Protein Dynamics in the Plant Extracellular Space
Abstract
:1. Introduction
1.1. The Extracellular Space or Apoplast
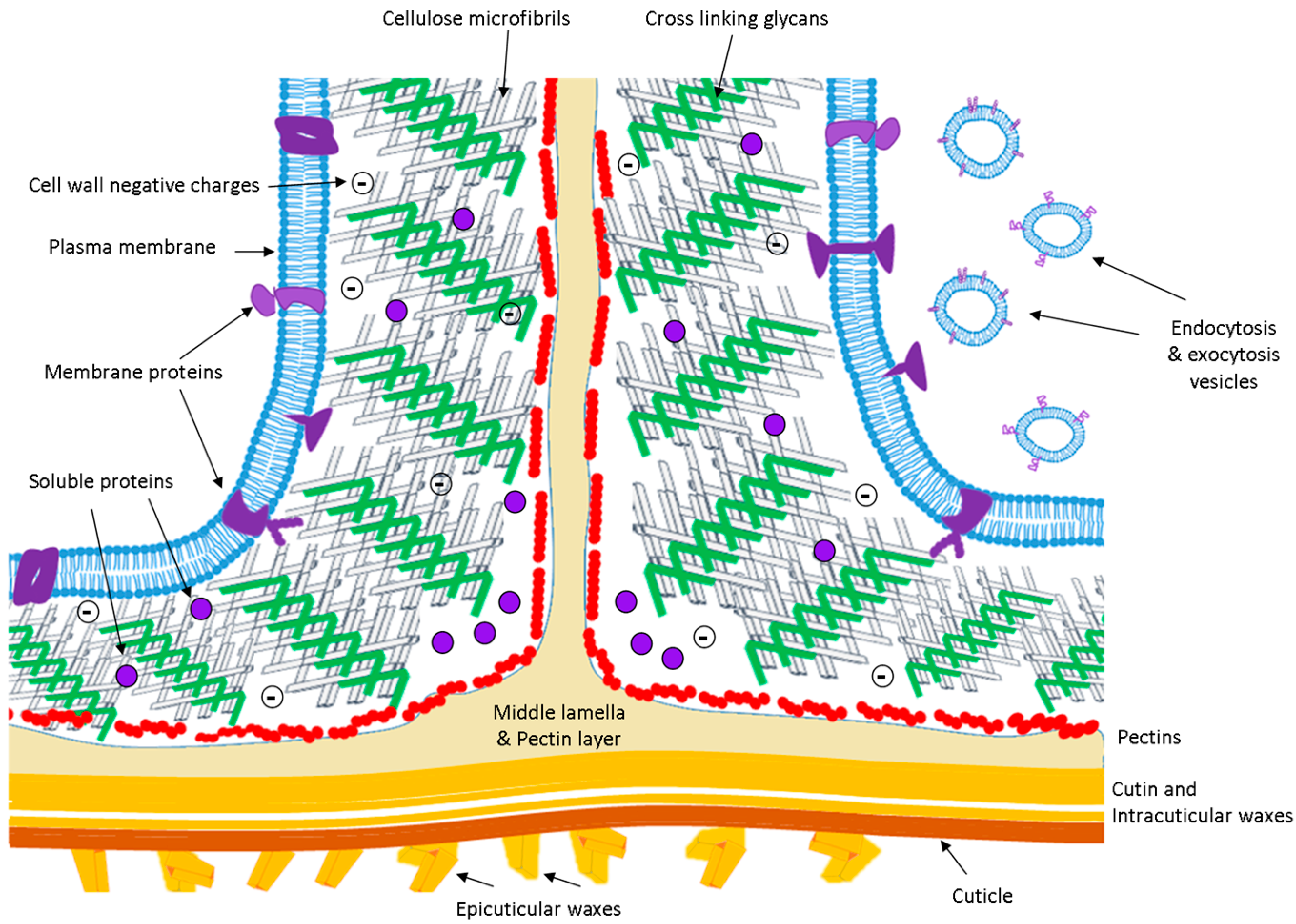
1.2. The Cell Wall
1.3. The Proteins of the APF
1.4. Proteins of the Plasma Membrane Related with the ECS
1.5. A Proteomics Approach to the ECS
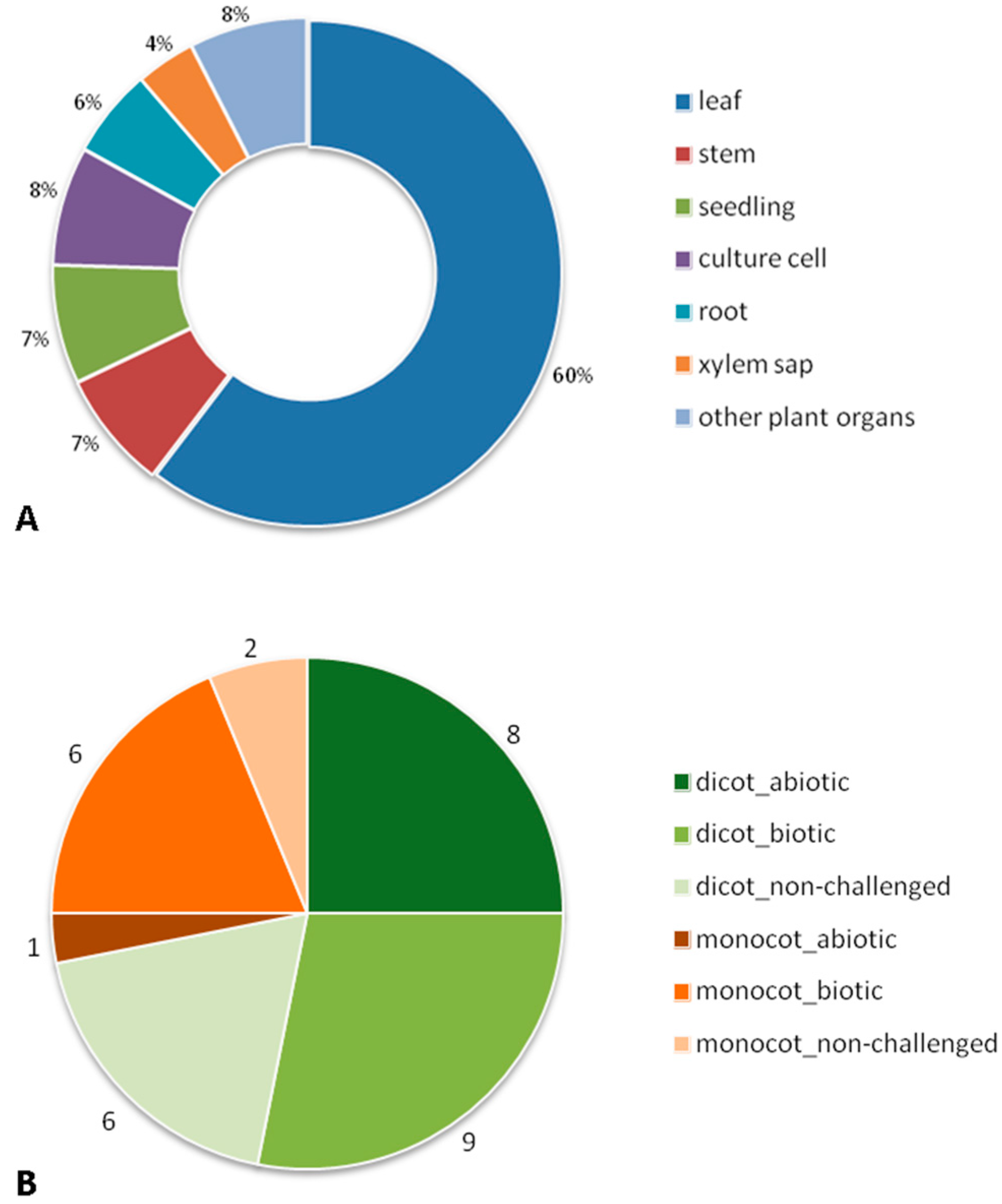
2. Bibliographic Search (Description of the Paper Dataset of the Review)
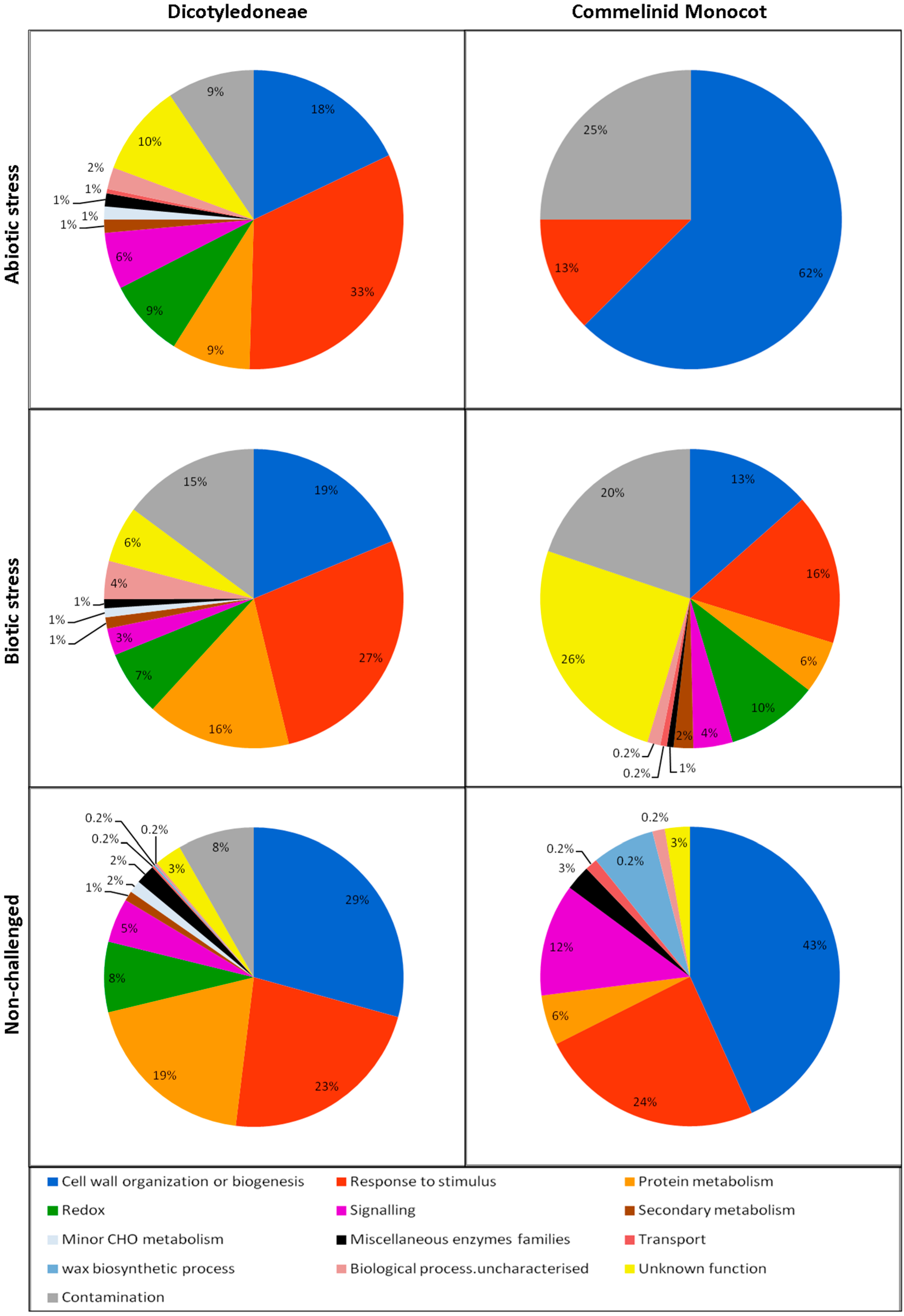
3. The Biological Processes Detected in the ECS
3.1. Cell Wall Organization or Biogenesis
3.2. Response to Stimulus
3.3. Protein Metabolism
3.4. Redox
3.5. Signaling
3.6. Other Metabolic Processes
4. Conclusions and Future Perspectives
- Increase the number of available publications with access to quantitative data, to have a better picture of the plant ECS both in commelinid monocots and dicot plants and in different tissues/organs.
- Despite all of the improvements of 2-DE associated with MS, technical limitations still persist for the detection of certain types of polypeptides. For instance, small apoplastic peptides which have fundamental signaling roles are lost during the common procedures and their detection requires a special 2-DE methodology [115]. Furthermore, woody plants (grapevine, poplar, coffee) present additional problems due to the difficulty in extracting and separating their proteins. Solubilization of the ECS proteins is also a matter of concern since these proteins could be tightly trapped into the extracellular matrix or be adsorbed to cell wall components. Specialist approaches of cell wall treatment could be attempted to solve the problem. On the other hand the use of fractionation techniques of the APF (e.g., by means of affinity columns) could provide additional information on certain types of proteins, isoform level, and regulatory components. Some of these problems of protein separation and analysis can be solved if high-throughput gel-free techniques can be used, which normally implies the availability of complete sequenced genomes.
- Another important point that needs improvement is the validation of the sub-cellular localization of the proteins. The eventual presence of cytoplasmic proteins in the ECS is a situation that should be dealt with care and technical precautions should be taken to reduce possible contamination. However, the presence of the leaderless or non-classically secreted proteins in the apoplast cannot be discarded. For instance, accumulated evidence from several plant species suggests the existence of exosome-like structures that carry and deliver specific proteins to the ECS, with still-undiscovered functions [27,116,117].
- Information stored on MS databases should be improved since proteins are sometimes difficult to identify. Examples of the problems encountered are the heterogeneity in biological function annotation that is a consequence of the complex cellular roles of the proteins; the poor representation of proteins less abundant in the cells; and the frequent lack of information on PTMs, such as glycosylation.
- In order to fully understand all the complexity of the processes that involve the extracellular proteins it would be necessary to build networks of interactions between the diversity of those proteins.
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a Systems Approach to Understanding Plant Cell Walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.; Rose, J. Blueprints for Building plant cell walls. Plant Physiol. 2010, 153, 365. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B.; Gruissen, W.; Jones, R.L. (Eds.) Biochemistry and Molecular Biology of Plants, 2nd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015.
- Lodish, H.; Arnold Berk, S.; Zipursky, L.; Matsudaira, P.; Baltimore, D.; Darnell, J. The Dynamic Plant Cell Wall. In Molecular Cell Biology, 4th ed.; WH Freeman: New York, NY, USA, 2000; Section 22.5. [Google Scholar]
- Keller, B. Structural cell wall proteins. Plant Physiol. 1993, 101, 1127–1130. [Google Scholar] [PubMed]
- Cassab, G.I. Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 281–309. [Google Scholar] [CrossRef] [PubMed]
- Basu, D.; Tian, L.; Wang, W.; Bobbs, S.; Herock, H.; Travers, A.; Showalter, A.M. A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol. 2015, 15, 295. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Basic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Canut, H.; Boudart, G.; Pont-Lezica, R.F. Cell wall proteins: A new insight through proteomics. Trends Plant Sci. 2006, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Brisson, L.F.; Tenhaken, R.; Lamb, C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 1994, 6, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Boudart, G.; Jamet, E.; Rossignol, M.; Lafitte, C.; Borderies, G.; Jauneau, A.; Esquerré-Tugayé, M.-T.; Pont-Lezica, R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 2005, 5, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Klement, Z. Method of obtaining fluid from the intercellular spaces of foliage and the fluid’s merit as substrate for phytobacterial pathogens. Phytopathology 1965, 55, 1033–1034. [Google Scholar]
- Rathmell, W.G.; Sequeira, L. Soluble peroxidase in fluid from the Intercellular spaces of tobacco leaves. Plant Physiol. 1974, 53, 317–318. [Google Scholar] [CrossRef] [PubMed]
- Ridge, I.; Osborn, D.J. Role of peroxidase when hydroxyproline-rich protein in plant cell walls is increased by ethylene. Nat. New Biol. 1971, 229, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Dahal, D.; Pich, A.; Braun, H.P.; Wydra, K. Analysis of cell wall proteins regulated in stem of susceptible and resistant tomato species after inoculation with Ralstonia solanacearum: A proteomic approach. Plant Mol. Biol. 2010, 73, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, C.; Ge, W.; Zhang, Y.; Burlingame, A.L.; Guo, Y. Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE. J. Proteom. 2011, 74, 1045–1067. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Guimarães, L.; Vieira, A.; Chaves, I.; Pinheiro, C.; Queiroz, V.; Renaut, J.; Ricardo, C.P. Effect of greenhouse conditions on the leaf apoplastic proteome of Coffea arabica plants. J. Proteom. 2014, 104, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva, M.D.C.; Cardoso, F.M.H.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pechanova, O.; Hsu, C.-Y.; Adams, J.P.; Pechan, T.; Vandervelde, L.; Drnevich, J.; Jawdy, S.; Adeli, A.; Suttle, J.C.; Lawrence, A.M.; et al. Apoplast proteome reveals that extracellular matrix contributes to multistress response in poplar. BMC Genom. 2010, 11, 674. [Google Scholar] [CrossRef] [PubMed]
- Soares, N.C.; Francisco, R.; Ricardo, C.P.; Jackson, P.A. Proteomics of ionically bound and soluble extracellular proteins in Medicago truncatula leaves. Proteomics 2007, 7, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Li, R.; Pan, J.; Ding, Z.; Lin, J. Endocytosis and its regulation in plants. Trends Plant Sci. 2015, 20, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, J.; Wang, J.; Stierhof, Y.-D.; Robinson, D.G.; Jiang, L. Unconventional protein secretion. Trends Plant Sci. 2012, 17, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Drakakaki, G.; Dandekar, A. Protein secretion: How many secretory routes does a plant cell have? Plant Sci. 2013, 203–204, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Robinson, D.G.; Jiang, L. Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 2014, 29, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ding, Y.; Wang, J.; Kung, C.-H.; Zhuang, X.; Yin, Z.; Xia, Y.; Robinson, D.G.; Shen, J.; Lin, H.-X.; et al. EXPO and Autophagosomes are Distinct Organelles in Plants. Plant Physiol. 2015, 169, 1917–1932. [Google Scholar] [CrossRef] [PubMed]
- San Clemente, H.; Jamet, E. WallProtDB, a database resource for plant cell wall proteomics. Plant Methods 2015, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Fernandez, A.G.; Fujita, M.; Himmelspach, R.; Borner, G.H.H.; Schindelman, G.; Song, S.; Baskin, T.I.; Dupree, P.; Wasteneys, G.O.; et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 2005, 17, 1749–1763. [Google Scholar] [CrossRef] [PubMed]
- Borner, G.H.H.; Lilley, K.S.; Stevens, T.J.; Dupree, P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003, 132, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Leiber, R.-M.; John, F.; Verhertbruggen, Y.; Diet, A.; Knox, J.P.; Ringli, C. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 2010, 22, 1898–1908. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C. Monitoring the outside: Cell wall-sensing mechanisms. Plant Physiol. 2010, 153, 1445–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harpaz-Saad, S.; McFarlane, H.E.; Xu, S.; Divi, U.K.; Forward, B.; Western, T.L.; Kieber, J.J. Cellulose synthesis via the FEI2 RLK/SOS5 pathway and CELLULOSE SYNTHASE 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J. 2011, 68, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in Action: Lectin Receptor-Like Kinases in Plant Development and Stress Responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Alexandersson, E.; Sandin, M.; Resjo, S.; Lenman, M.; Hedley, P.; Levander, F.; Andreasson, E. Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions. BMC Genom. 2014, 15, 497. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, T.; Weighill, D.; Proux-Wéra, E.; Levander, F.; Resjö, S.; Burra, D.D.; Moushib, L.I.; Hedley, P.E.; Liljeroth, E.; Jacobson, D.; et al. Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genom. 2014, 15, 315. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, N.; Hernandez, M.; Casado-Vela, J.; Bru, R.; Elortza, F.; Hedden, P.; Olmos, E. Changes to the proteome and targeted metabolites of xylem sap in Brassica oleracea in response to salt stress. Plant Cell Environ. 2011, 34, 821–836. [Google Scholar] [CrossRef] [PubMed]
- Floerl, S.; Majcherczyk, A.; Possienke, M.; Feussner, K.; Tappe, H.; Gatz, C.; Feussner, I.; Kües, U.; Polle, A. Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana. PLoS ONE 2012, 7, e31435. [Google Scholar] [CrossRef] [PubMed]
- Fuhrs, H.; Gotze, S.; Specht, A.; Erban, A.; Gallien, S.; Heintz, D.; Van Dorsselaer, A.; Kopka, J.; Braun, H.-P.; Horst, W.J. Characterization of leaf apoplastic peroxidases and metabolites in Vigna unguiculata in response to toxic manganese supply and silicon. J. Exp. Bot. 2009, 60, 1663–1678. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [PubMed]
- Novo, J.V.J. Scientific standards and MIAPEs in plant proteomics research and publications. Front. Plant Sci. 2015, 6, 473. [Google Scholar]
- Minic, Z.; Jamet, E.; Négroni, L.; Arsene Der Garabedian, P.; Zivy, M.; Jouanin, L. A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 2007, 58, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that “cut and paste” plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [PubMed]
- Cavalier, D.M.; Lerouxel, O.; Neumetzler, L.; Yamauchi, K.; Reinecke, A.; Freshour, G.; Zabotina, O.A.; Hahn, M.G.; Burgert, I.; Pauly, M.; et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 2008, 20, 1519–1537. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Update on Cell Growth Cell Wall Loosening by Expansins 1. Plant Physiol. 1998, 118, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. A Revised Architecture of Primary Cell Walls Based on Biomechanical Changes Induced by Substrate-Specific Endoglucanases. Plant Physiol. 2012, 158, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Kalunke, R.M.; Tundo, S.; Benedetti, M.; Cervone, F.; De Lorenzo, G.; D’Ovidio, R. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 2015, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef]
- Battaglia, M.; Cuellar-ortiz, S.M.; Battaglia, M.; Solórzano, R.M.; Hernández, M.; Blanca, S.C.; Márquez, G.J.; Covarrubias, A.A. Proline-rich cell wall proteins accumulate in growing regions and phloem tissue in response to water deficit in common bean seedlings. Planta 2007, 5, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; Rubio, M.; Mesonero, V.; Periago, P.M.; Ros Barceló, A.; Martínez-Gómez, P.; Hernández, J.A. The apoplastic antioxidant system in Prunus: Response to long-term plum pox virus infection. J. Exp. Bot. 2006, 57, 3813–3824. [Google Scholar] [CrossRef] [PubMed]
- Szabó, E.; Szatmári, Á.; Hunyadi-Gulyás, É.; Besenyei, E.; Zsiros, L.R.; Bozsó, Z.; Ott, P.G. Changes in apoplast protein pattern suggest an early role of cell wall structure remodelling in flagellin-triggered basal immunity. Biol. Plant. 2012, 56, 551–559. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Vieira, A.; Chaves, I.; Queiroz, V.; Pinheiro, C.; Renaut, J.; Silva, L.; Zambolim, L.; Ricardo, C.; Silva, M.C. Integrated Cytologic and Proteomic Analysis of Coffea arabica—Hemileia vastatrix Interactions. In Proceedings of 24th International Conference on Coffee Science (ASIC), San Jose, Costa Rica, 11–16 November 2012; pp. 1414–1418.
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jordá, L.; Maldonado-Alconada, A.M.; Sarioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contrasting Roles of the Apoplastic Aspartyl Protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis Systemic Acquired Resistance. Plant Physiol. 2014, 165, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Petriccione, M.; Salzano, A.M.; Di Cecco, I.; Scaloni, A.; Scortichini, M. Proteomic analysis of the Actinidia deliciosa leaf apoplast during biotrophic colonization by Pseudomonas syringae pv. actinidiae. J. Proteom. 2014, 101, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Várzea, V.; Guerra-guimarães, L.; Azinheira, H.G.; Fernandez, D.; Petitot, A.-S.; Bertrand, B.; Lashermes, P.; Nicole, M. Coffee resistance to the main diseases: Leaf rust and coffee berry disease. Braz. J. Plant Pgysiol. 2006, 18, 119–147. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Silva, M.C.; Struck, C.; Loureiro, A.; Nicole, M.; Rodrigues, C.J.; Ricardo, C.P.P. Chitinases of Coffea arabica genotypes resistant to orange rust Hemileia vastatrix. Biol. Plant. 2009, 53, 702–706. [Google Scholar] [CrossRef]
- Leah, R.; Tommerup, H.; Svendsen, I.; Mundy, J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 1991, 266, 1564–1573. [Google Scholar] [PubMed]
- Mauch, F.; Mauch-Mani, B.; Boller, T. Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988, 88, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Maher, E.A.; Masoud, S.; Dixon, R.A.; Lamb, C.J. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Nat. Biotechnol. 1994, 12, 807–812. [Google Scholar] [CrossRef]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Proost, P.; Swennen, R.L.; Van Damme, E.J.M. The abundant class III chitinase homolog in young developing banana fruits behaves as a transient vegetative storage protein and most probably serves as an important supply of amino acids for the synthesis of ripening-associated proteins. Plant Physiol. 2002, 130, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Grenier, J.; Potvin, C.; Trudel, J.; Asselin, A. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 1999, 19, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.K.; Selitrennikoff, C.P. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J. Gen. Microbiol. 1990, 136, 1771–1778. [Google Scholar] [CrossRef]
- Godfrey, D.; Able, A.J.; Dry, I.B. Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: A possible role in defense? Mol. Plant-Microbe Interact. 2007, 20, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M.; Gibbings, J.G.; Mahmood, T.; Naqvi, S.M.S. Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 2008, 27, 242–375. [Google Scholar] [CrossRef]
- Rietz, S.; Bernsdorff, F.E.M.; Cai, D. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 2012, 63, 5507–5519. [Google Scholar] [CrossRef] [PubMed]
- Goulet, C.; Goulet, C.; Goulet, M.; Michaud, D. 2-DE proteome maps for the leaf apoplast of Nicotiana benthamiana. Proteomics 2010, 10, 2536–2544. [Google Scholar] [CrossRef] [PubMed]
- Delaunois, B.; Colby, T.; Belloy, N.; Conreux, A.; Harzen, A.; Baillieul, F.; Clément, C.; Schmidt, J.; Jeandet, P.; Cordelier, S. Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biol. 2013, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Trentin, A.R.; Pivato, M.; Mehdi, S.M.M.; Barnabas, L.E.; Giaretta, S.; Fabrega-Prats, M.; Prasad, D.; Arrigoni, G.; Masi, A. Proteome readjustments in the apoplastic space of Arabidopsis thaliana ggt1 mutant leaves exposed to UV-B radiation. Front. Plant Sci. 2015, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H.; Shade, R.E.; Zhu-salzman, K.; Paino, M.; Urzo, D.; Murdock, L.L.; Bressan, R.A.; Hisashi, P.M. A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Lett. 2000, 471, 67–70. [Google Scholar] [CrossRef]
- van der Linde, K.; Hemetsberger, C.; Kastner, C.; Kaschani, F.; van der Hoorn, R.A.L.; Kumlehn, J.; Doehlemann, G. A Maize Cystatin Suppresses Host Immunity by Inhibiting Apoplastic Cysteine Proteases. Plant Cell 2012, 24, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, W.; Wang, W.; Li, F.; Wang, Q.; Xu, Y.; Wang, S. Overexpression of a cysteine proteinase inhibitor gene from Jatropha curcas confers enhanced tolerance to salinity stress. Electron. J. Biotechnol. 2015, 18, 368–375. [Google Scholar] [CrossRef]
- Falco, M.C.; Silva-Filho, M.C. Expression of soybean proteinase inhibitors in transgenic sugarcane plants: Effects on natural defense against Diatraea saccharalis. Plant Physiol. Biochem. 2003, 41, 761–766. [Google Scholar] [CrossRef]
- Lee, J.; Feng, J.; Campbell, K.B.; Scheffler, B.E.; Garrett, W.M.; Thibivilliers, S.; Stacey, G.; Naiman, D.Q.; Tucker, M.L.; Pastor-Corrales, M.A.; et al. Quantitative proteomic analysis of bean plants infected by a virulent and avirulent obligate rust fungus. Mol. Cell. Proteom. 2009, 8, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Delaunois, B.; Jeandet, P.; Clément, C.; Baillieul, F.; Dorey, S.; Cordelier, S. Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant Sci. 2014, 5, 249. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Merops database. Available online: https://merops.sanger.ac.uk (accessed on 12 March 2016).
- Rawlings, N.D. Protease families, evolution and mechanism of action. In Proteases: Structure and Function; Brix, K., Stöcker, W., Eds.; Springer-Verlag: Vienna, Austria, 2013; pp. 1–36. [Google Scholar]
- Van der Hoorn, R.A.L. Determining the fate of proteins. Int. Innov. 2014, 141, 22–24. [Google Scholar]
- Vartapetian, A.B.; Tuzhikov, A.I.; Chichkova, N.V.; Taliansky, M.; Wolpert, T.J. A plant alternative to animal caspases: Subtilisin-like proteases. Cell Death Differ. 2011, 18, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M.A. Subtilisin-like proteases in planta pathogen recognition and immune priming: A perspective. Front. Plant Sci. 2014, 5, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Bondili, J.S.; Schoberer, J.; Svoboda, B.; Liebminger, E.; Glossl, J.; Altmann, F.; Steinkellner, H.; Mach, L. Enzymatic properties and subcellular localization of Arabidopsis beta-N-acetylhexosaminidases. Plant Physiol. 2007, 145, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Liebminger, E.; Veit, C.; Pabst, M.; Batoux, M.; Zipfel, C.; Altmann, F.; Mach, L.; Strasser, R. Beta-N-acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 10793–10802. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Meli, V.S.; Kumar, A.; Thakur, A.; Chakraborty, N.; Chakraborty, S.; Datta, A. The N-glycan processing enzymes α-mannosidase and β-d-N-acetylhexosaminidase are involved in ripening-associated softening in the non-climacteric fruits of capsicum. J. Exp. Bot. 2011, 62, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox signaling in plants. Antioxid. Redox Signal. 2013, 18, 2087–2090. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, E.; Marmey, P.; Jalloul, A.; Etienne, H.; Nicole, M. Molecular Analysis of Class III Peroxidases from Cotton. J. Cotton Sci. 2006, 10, 53–60. [Google Scholar]
- Luthje, S.; Meisrimler, C.-N.; Hopff, D.; Moller, B. Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry 2011, 72, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Lazzarotto, F.; Turchetto-Zolet, A.C.; Margis-Pinheiro, M. Revisiting the Non-Animal Peroxidase Superfamily. Trends Plant Sci. 2015, 20, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Guerra-Guimarães, L.; Loureiro, A.; Nicole, M.R. Involvement of peroxidases in the coffee resistance to orange rust (Hemileia vastatrix). Physiol. Mol. Plant Pathol. 2008, 72, 29–38. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Cardoso, S.; Martins, I.; Loureiro, A.; Bernardes, A.S.; Varzea, V.; Silva, M.C. Differential Induction of Superoxide Dismutase in Coffea arabica—Hemileia vastatrix Interactions. In Proceedings of the 22th International Conference on Coffee Science (ASIC), Campinas, Brazil, 14–19 September 2008; pp. 1036–1039.
- Matsubayashi, Y.; Sakagami, Y. Peptide Hormones in Plants. Annu. Rev. Plant Biol. 2006, 57, 649–674. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y. Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 2014, 65, 385–413. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Florance, H.V.; Johns, A.; Smirnoff, N. Ascorbate deficiency influences the leaf cell wall glycoproteome in Arabidopsis thaliana. Plant. Cell Environ. 2015, 38, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Matsushima, N.; Miyashita, H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules 2012, 2, 288–311. [Google Scholar] [CrossRef] [PubMed]
- Eudes, A.; Mouille, G.; Thévenin, J.; Goyallon, A.; Minic, Z.; Jouanin, L. Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Witzel, K.; Shahzad, M.; Matros, A.; Mock, H.-P.; Mühling, K.H. Comparative evaluation of extraction methods for apoplastic proteins from maize leaves. Plant Methods 2011, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Roitsch, T.; González, M. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Roitsch, T.; Balibrea, M.E.; Hofmann, M.; Proels, R.; Sinha, A.K. Extracellular invertase: Key metabolic enzyme and PR protein. J. Exp. Bot. 2003, 54, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.; Pavkov-Keller, T.; Steiner, B.; Dordic, A.; Gutmann, A.; Nidetzky, B.; Sensen, C.W.; van der Graaff, E.; Wallner, S.; Gruber, K.; et al. Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Plant Cell Wall Metabolism. J. Biol. Chem. 2015, 290, 18770–18781. [Google Scholar] [CrossRef] [PubMed]
- Trabucco, G.M.; Matos, D.A.; Lee, S.J.; Saathoff, A.J.; Priest, H.D.; Mockler, T.C.; Sarath, G.; Hazen, S.P. Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O-methyltransferase in Brachypodium distachyon. BMC Biotechnol. 2013, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Yan, S.; Tan, R.; Zhang, Z.; Wang, Z.; Chen, J. Characterization and expression of a GDSL-like lipase gene from Brassica napus in Nicotiana benthamiana. Protein J. 2014, 33, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gui, S.; Yang, T.; Walk, T.; Wang, X.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2012, 109, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Kaffarnik, F.A.; Jones, A.M.; Rathjen, J.P.; Peck, S.C. Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant Arabidopsis thaliana. Mol. Cell. Proteom. 2009, 8, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.K.; Yokoyama, R.; Nishitani, K. A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 2005, 46, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Carrilho, D.M.; Duarte, I.C.; Francisco, R.; Ricardo, C.P.P.; Duque-Magalhães, M.C. Discovery of novel plant peptides as strong inhibitors of metalloproteinases. Protein Pept. Lett. 2009, 16, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Regente, M.; Pinedo, M.; Elizalde, M.; de la Canal, L. Apoplastic exosome-like vesicles: A new way of protein secretion in plants? Plant Signal. Behav. 2012, 7, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerra-Guimarães, L.; Pinheiro, C.; Chaves, I.; Barros, D.R.; Ricardo, C.P. Protein Dynamics in the Plant Extracellular Space. Proteomes 2016, 4, 22. https://doi.org/10.3390/proteomes4030022
Guerra-Guimarães L, Pinheiro C, Chaves I, Barros DR, Ricardo CP. Protein Dynamics in the Plant Extracellular Space. Proteomes. 2016; 4(3):22. https://doi.org/10.3390/proteomes4030022
Chicago/Turabian StyleGuerra-Guimarães, Leonor, Carla Pinheiro, Inês Chaves, Danielle R. Barros, and Cândido P. Ricardo. 2016. "Protein Dynamics in the Plant Extracellular Space" Proteomes 4, no. 3: 22. https://doi.org/10.3390/proteomes4030022





