halliwell, B., Eds.; OICA International: Santa Lucia and London, UK, 1998; pp. 3–26. [Google Scholar]
- Sonntag, V. The Chemical Basis of Radiation Biology; Taylor and Francis: New York, NY, USA, 1987. [Google Scholar]
- Douki, T. The variety of UV-induced pyrimidine dimeric photoproducts in DNA as shown by chromatographic quantification methods. Photochem. Photobiol. Sci. 2013. [Google Scholar]
- Cooper, S. Checkpoints and restriction points in bacteria and eukaryotic cells. BioEssays 2006, 28, 1035–1039. [Google Scholar] [CrossRef]
- Aldsworth, T.G.; Sharman, R.L.; Dodd, C.E.R. Bacterial suicide through stress. Cell. Mol. Life Sci. 1999, 56, 378–383. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.W.; Kemner, K.M.; et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007, 5, e92. [Google Scholar] [CrossRef]
- Santos, A.L.; Oliveira, V.; Baptista, I.; Henriques, I.; Gomes, N.C.; Almeida, A.; Correia, A.; Cunha, Â. Wavelength dependence of biological damage induced by UV radiation on bacteria. Arch. Microbiol. 2013, 195, 63–74. [Google Scholar] [CrossRef]
- Sharma, C.M.; Hoffmann, S.; Darfeuille, F.; Reignier, J.; Findeiss, S.; Sittka, A.; Chabas, S.; Reiche, K.; Hackermüller, J.; Reinhardt, R.; et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010, 464, 250–255. [Google Scholar] [CrossRef]
- Filiatrault, M.J.; Stodghill, P.V.; Bronstein, P.A.; Moll, S.; Lindeberg, M.; Grills, G.; Schweitzer, P.; Wang, W.; Schroth, G.P.; Luo, S.; et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 2010, 192, 2359–2372. [Google Scholar] [CrossRef]
- Kogenaru, S.; Qing, Y.; Guo, Y.; Wang, N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 2012, 13, e629. [Google Scholar] [CrossRef]
- Yuan, M.; Chen, M.; Zhang, W.; Lu, W.; Wang, J.; Yang, M.; Zhao, P.; Tang, R.; Li, X.; Hao, Y.; et al. Genome sequence and transcriptome analysis of the radioresistant bacterium Deinococcus gobiensis: Insights into the extreme environmental adaptations. PLoS One 2012, 7, e34458. [Google Scholar] [CrossRef]
- Fu, X.; Fu, N.; Guo, S.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Chen, W.; Li, Y.; Zeng, R.; et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 2009, 10, e161. [Google Scholar]
- Van den Bergh, G.; Arckens, L. Fluorescent two-dimensional difference gel electrophoresis unveils the potential of gel-based proteomics. Curr. Opin. Biotechnol. 2004, 15, 38–43. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rist, B.; Gerber, S.A.; Turecek, F.; Gelb, M.H.; Aebersold, R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999, 17, 994–999. [Google Scholar] [CrossRef]
- Krijgsveld, J.; Ketting, R.F.; Mahmoudi, T.; Johansen, J.; Artal-Sanz, M.; Verrijzer, C.P.; Plasterk, R.H.A.; Heck, A.J.R. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 2003, 21, 927–931. [Google Scholar] [CrossRef]
- Zhong, H.; Marcus, S.L.; Li, L. Two-dimensional mass spectra generated from the analysis of 15N-labeled and unlabeled peptides for efficient protein identification and de novo peptide sequencing. J. Proteome Res. 2004, 3, 1155–1163. [Google Scholar] [CrossRef]
- Leroy, B.; Rosier, C.; Erculisse, V.; Leys, N.; Mergeay, M.; Wattiez, R. Differential proteomic analysis using isotope-coded protein-labeling strategies: Comparison, improvements and application to simulated microgravity effect on Cupriavidus metallidurans CH34. Proteomics 2010, 10, 2281–2291. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Leroy, B.; Wattiez, R. Shotgun proteomics: Concept, key points and data mining. Exp. Rev. Proteomics 2010, 7, 5–7. [Google Scholar] [CrossRef]
- Goshe, M.B.; Smith, R.D. Stable isotope-coded proteomic mass spectrometry. Curr. Opin. Biotechnol. 2003, 14, 101–109. [Google Scholar] [CrossRef]
- Oda, Y.; Huang, K.; Cross, F.R.; Cowburn, D.; Chait, B.T. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci.USA 1999, 96, 6591–6596. [Google Scholar] [CrossRef]
- Conrads, T.P.; Alving, K.; Veenstra, T.D.; Belov, M.E.; Anderson, G.A.; Anderson, D.J.; Lipton, M.S.; Pasa-Tolic, L.; Udseth, H.R.; Chrisler, W.B.; et al. Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal. Chem. 2001, 73, 2132–2139. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Omelchenko, M.V.; Beliaev, A.S.; Venkateswaran, A.; Stair, J.; Wu, L.; Thompson, D.K.; Xu, D.; Rogozin, I.B.; et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 2003, 100, 4191–4196. [Google Scholar] [CrossRef]
- Tanaka, M.; Earl, A.M.; Howell, H.A.; Park, M.J.; Eisen, J.A.; Peterson, S.N.; Battista, J.R. Analysis of Deinococcus radiodurans’s transcriptional response to ionizing radiation and dessication reveals novel proteins that contribute to extreme radioresistance. Genetics 2004, 168, 21–33. [Google Scholar] [CrossRef]
- Zhang, C.; Jianfeng, W.; Zhiguo, Z.; Nanjiao, Y.; Duohong, S.; Yuejin, H. Proteomic analysis of Deinococcus radiodurans recovering from gamma-radiation. Proteomics 2005, 5, 138–143. [Google Scholar] [CrossRef]
- Lipton, M.S.; Paša-Tolić, L.; Anderson, G.A.; Anderson, D.J.; Auberry, D.L.; Battista, J.R.; Daly, M.J.; Fredrickson, J.; Hixson, K.K.; Kostandarithes, H.; et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 2002, 99, 11049–11054. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Schulz, S.; Wait, R.; Görg, A.; Scherer, S. The UV-B stimulon of the terrestrial cyanobacterium Nostoc commune comprises early shock proteins and late acclimation proteins. Mol. Microbiol. 2002, 46, 827–843. [Google Scholar] [CrossRef]
- Groot, A.; Dulermo, R.; Ortet, P.; Blanchard, L.; Guérin, P.; Fernandez, B.; Vacherie, B.; Dossat, C.; Jolivet, E.; Siguier, P.; et al. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 2009, 5, e1000434. [Google Scholar] [CrossRef]
- Christie-Oleza, J.A.; Fernandez, B.; Nogales, B.; Bosch, R.; Armengaud, J. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J. 2012, 6, 124–135. [Google Scholar] [CrossRef]
- Kolker, E.; Picone, A.F.; Galperin, M.Y.; Romine, M.F.; Higdon, R.; Makarova, K.S.; Kolker, N.; Anderson, G.A.; Qiu, X.; Auberry, K.J.; et al. Global profiling of Shewanella oneidensis MR-1: Expression of hypothetical genes and improved functional annotations. Proc. Natl. Acad. Sci.USA 2005, 102, 2099–2104. [Google Scholar] [CrossRef]
- Qiu, X.; Sundin, G.W.; Wu, L.; Zhou, J.; Tiedje, J.M. Comparative analysis of differentially expressed genes in Shewanella oneidensis MR-1 following exposure to UVC, UVB, and UVA radiation. J. Bacteriol. 2005, 187, 3556–3564. [Google Scholar] [CrossRef]
- Qiu, X.; Daly, M.J.; Vasilenko, A.; Omelchenko, M.V.; Gaidamakova, E.K.; Wu, L.; Zhou, J.; Sundin, G.W.; Tiedje, J.M. Transcriptome analysis applied to survival of Shewanella oneidensis MR-1 exposed to ionizing radiation. J. Bacteriol. 2006, 188, 1199–1204. [Google Scholar] [CrossRef]
- Berney, M.; Weilenmann, H.U.; Ihssen, J.; Bassin, C.; Egli, T. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl. Environ. Microbiol. 2006, 72, 2586–2593. [Google Scholar] [CrossRef]
- Vaishampayan, P.A.; Rabbow, E.; Horneck, G.; Venkateswaran, K.J. Survival of Bacillus pumilus spores for a prolonged period of time in real space conditions. Astrobiology 2012, 12, 487–497. [Google Scholar] [CrossRef]
- Mastroleo, F.; van Houdt, R.; Leroy, B.; Benotmane, M.A.; Janssen, A.; Mergeay, M.; Vanhavere, F.; Hendrickx, L.; Wattiez, R.; Leys, N. Experimental design and environmental parameters affect Rhodospirillum rubrum S1H response to space flight. ISME J. 2009, 3, 1402–1419. [Google Scholar] [CrossRef]
- Steglich, C.; Futschik, M.; Rector, T.; Steen, R.; Chisholm, S.W. Genome-wide analysis of light sensing in Prochlorococcus. J.Bacteriol. 2006, 188, 7796–7806. [Google Scholar] [CrossRef]
- Pandhal, J.; Wright, P.C.; Biggs, C.A. A quantitative proteomic analysis of light adaptation in a globally significant marine cyanobacterium Prochlorococcus marinus MED4. J. Proteome Res. 2007, 6, 996–1005. [Google Scholar] [CrossRef]
- Hihara, Y.; Kamei, A.; Kanehisa, M.; Kaplan, A.; Ikeuchi, M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001, 13, 793–806. [Google Scholar]
- Huang, L.; McCluskey, M.P.; Ni, H.; LaRossa, R.A. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 2002, 184, 6845–6858. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Joux, F.; Wattiez, R.; Lebaron, P. Proteome analysis of the UVB resistant marine bacterium Photobacterium angustum S14. PLoS One 2012, 7, e42299. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Joux, F.; Raftery, M.J.; Cavicchioli, R. The response of the marine bacterium Sphingopyxis alaskensis to solar radiation assessed by quantitative proteomics. Environ. Microbiol. 2009, 11, 2660–2675. [Google Scholar] [CrossRef]
- Narumi, I.; Satoh, K.; Cui, S.; Funayama, T.; Kitayama, S.; Watanabe, H. PprA: A novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol. Microbiol. 2004, 54, 278–285. [Google Scholar] [CrossRef]
- Ansong, C.; Purvine, S.O.; Adkins, J.N.; Lipton, M.S.; Smith, R.D. Proteogenomics: Needs and roles to be filled by proteomics in genome annotation. Briefs Funct. Genomics Proteomics 2008, 7, 50–62. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Venkateswaran, A.; Hess, M.; Omelchenko, M.V.; Kostandarithes, H.M.; Makarova, K.S.; et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 2004, 306, 1025–1028. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Paulsen, I.T.; Nelson, K.E.; Gaidos, R.J.; Nelson, W.C.; Read, T.D.; Eisen, J.A.; Seshadri, R.; Ward, N.; Methe, B.; et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002, 20, 1118–1123. [Google Scholar] [CrossRef]
- Santos, A.L.; Gomes, N.C.M.; Henriques, I.; Almeida, A.; Correia, A.; Cunha, A. Role of transition metals in UVB-induced damage to bacteria. Photochem. Photobiol. 2013, 89, 640–648. [Google Scholar] [CrossRef]
- Hollaender, A. Effect of long ultraviolet and short visible radiation on Escherichia coli. J. Bacteriol. 1943, 46, 531–541. [Google Scholar]
- Quillardet, P.; Rouffaud, M.A.; Bouige, P. DNA array analysis of gene expression in response to UV irradiation in Escherichia coli. Res. Microbiol. 2003, 154, 559–572. [Google Scholar] [CrossRef]
- Hendrickx, L.; de Wever, H.; Hermans, V.; Mastroleo, F.; Morin, N.; Wilmotte, A.; Janssen, P.; Mergeay, M. Microbial ecology of the closed artificial ecosystem MELiSSA (Micro-Ecological Life Support System Alternative): Reinventing and compartmentalizing the Earth’s food and oxygen regeneration system for long-haul space exploration missions. Res. Microbiol. 2006, 157, 77–86. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- Tedetti, M.; Sempéré, R. Penetration of UV radiation in the marine environment: A review. Photochem. Photobiol. 2006, 82, 89–397. [Google Scholar]
- Madronich, S.; McKenzie, R.L.; Björn, L.O.; Caldwell, M.M. Changes in biologically active ultraviolet radiation reaching the Earth’s surface. J. Photochem. Photobiol. B 1998, 46, 5–19. [Google Scholar] [CrossRef]
- Mary, I.; Tu, C.J.; Grossman, A.; Vaulot, D. Effects of high light on transcripts of stress-associated genes for the cyanobacteria Synechocystis sp. PCC 6803 and Prochlorococcus MED4 and MIT9313. Microbiology 2004, 150, 1271–1281. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Ferreri, C.; Torreggiani, A.; Salzano, A.M.; Renzone, G.; Scaloni, A. Radiation-induced reductive modifications of sulfur-containing amino acids within peptides and proteins. J. Proteomics 2011, 74, 2264–2273. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Milzani, A.; Colombo, R. Protein carbonyls groups as biomarkers of oxidative stress. Clin. Chim. Acta 2003, 329, 23–38. [Google Scholar] [CrossRef]
- Dukan, S.; Farewell, A.; Ballesteros, M.; Taddei, F.; Radman, M.; Nyström, T. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 2000, 97, 5746–5749. [Google Scholar] [CrossRef]
- Ballesteros, M.; Fredriksson, A.; Henriksson, J.; Nyström, T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001, 20, 5280–5289. [Google Scholar] [CrossRef]
- Bota, D.A.; Davies, K.J. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell. Biol. 2002, 4, 674–680. [Google Scholar] [CrossRef]
- Grune, T.; Merker, K.; Sandig, G.; Davies, K.J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003, 305, 709–718. [Google Scholar] [CrossRef]
- Grune, T.; Jung, T.; Merker, K.; Davies, K.J.A. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and “aggresomes” during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 2004, 36, 2519–2530. [Google Scholar] [CrossRef]
- Levine, R.L. Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J. Biol. Chem. 1983, 258, 11823–11827. [Google Scholar]
- Levine, R.L. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic.Biol. Med. 2002, 32, 790–796. [Google Scholar] [CrossRef]
- Requena, J.R.; Levine, R.L.; Stadtman, E.R. Recent advances in the analysis of oxidized proteins. Amino Acids 2003, 25, 221–226. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Protein and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci.USA 2010, 107, 14373–14377. [Google Scholar] [CrossRef]
- Desnues, B.; Gregori, G.; Dukan, S.; Aguilaniu, H.; Nyström, T. Differential oxidative damage and expression of stress regulons in culturable and nonculturable cells of Escherichia coli. EMBO Rep. 2003, 4, 400–440. [Google Scholar] [CrossRef]
- Bosshard, F.; Riedel, K.; Schneider, T.; Geiser, C.; Bucheli, M.; Egli, T. Protein oxidation and aggregation in UVA-irradiated Escherichia coli cells as signs of accelerated cellular senescence. Environ. Microbiol. 2010, 12, 2931–2945. [Google Scholar] [CrossRef]
- Matallana-Surget, S.; Cavicchioli, R.; Fauconnier, C.; Wattiez, R.; Leroy, B.; Joux, F.; Raftery, M.J.; Lebaron, P. Shotgun redox proteomics: identification and quantitation of carbonylated proteins in the UVB resistant marine bacterium, Photobacterium angustum S14. PLoS One 2013, 8, e68112. [Google Scholar] [CrossRef]
- Tamarit, J.; Cabiscol, E.; Ros, J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J. Biol. Chem. 1998, 273, 3027–3032. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Matallana-Surget, S.; Wattiez, R. Impact of Solar Radiation on Gene Expression in Bacteria. Proteomes 2013, 1, 70-86. https://doi.org/10.3390/proteomes1020070
Matallana-Surget S, Wattiez R. Impact of Solar Radiation on Gene Expression in Bacteria. Proteomes. 2013; 1(2):70-86. https://doi.org/10.3390/proteomes1020070
Chicago/Turabian StyleMatallana-Surget, Sabine, and Ruddy Wattiez. 2013. "Impact of Solar Radiation on Gene Expression in Bacteria" Proteomes 1, no. 2: 70-86. https://doi.org/10.3390/proteomes1020070
APA StyleMatallana-Surget, S., & Wattiez, R. (2013). Impact of Solar Radiation on Gene Expression in Bacteria. Proteomes, 1(2), 70-86. https://doi.org/10.3390/proteomes1020070




