Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model
Abstract
:1. Introduction
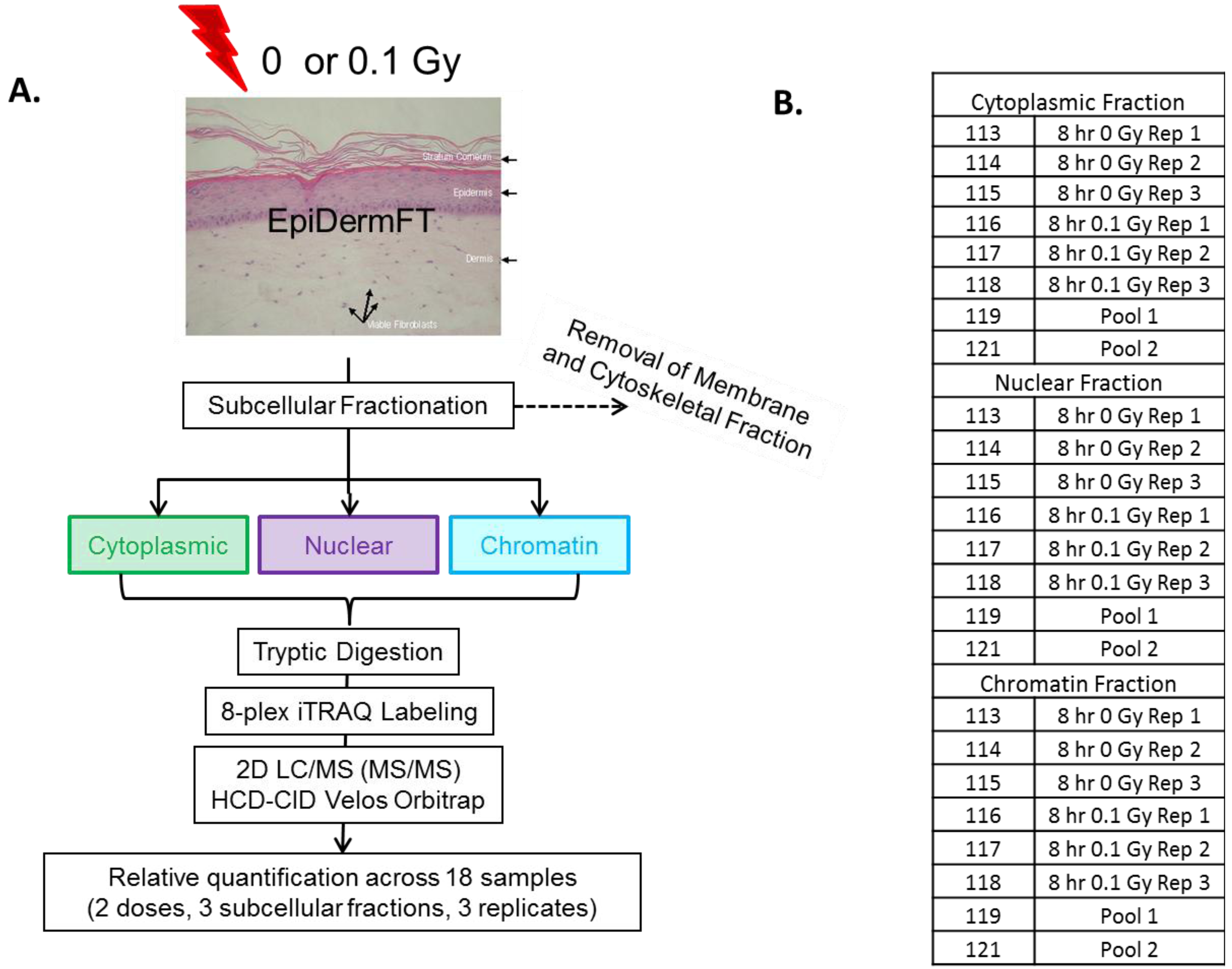
2. Experimental
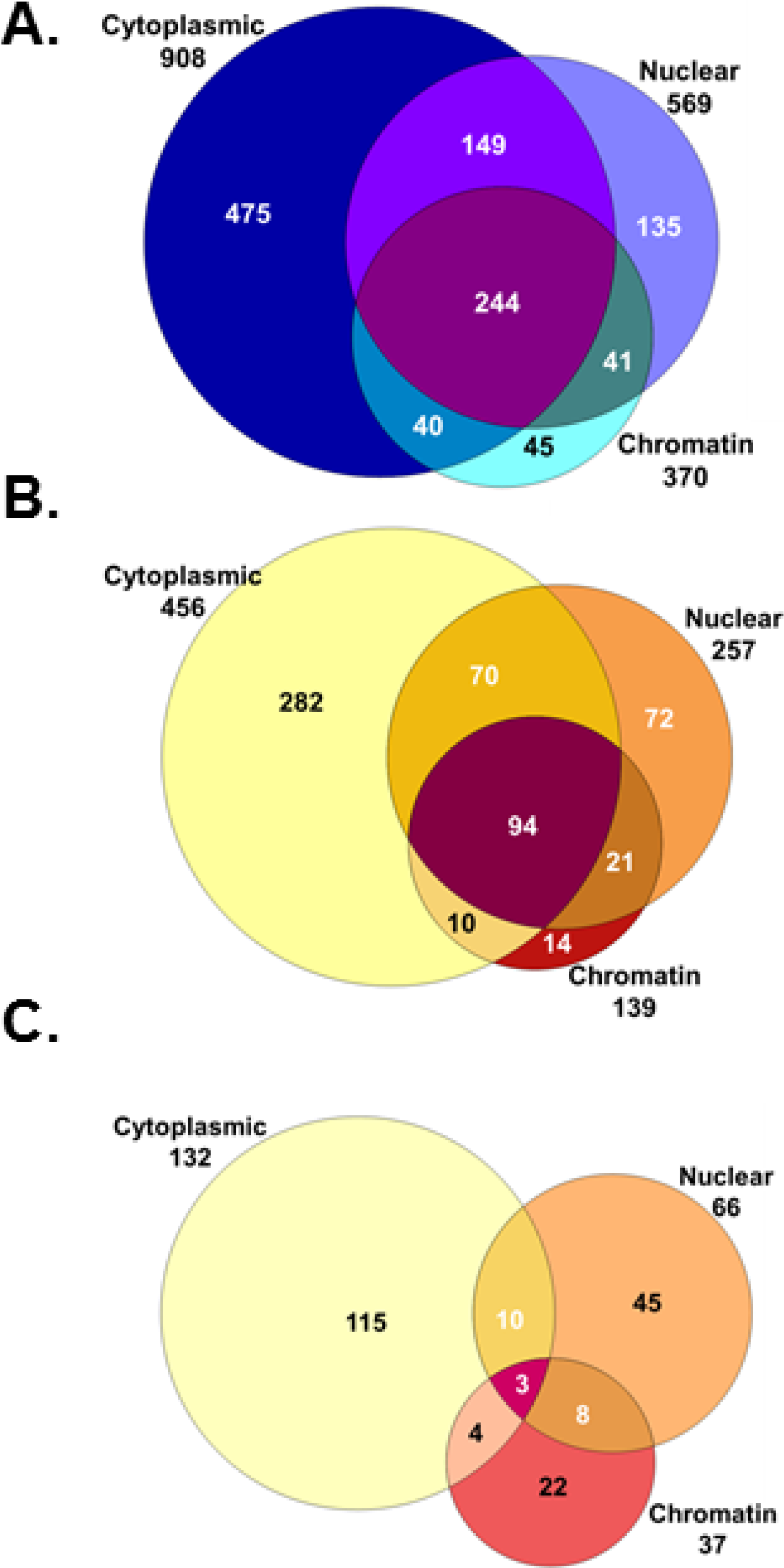
3. Results and Discussion
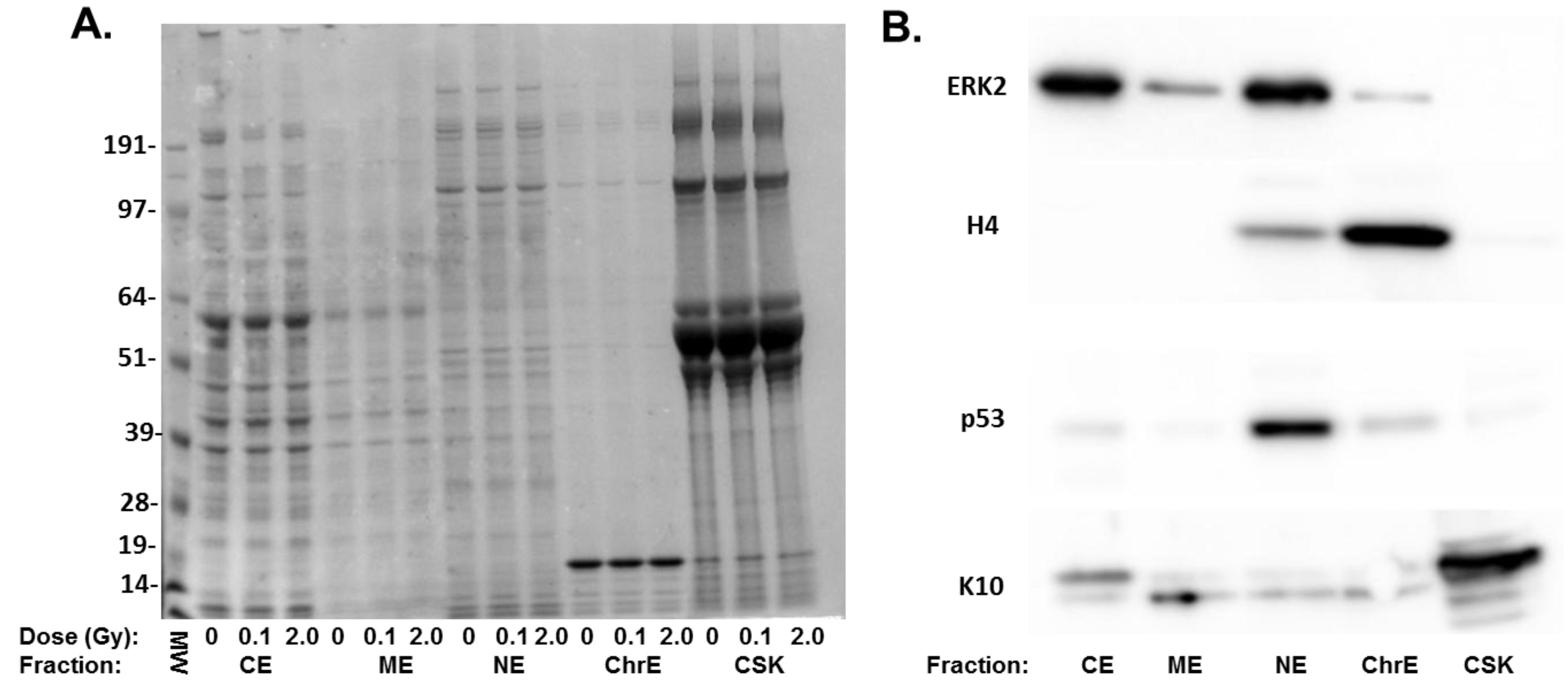
| Protein ID | Fraction | Change | p-value | Gene ID | Protein Name |
|---|---|---|---|---|---|
| FKB1A_HUMAN | Chromatin | −1.83 | 6.6E-05 | FKBP1A | FK506 binding protein 1A, 12 kDa/FKBP12 |
| PGS2_HUMAN | Cytoplasm | −1.67 | 1.7E-03 | DCN | decorin |
| ODO2_HUMAN | Cytoplasm | −1.43 | 1.7E-03 | DLST | dihydrolipoamide S-succinyltransferase |
| DBPA_HUMAN | Chromatin | −1.20 | 2.2E-02 | CSDA | cold shock domain protein A |
| ATPA_HUMAN | Chromatin | −1.19 | 4.9E-02 | ATP5A1 | ATP synthase alpha subunit 1 |
| FBLN2_HUMAN | Chromatin | −1.17 | 4.0E-02 | FBLN2 | fibulin 2 |
| KLK10_HUMAN | Chromatin | −1.01 | 1.8E-02 | KLK10 | kallikrein-related peptidase 10 |
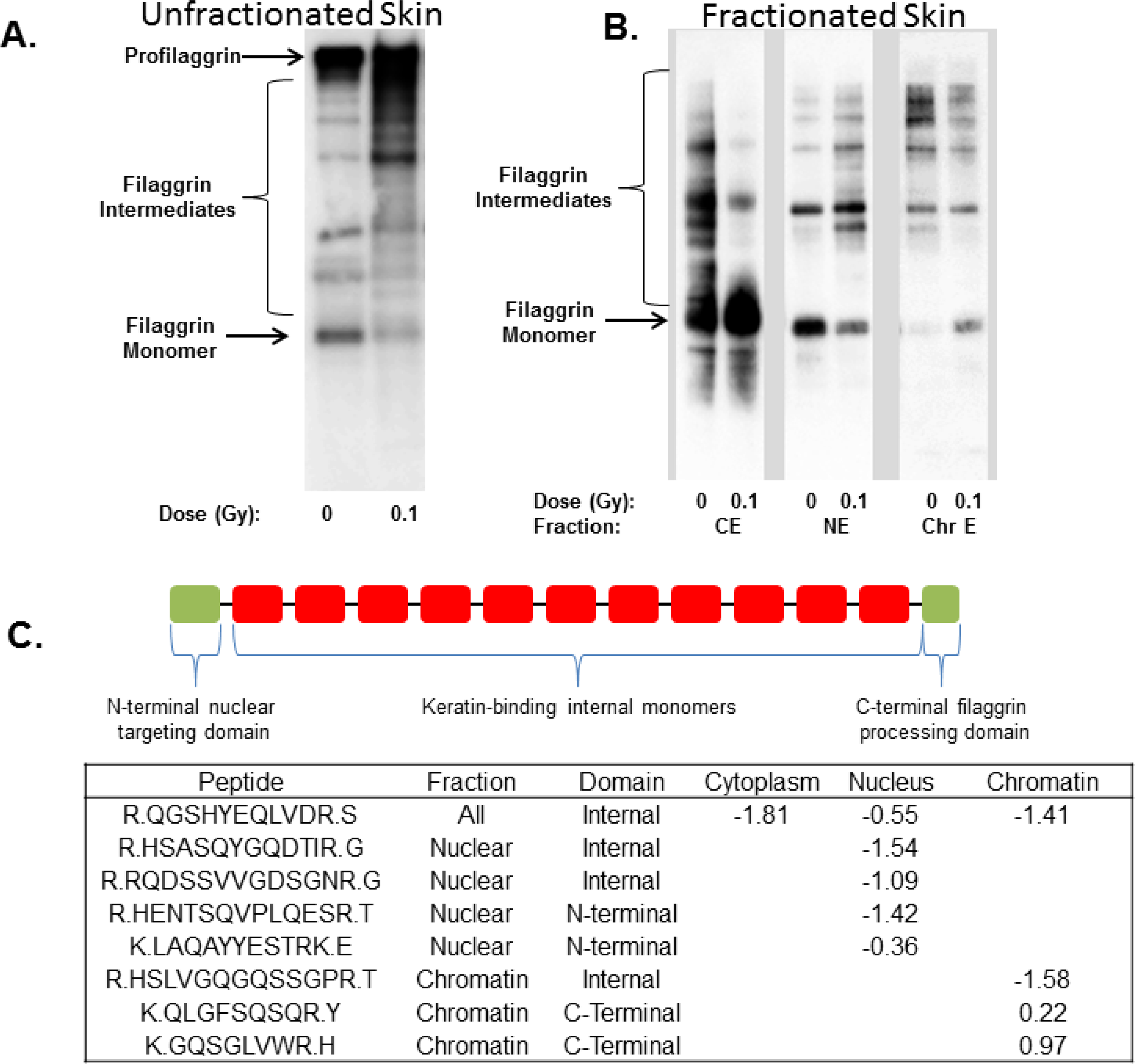
| FILA_HUMAN | Nucleus | −0.99 | 9.8E-05 | FLG | filaggrin |
| HUTH_HUMAN | Nucleus | −0.99 | 1.1E-03 | HAL | histidine ammonia-lyase |
| HSP72_HUMAN | Cytoplasm | −0.98 | 1.4E-02 | HSPA2 | heat shock 70 kDa protein 2 |
| SYAC_HUMAN | Cytoplasm | −0.96 | 2.0E-03 | AARS | alanyl-tRNA synthetase |
| THIL_HUMAN | Nucleus | −0.93 | 4.1E-02 | ACAT1 | acetyl-CoA acetyltransferase 1 |
| K22E_HUMAN | Nucleus | −0.91 | 4.5E-03 | KRT2 | keratin 2 |
| SPR1B_HUMAN | Cytoplasm | −0.89 | 1.7E-03 | SPRR1B | small proline-rich protein 1B |
| CAPZB_HUMAN | Cytoplasm | −0.88 | 1.7E-02 | CAPZB | capping protein Z-line, beta |
| LAMC1_HUMAN | Chromatin | −0.85 | 3.5E-02 | LAMC1 | laminin, gamma 1 |
| SFPQ_HUMAN | Nucleus | −0.85 | 3.3E-04 | SFPQ | splicing factor proline/glutamine-rich |
| CYTB_HUMAN | Cytoplasm | −0.84 | 9.3E-04 | CSTB | cystatin B (stefin B) |
| S10A8_HUMAN | Cytoplasm | −0.83 | 7.0E-07 | S100A8 | S100 calcium binding protein A8 |
| MVP_HUMAN | Chromatin | −0.82 | 2.7E-03 | MVP | major vault protein |
| Protein ID | Fraction | Change | p-value | Gene ID | Protein Name |
|---|---|---|---|---|---|
| FMOD_HUMAN | Chromatin | 1.67 | 9.9E-04 | FMOD | fibromodulin |
| TCPB_HUMAN | Cytoplasm | 1.49 | 4.0E-03 | CCT2 | chaperonin containing TCP1, subunit 2 |
| TCO1_HUMAN | Cytoplasm | 1.36 | 1.0E-02 | TCN1 | transcobalamin I |
| ELAF_HUMAN | Chromatin | 1.10 | 2.2E-03 | PI3 | peptidase inhibitor 3, skin-derived |
| TIMP1_HUMAN | Chromatin | 1.07 | 3.5E-03 | TIMP1 | TIMP metallopeptidase inhibitor 1 |
| MMP2_HUMAN | Chromatin | 0.94 | 1.1E-03 | MMP2 | matrix metallopeptidase 2 |
| RLA1_HUMAN | Cytoplasm | 0.91 | 7.7E-05 | RPLP1 | ribosomal protein, large, P1 |
| TSP1_HUMAN | Chromatin | 0.90 | 4.1E-07 | THBS1 | thrombospondin 1 |
| ACTN1_HUMAN | Nucleus | 0.88 | 1.5E-05 | ACTN1 | actinin, alpha 1 |
| COF1_HUMAN | Cytoplasm | 0.77 | 7.4E-05 | CFL1 | cofilin 1 |
| RL23A_HUMAN | Nucleus | 0.75 | 3.3E-02 | RPL23A | ribosomal protein L23a |
| CD109_HUMAN | Cytoplasm | 0.74 | 1.3E-02 | CD109 | CD109 molecule |
| TPM3L_HUMAN | Chromatin | 0.72 | 1.6E-02 | TPM3 | Tropomyosin 3 |
| ACON_HUMAN | Chromatin | 0.71 | 2.1E-04 | ACO2 | aconitase 2 |
| PEDF_HUMAN | Chromatin | 0.71 | 3.6E-08 | SERPINF1 | serpin peptidase inhibitor, clade F |
| CD59_HUMAN | Nucleus | 0.69 | 6.8E-05 | CD59 | CD59 molecule |
| K1C10_HUMAN | Cytoplasm | 0.69 | 1.5E-02 | KRT10 | keratin 10 |
| GRP78_HUMAN | Chromatin | 0.69 | 6.0E-03 | HSPA5 | heat shock 70 kDa protein 5 |
| UGDH_HUMAN | Cytoplasm | 0.67 | 4.1E-02 | UGDH | UDP-glucose 6-dehydrogenase |
| NAGK_HUMAN | Cytoplasm | 0.67 | 1.2E-02 | NAGK | N-acetylglucosamine kinase |
| Protein ID | Chromatin | Nucleus | Cytoplasm | Gene ID | Protein Name |
|---|---|---|---|---|---|
| GBLP_HUMAN | 0.616 | 0.405 | −0.335 | GNB2L1 | RACK1 |
| MDHM_HUMAN | 0.508 | 0.385 | 0.249 | MDH2 | Malate Dehydrogenase 2 |
| LAMC1_HUMAN | −0.854 | 0.193 | −0.455 | LAMC1 | Laminin gamma 1 |
| ISK5_HUMAN | 0.292 | 0.501 | SPINK5 | Serine peptidase Inhibitor | |
| HSPB1_HUMAN | −0.475 | −0.535 | HSPB1 | Heat shock 27 kDa protein 1 | |
| ACTN4_HUMAN | 0.432 | −0.362 | ACTN4 | Actinin, alpha4 | |
| ANXA2_HUMAN | 0.350 | −0.404 | ANXA2 | Annexin A2 | |
| CH60_HUMAN | 0.414 | −0.196 | HSPD1 | 60 kDa chaperonin | |
| COF1_HUMAN | 0.394 | 0.768 | CFL1 | Cofilin 1 | |
| PARK7_HUMAN | 0.296 | −0.394 | PARK7 | Parkinson Disease 7 | |
| HS90A_HUMAN | 0.265 | −0.743 | HSP90AA1 | HSP90 alpha 1 | |
| LDHA_HUMAN | −0.440 | 0.358 | LDHA | Lactate dehydrogenase | |
| POSTN_HUMAN | −0.589 | −0.464 | POSTN | Periostin | |
| LUM_HUMAN | 0.471 | 0.247 | LUM | Lumican | |
| TCPQ_HUMAN | 0.343 | −0.302 | CCT8 | chaperonin containing TCP1, subunit 8 | |
| EMIL1_HUMAN | −0.596 | −0.812 | EMILIN1 | Elastin microfibril interfacer 1 | |
| CO6A2_HUMAN | −0.727 | 0.336 | COL6A2 | Collagen, Type VI, alpha 2 | |
| FMOD_HUMAN | 1.667 | 0.474 | FMOD | Fibromodulin | |
| GRP78_HUMAN | 0.687 | 0.250 | HSPA5 | heat shock 70 kDa protein 5 | |
| MMP1_HUMAN | 0.599 | 0.589 | MMP1 | Matrix metallopeptidase 1 | |
| RINI_HUMAN | 0.544 | −0.213 | RNH1 | Ribonuclease inhibitor 1 | |
| LEG3_HUMAN | 0.485 | 0.171 | LGALS3 | Lectin 3 | |
| A2ML1_HUMAN | 0.386 | 0.245 | A2ML1 | Alpha-2-macroglobulin-like 1 | |
| DESP_HUMAN | −0.701 | 0.594 | DSP | Desmoplakin | |
| DBPA_HUMAN | −1.203 | −0.588 | CSDA | Cold shock domain protein A |
| Pathway | p-Value | Affected Proteins |
|---|---|---|
| Protein Ubiquitination Pathway | 4.17E-7 | PSMA6,PSMA1,HSPD1,HSPA5,HSPA2,PSMD8,HSPA8,PSMD11, SP90B1,PSME1,PSMA5,HSP90AA1,PSMC3,HSPB6,HSPB1 |
| Glycolysis I | 8.02E-6 | ENO1,TPI1,PKM,GAPDH,PFKP |
| EIF2 Signaling | 7.24E-5 | RPL4,EIF3B,RPL5,EIF4A1,RPS8,RPL23A,RPS3,RPS23, RPLP1,RPL13 |
| TCA Cycle II | 1.44E-4 | CS,ACO2,DLST,MDH2 |
| Pentose Phosphate Pathway | 2.55E-4 | PGD,TKT,G6PD |
| Role of IL-17A in Psoriasis | 4.35E-4 | S100A7,S100A9,S100A8 |
| Aldosterone Signaling in Epithelial Cells | 4.75E-4 | HSPA8,HSP90B1,HSP90AA1,HSPD1,HSPA5,HSPA2, HSPB6,HSPB1 |
| Inhibition of Matrix Metalloproteases | 1.15E-3 | HSPG2,TIMP1,MMP2,MMP1 |
| Tryptophan Degradation X | 1.19E-3 | AKR1A1,ALDH1A1,ALDH7A1 |
| Remodeling of Epithelial Adherens Junctions | 1.29E-3 | ARPC2,ARPC3,VCL,ACTN4,ACTN1 |
| Pentose Phosphate Pathway | 1.38E-3 | PGD,G6PD |
| ILK Signaling | 2.05E-3 | CFL1,MYL6,MYH9,VIM,ACTN4,ACTN1,DSP,NACA |
| Epithelial Adherens Junction Signaling | 2.05E-3 | MYL6,ARPC2,MYH9,ARPC3,VCL,ACTN4,ACTN1 |
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hall, E.J.; Brenner, D.J. Cancer risks from diagnostic radiology. Br. J. Radiol. 2008, 81, 362–378. [Google Scholar] [CrossRef]
- Wakeford, R. Radiation in the workplace—A review of studies of the risks of occupational exposure to ionising radiation. J. Radiol. Prot. 2009, 29, A61–A79. [Google Scholar] [CrossRef]
- Chao, N.J. Accidental or intentional exposure to ionizing radiation: Biodosimetry and treatment options. Exp. Hematol. 2007, 35, 24–27. [Google Scholar] [CrossRef]
- Morgan, W.F.; Sowa, M.B. Non-targeted effects of ionizing radiation: Implications for risk assessment and the radiation dose response profile. Health Phys. 2009, 97, 426–432. [Google Scholar] [CrossRef]
- Dauer, L.T.; Brooks, A.L.; Hoel, D.G.; Morgan, W.F.; Stram, D.; Tran, P. Review and evaluation of updated research on the health effects associated with low-dose ionising radiation. Radiat. Prot. Dosim. 2010, 140, 103–136. [Google Scholar] [CrossRef]
- Brenner, D.J.; Sachs, R.K. Estimating radiation-induced cancer risks at very low doses: Rationale for using a linear no-threshold approach. Radiat. Environ. Biophys. 2006, 44, 253–256. [Google Scholar] [CrossRef]
- Brooks, A.L.; Hui, T.E.; Couch, L.A. Very large amounts of radiation are required to produce cancer. Dose Response 2007, 5, 263–274. [Google Scholar] [CrossRef]
- Goldberg, Z.; Rocke, D.M.; Schwietert, C.; Berglund, S.R.; Santana, A.; Jones, A.; Lehmann, J.; Stern, R.; Lu, R.; Hartmann Siantar, C. Human in vivo dose-response to controlled, low-dose low linear energy transfer ionizing radiation exposure. Clin. Cancer Res. 2006, 12, 3723–3729. [Google Scholar] [CrossRef]
- Mezentsev, A.; Amundson, S.A. Global gene expression responses to low- or high-dose radiation in a human three-dimensional tissue model. Radiat. Res. 2011, 175, 677–688. [Google Scholar] [CrossRef]
- Albrecht, H.; Durbin-Johnson, B.; Yunis, R.; Kalanetra, K.M.; Wu, S.; Chen, R.; Stevenson, T.S.; Rocke, D.M. Transcriptional response of ex vivo human skin to ionizing radiation: Comparison between low- and high-dose effects. Radiat. Res. 2012, 177, 69–83. [Google Scholar] [CrossRef]
- Von Neubeck, C.; Shankaran, H.; Karin, N.J.; Kauer, P.M.; Chrisler, W.B.; Wang, X.; Robinson, R.J.; Waters, K.M.; Tilton, S.C.; Sowa, M.B. Cell type-dependent gene transcription profile in a three-dimensional human skin tissue model exposed to low doses of ionizing radiation: Implications for medical exposures. Environ. Mol. Mutagen. 2012, 53, 247–259. [Google Scholar] [CrossRef]
- Leszczynski, D. Radiation proteomics: A brief overview. Proteomics 2014, 14, 481–488. [Google Scholar] [CrossRef]
- Pluder, F.; Barjaktarovic, Z.; Azimzadeh, O.; Mortl, S.; Kramer, A.; Steininger, S.; Sarioglu, H.; Leszczynski, D.; Nylund, R.; Hakanen, A.; et al. Low-dose irradiation causes rapid alterations to the proteome of the human endothelial cell line EA.hy926. Radiat. Environ. Biophys. 2011, 50, 155–166. [Google Scholar] [CrossRef]
- Berglund, S.R.; Santana, A.R.; Li, D.; Rice, R.H.; Rocke, D.M.; Goldberg, Z. Proteomic analysis of low dose arsenic and ionizing radiation exposure on keratinocytes. Proteomics 2009, 9, 1925–1938. [Google Scholar] [CrossRef]
- Bakshi, M.V.; Barjaktarovic, Z.; Azimzadeh, O.; Kempf, S.J.; Merl, J.; Hauck, S.M.; Eriksson, P.; Buratovic, S.; Atkinson, M.J.; Tapio, S. Long-term effects of acute low-dose ionizing radiation on the neonatal mouse heart: A proteomic study. Radiat. Environ. Biophys. 2013, 52, 451–461. [Google Scholar] [CrossRef]
- Yang, F.; Waters, K.M.; Webb-Robertson, B.J.; Sowa, M.B.; von Neubeck, C.; Aldrich, J.T.; Markillie, L.M.; Wirgau, R.M.; Gritsenko, M.A.; Zhao, R.; et al. Quantitative phosphoproteomics identifies filaggrin and other targets of ionizing radiation in a human skin model. Exp. Dermatol. 2012, 21, 352–357. [Google Scholar] [CrossRef]
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294. [Google Scholar] [CrossRef]
- Maiolica, A.; Borsotti, D.; Rappsilber, J. Self-made frits for nanoscale columns in proteomics. Proteomics 2005, 5, 3847–3850. [Google Scholar] [CrossRef]
- Kim, S.; Gupta, N.; Pevzner, P.A. Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res. 2008, 7, 3354–3363. [Google Scholar] [CrossRef]
- Monroe, M.E.; Shaw, J.L.; Daly, D.S.; Adkins, J.N.; Smith, R.D. MASIC: A software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 2008, 32, 215–217. [Google Scholar] [CrossRef]
- Zhang, Y.; Askenazi, M.; Jiang, J.; Luckey, C.J.; Griffen, J.D.; Marto, J.A. A robust error model for iTRAQ quantification reveals divergent signaling between oncogenic FLT3 mutants in acute myeloid leukemia. Mol. Cell. Proteomics 2010, 9, 780–790. [Google Scholar] [CrossRef]
- Pacific Northwest National Laboratory Pan-Omics Research website. Available online: http://Omics.pnl.gov (accessed on 24 July 2014).
- DAVID Bioinformatics Resources 6.7. Available online: http://david.abcc.ncifcrf.gov (accessed on 24 July 2014).
- Romano, S.; Di Pace, A.; Sorrentino, A.; Bisogni, R.; Sivero, L.; Romano, M.F. FK506 binding proteins as targets in anticancer therapy. Anticancer Agents Med. Chem. 2010, 10, 651–656. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Zipori, D. PSF and p54(nrb)/NonO—Multi-functional nuclear proteins. FEBS Lett. 2002, 531, 109–114. [Google Scholar] [CrossRef]
- Ha, K.; Takeda, Y.; Dynan, W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst.) 2011, 10, 252–259. [Google Scholar] [CrossRef]
- Li, S.; Kuhne, W.W.; Kulharya, A.; Hudson, F.Z.; Ha, K.; Cao, Z.; Dynan, W.S. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009, 37, 6746–6753. [Google Scholar] [CrossRef]
- Oldberg, A.; Antonsson, P.; Lindblom, K.; Heinegard, D. A collagen-binding 59-kD protein (fibromodulin) is structurally related to the small interstitial proteoglycans PG-S1 and PG-S2 (decorin). EMBO J. 1989, 8, 2601–2604. [Google Scholar]
- Lee, Y.H.; Schiemann, W.P. Fibromodulin suppresses nuclear factor-kappaB activity by inducing the delayed degradation of IKBA via a JNK-dependent pathway coupled to fibroblast apoptosis. J. Biol. Chem. 2011, 286, 6414–6422. [Google Scholar] [CrossRef]
- Weber, T.J.; Opresko, L.K.; Waisman, D.M.; Newton, G.J.; Quesenberry, R.D.; Bollinger, N.; Moore, R.J.; Smith, R.D. Regulation of the low-dose radiation paracrine-specific anchorage-independent growth response by annexin A2. Radiat. Res. 2009, 172, 96–105. [Google Scholar] [CrossRef]
- Waters, K.M.; Stenoien, D.L.; Sowa, M.B.; von Neubeck, C.; Chrisler, W.B.; Tan, R.; Sontag, R.L.; Weber, T.J. Annexin A2 modulates radiation-sensitive transcriptional programming and cell fate. Radiat. Res. 2013, 179, 53–61. [Google Scholar] [CrossRef]
- Schechtman, D.; Mochly-Rosen, D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene 2001, 20, 6339–6347. [Google Scholar] [CrossRef]
- Mildner, M.; Jin, J.; Eckhart, L.; Kezic, S.; Gruber, F.; Barresi, C.; Stremnitzer, C.; Buchberger, M.; Mlitz, V.; Ballaun, C.; et al. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J. Investig. Dermatol. 2010, 130, 2286–2294. [Google Scholar] [CrossRef]
- Ovaere, P.; Lippens, S.; Vandenabeele, P.; Declercq, W. The emerging roles of serine protease cascades in the epidermis. Trends Biochem. Sci. 2009, 34, 453–463. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Takahashi, H.; Presland, R.B.; Dale, B.A.; Iizuka, H. Translocation of profilaggrin N-terminal domain into keratinocyte nuclei with fragmented DNA in normal human skin and loricrin keratoderma. Lab. Invest. 1998, 78, 1245–1253. [Google Scholar]
- Pearton, D.J.; Dale, B.A.; Presland, R.B. Functional analysis of the profilaggrin N-terminal peptide: Identification of domains that regulate nuclear and cytoplasmic distribution. J. Invest. Dermatol. 2002, 119, 661–669. [Google Scholar] [CrossRef]
- Yang, F.; Stenoien, D.L.; Strittmatter, E.F.; Wang, J.; Ding, L.; Lipton, M.S.; Monroe, M.E.; Nicora, C.D.; Gritsenko, M.A.; Tang, K.; et al. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J. Proteome Res. 2006, 5, 1252–1260. [Google Scholar] [CrossRef]
- Yang, F.; Waters, K.M.; Miller, J.H.; Gritsenko, M.A.; Zhao, R.; Du, X.; Livesay, E.A.; Purvine, S.O.; Monroe, M.E.; Wang, Y.; et al. Phosphoproteomics profiling of human skin fibroblast cells reveals pathways and proteins affected by low doses of ionizing radiation. PLoS One 2010, 5, e14152. [Google Scholar] [CrossRef]
- Zhang, Q.; Dai, T.; Zhang, L.; Zhang, M.; Xiao, X.; Hu, H.; Zou, P.; Liu, X.; Xiang, Q.; Su, Z.; et al. Identification of potential biomarkers for predicting acute dermal irritation by proteomic analysis. J. Appl. Toxicol. 2011, 31, 762–772. [Google Scholar] [CrossRef]
- QIAGEN’s Ingenuity Pathway Analysis (IPA). Available online: http://www.qiagen.com/ingenuity (accessed on 24 July 2014).
- Hu, Z.P.; Kim, Y.M.; Sowa, M.B.; Robinson, R.J.; Gao, X.; Metz, T.O.; Morgan, W.F.; Zhang, Q. Metabolomic response of human skin tissue to low dose ionizing radiation. Mol. Biosyst. 2012, 8, 1979–1986. [Google Scholar] [CrossRef]
- Sriharshan, A.; Boldt, K.; Sarioglu, H.; Barjaktarovic, Z.; Azimzadeh, O.; Hieber, L.; Zitzelsberger, H.; Ueffing, M.; Atkinson, M.J.; Tapio, S. Proteomic analysis by SILAC and 2D-DIGE reveals radiation-induced endothelial response: Four key pathways. J. Proteomics 2012, 75, 2319–2330. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Z.; Zhang, X.; Qiao, Y.; Wu, S.; Dong, F.; Chen, Y. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat. Prot. Dosim. 2013, 154, 9–17. [Google Scholar] [CrossRef]
- Bantscheff, M.; Boesche, M.; Eberhard, D.; Matthieson, T.; Sweetman, G.; Kuster, B. Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 2008, 7, 1702–1713. [Google Scholar] [CrossRef]
- Wenger, C.D.; Lee, M.V.; Hebert, A.S.; McAlister, G.C.; Phanstiel, D.H.; Westphall, M.S.; Coon, J.J. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 2011, 8, 933–935. [Google Scholar] [CrossRef]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of ionizing radiation on biological molecules-mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef]
- Tapio, S. Ionizing radiation effects on cells, organelles and tissues on proteome level. Adv. Exp. Med. Biol. 2013, 990, 37–48. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hengel, S.M.; Aldrich, J.T.; Waters, K.M.; Pasa-Tolic, L.; Stenoien, D.L. Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model. Proteomes 2014, 2, 382-398. https://doi.org/10.3390/proteomes2030382
Hengel SM, Aldrich JT, Waters KM, Pasa-Tolic L, Stenoien DL. Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model. Proteomes. 2014; 2(3):382-398. https://doi.org/10.3390/proteomes2030382
Chicago/Turabian StyleHengel, Shawna M., Joshua T. Aldrich, Katrina M. Waters, Ljiljana Pasa-Tolic, and David L. Stenoien. 2014. "Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model" Proteomes 2, no. 3: 382-398. https://doi.org/10.3390/proteomes2030382
APA StyleHengel, S. M., Aldrich, J. T., Waters, K. M., Pasa-Tolic, L., & Stenoien, D. L. (2014). Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model. Proteomes, 2(3), 382-398. https://doi.org/10.3390/proteomes2030382





