A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda
Abstract
1. Introduction
2. Materials and Methods
2.1. Fruits, Seeds, and Embryos
2.2. Histochemistry
2.2.1. Inclusion in Historesin
2.2.2. Inclusion in Agarose
2.2.3. Double Staining with Naphthol Blue Black and Periodic Acid-Schiff Reagent
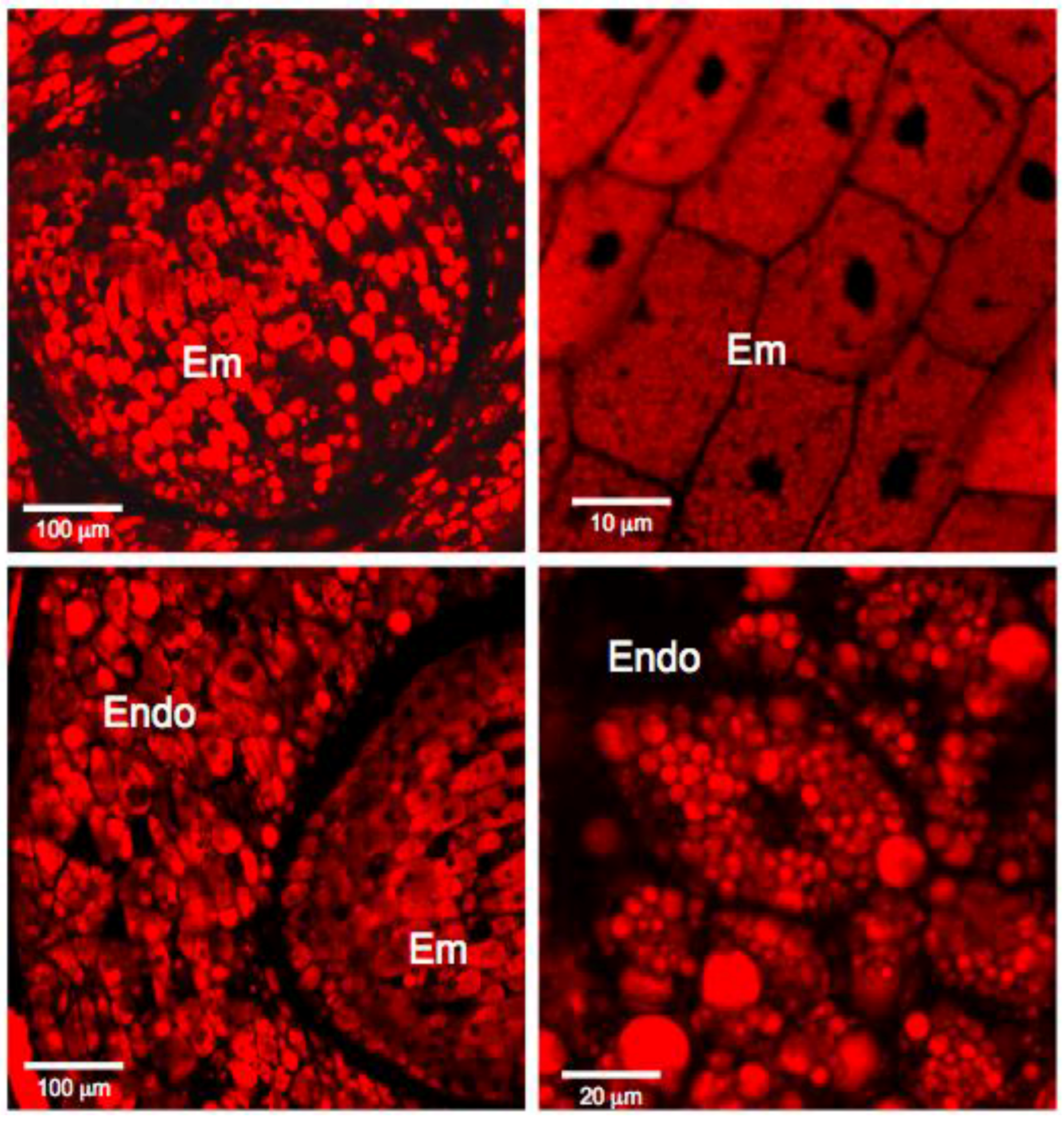
2.2.4. Staining with Nile Red
2.3. Seed Desiccation Tolerance
2.4. Preparation of Protein Extracts
2.5. Shotgun Proteomics
3. Results
3.1. Histochemistry
3.2. Desiccation Tolerance of the Mature Amborella Seeds
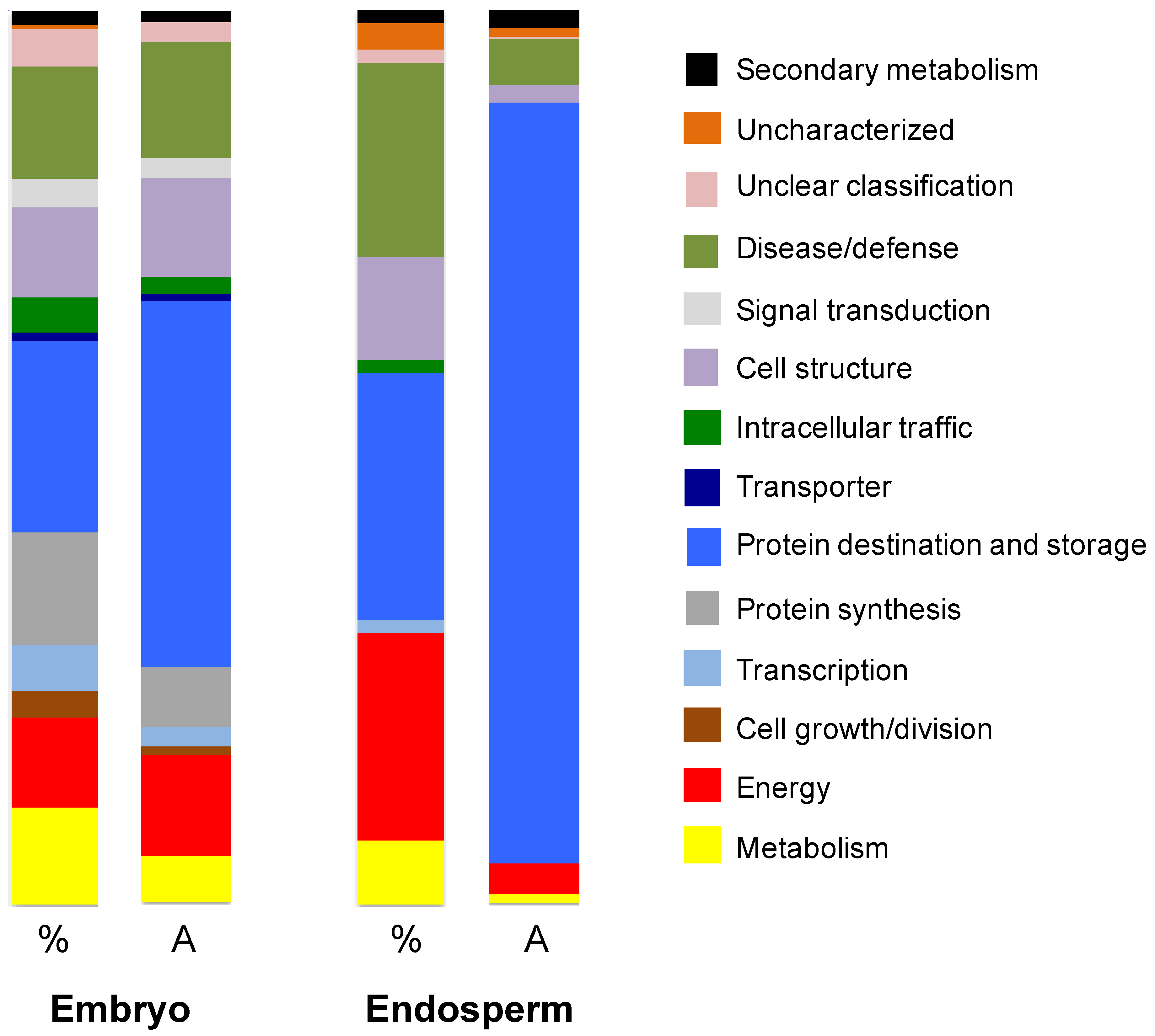
3.3. Characterization of the Amborella Seed Proteins by Shotgun Proteomics
3.3.1. Endosperm Proteins
3.3.2. Embryo Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mathews, S.; Donoghue, M.J. The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science 1999, 286, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, C.L.; Adams, K.L.; Palmer, J.D. Multigene analyses identify the three earliest lineages of extant flowering plants. Curr. Biol. 1999, 9, 1485–1488. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E.; Chase, M.W. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 1999, 402, 402–404. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-L.; Lee, J.; Bernasconi-Quadroni, F.; Soltis, D.E.; Soltis, P.S.; Zanis, M.; Zimmer, E.A.; Chen, Z.; Savolainen, V.; Chase, M.W. The earliest angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature 1999, 402, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, V.; Chase, M.W.; Hoot, S.B.; Morton, C.M.; Soltis, D.E.; Bayer, C.; Fay, M.F.; de Bruijn, A.Y.; Sullivan, S.; Qiu, Y.-L. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst. Biol. 2000, 49, 306–362. [Google Scholar] [CrossRef] [PubMed]
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar]
- Carlquist, S.; Schneider, E.L. Vegetative anatomy of the New Caledonian endemic Amborella trichopoda: Relationships with the Illiciales and implications for vessel origin. Pac. Sci. 2001, 55, 305–312. [Google Scholar] [CrossRef]
- Bobrov, A.V.F.C.; Endress, P.K.; Melikian, A.P.; Romanov, M.S.; Sorokin, A.N.; Bejerano, A.P. Fruit structure of Amborella trichopoda (Amborellaceae). Bot. J. Linn. Soc. 2005, 148, 265–274. [Google Scholar] [CrossRef]
- Tobe, H.; Jaffré, T.; Raven, P.H. Embryology of Amborella (Amborellaceae): Descriptions and polarity of character states. J. Plant Res. 2000, 113, 271–280. [Google Scholar] [CrossRef]
- Floyd, S.K.; Friedman, W.E. Developmental evolution of endosperm in basal angiosperms: Evidence from Amborella (Amborellaceae), Nuphar (Nymphaeaceae), and Illicium (Illiciaceae). Plant Syst. Evol. 2001, 228, 153–169. [Google Scholar] [CrossRef]
- Friedman, W.E. Embryological evidence for developmental lability during early angiosperm evolution. Nature 2006, 441, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Feild, T.S.; Zweiniecki, M.A.; Brodribb, T.; Jaffré, T.; Donoghue, M.J.; Holbrook, M.N. Structure and function of tracheary elements in Amborella trichopoda. Int. J. Plant Sci. 2000, 161, 705–712. [Google Scholar] [CrossRef]
- Poncet, V.; Scutt, C.; Tournebize, R.; Villegente, M.; Cueff, G.; Rajjou, L.; Balliau, T.; Zivy, M.; Fogliani, B.; Job, C.; et al. The Amborella vacuolar processing enzyme family. Front. Plant Sci. 2015, 6, 618. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.V.; Pressman, E.A. Structural study of germination in celery (Apium graveolens L.) seed with emphasis on endosperm breakdown. Planta 1979, 144, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Hepher, A.; Roberts, J.A. The control of seed germination in Trollius ledebouri: A model of seed dormancy. Planta 1985, 166, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Sansberro, P.A.; Rey, H.Y.; Mroginski, L.A.; Collavino, M.M. In Vitro culture of rudimentary embryos of Ilex paraguariensis: Responses to exogenous cytokinins. J. Plant Growth Regul. 1998, 17, 101–105. [Google Scholar] [CrossRef]
- Forbis, T.; Floyd, S.K.; de Queiroz, A. The evolution of embryo size in angiosperms and other seed plants: Implications for the evolution of seed dormancy. Evolution 2002, 56, 2112–2125. [Google Scholar] [CrossRef] [PubMed]
- Vandelook, F.; Bolle, N.; Van Assche, J.A. Seed dormancy and germination of the European Chaerophyllum temulum (Apiaceae), a member of a trans-Atlantic genus. Ann. Bot. 2007, 100, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Alves-Da-Silva, D.; Borghetti, F.; Thompson, K.; Pritchard, H.; Grime, J.P. Underdeveloped embryos and germination in Aristolochia galeata seeds. Plant Biol. 2011, 13, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-T.; Chen, S.-Y.; Tsai, C.-C.; Baskin, J.M.; Baskin, C.C.; Kuo-Huang, L.-L. Deep simple epicotyl morphophysiological dormancy in seeds of two Viburnum species, with special reference to shoot growth and development inside the seed. Ann. Bot. 2011, 108, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Fogliani, B.; Gâteblé, G.; Villegente, M.; Fabre, I.; Klein, N.; Anger, N.; Baskin, C.C.; Scutt, C.P. The morphophysiological dormancy in Amborella trichopoda seeds is a pleisiomorphic trait in angiosperms. Ann. Bot. 2017, 119, 581–590. [Google Scholar] [PubMed]
- Fourcade, F.; Pouteau, R.; Jaffré, T.; Marmey, P. In Situ observations of the basal angiosperm Amborella trichopoda reveal a long fruiting cycle overlapping two annual flowering periods. J. Plant Res. 2015, 128, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Puchtler, H.; Meloan, S.N.; Brewton, B.R. On the history of basic fuchsin and aldehyde-Schiff reactions from 1862 to 1935. Histochemistry 1975, 41, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Mathe, C.; Vieillescazes, C. Compréhension des mécanismes de coloration des liants protéiques picturaux à l’aide du Noir Amide 10B. L’Act. Chim. 2002, 7, 11–14. [Google Scholar]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Fogliani, B.; Klein, N.; Gâteblé, G.; Scutt, C.P. Seed biology and germination of the basal angiosperm Amborella trichopoda. In Proceedings of the Third International Society for Seed Science Meeting on Seeds and the Environment, Seeds and Change, Seed Ecology III, Salt Lake City, UT, USA, 20–24 June 2010; pp. 51–52. [Google Scholar]
- Dussert, S.; Chabrillange, N.; Engelmann, F.; Hamon, S. Quantitative estimation of seed desiccation using a quantal response model: Application to nine species of the genus Coffea L. Seed Sci. Res. 1999, 9, 135–144. [Google Scholar]
- Rajjou, L.; Lovigny, Y.; Groot, S.P.C.; Belghazi, M.; Job, C.; Job, D. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008, 148, 620–641. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bjornson, R.D.; Carriero, N.J.; Colangelo, C.; Shifman, M.; Cheung, K.H.; Miller, P.L.; Williams, K. X!!Tandem, an improved method for running Xtandem in parallel on collections of commodity computers. J. Proteome Res. 2008, 7, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Langella, O.; Valot, B.; Balliau, T.; Blein-Nicolas, M.; Bonhomme, L.; Zivy, M. X!TandemPipeline: A tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res. 2017, 16, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.; Bancroft, I.; Bent, E.; Love, K.; Goodman, H.; Dean, C.; Bergkamp, R.; Dirkse, W.; Van Staveren, M.; Stiekema, W.; et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 1998, 391, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, G.I.; Mundy, J.; Tzen, J.T. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2001, 112, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Poxleitner, M.; Rogers, S.W.; Lacey, S.A.; Browse, J.; Rogers, J.C. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 2006, 47, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed]
- Friedman, W.E. Hydatellaceae are water lilies with gymnospermous tendencies. Nature 2008, 453, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Rudall, P.J.; Eldridge, T.; Tratt, J.; Ramsay, M.M.; Tuckett, R.E.; Smith, S.Y.; Collinson, M.E.; Remizowa, M.V.; Sokoloff, D.D. Seed fertilization, development, and germination in Hydatellaceae (Nymphaeales): Implications for endosperm evolution in early angiosperms. Am. J. Bot. 2009, 96, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Tuckett, R.E.; Merritt, D.J.; Rudall, P.J.; Hay, F.; Hopper, S.D.; Baskin, C.C.; Baskin, J.M.; Tratt, J.; Dixon, K.W. A new type of specialized morphophysiological dormancy and seed storage behaviour in Hydatellaceae, an early-divergent angiosperm family. Ann. Bot. 2010, 105, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Kermode, A.R.; Finch-Savage, W.E. Desiccation sensitivity in orthodox and recalcitrant seeds in relation to development. In Desiccation and Survival in Plants: Drying Without Dying; Black, M., Pritchard, H.W., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 149–184. [Google Scholar]
- Berjak, P.; Pammenter, N.W. Implications of the lack of desiccation tolerance in recalcitrant seeds. Front. Plant Sci. 2013, 4, 478. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Bih, F.Y.; Learn, G.H.; Ting, J.T.L.; Sellers, C.; Huang, A.H.C. Oleosins in the gametophytes of Pinus and Brassica and their phylogenetic relationship with those in the sporophytes of various species. Planta 1994, 193, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Leprince, O.; Aelst, A.C.; van Pritchard, H.W.; Murphy, D.J. Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta 1998, 204, 109–119. [Google Scholar] [CrossRef]
- Guilloteau, M.; Laloi, M.; Blais, D.; Crouzillat, D.; Mc Carthy, J. Oil bodies in Theobroma cacao seeds: Cloning and characterization of cDNA encoding the 15.8 and 16.9 kDa oleosins. Plant Sci. 2003, 164, 597–606. [Google Scholar] [CrossRef]
- Bewley, J.D.; Black, M. Seeds: Physiology of Development and Germination; Springer Science & Business Media: Berlin, Germany, 1994; p. 445. [Google Scholar]
- Penfield, S.; Graham, S.; Graham, I.A. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 2005, 33, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Strub, J.-M.; Job, C.; Van Dorsselaer, A.; Job, D. Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. USA 2008, 105, 10262–10267. [Google Scholar] [CrossRef] [PubMed]
- Rylott, E.L.; Hooks, M.A.; Graham, I.A. Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem. Soc. Trans. 2001, 29, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Meinhard, J.; Job, C.; Strub, J.-M.; Fischer, U.; Pestsova, E.; Westhoff, P.; Van Dorsselaer, A.; Job, D. Proteomics reveals potential biomarkers of seed vigor in sugarbeet. Proteomics 2011, 11, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Ravanel, S.; Gakière, B.; Job, D.; Douce, R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 7805–7812. [Google Scholar] [CrossRef] [PubMed]
- Alban, C.; Job, D.; Douce, R. Biotin metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Kagan, R.M.; McFadden, H.J.; McFadden, P.N.; O’Connor, C.; Clarke, S. Molecular phylogenetics of a protein repair methyltransferase. Comp. Biochem. Physiol. 1997, 117B, 379–385. [Google Scholar] [CrossRef]
- Mudgett, M.B.; Lowenson, J.D.; Clarke, S. Protein repair l-isoaspartyl methyltransferase in plants. Phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiol. 1997, 115, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Thapar, N.; Kim, A.-K.; Clarke, S. Distinct patterns of expression but similar biochemical properties of protein L-isoaspartyl methyltransferase in higher plants. Plant Physiol. 2001, 125, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Ogé, L.; Bourdais, G.; Bove, J.; Collet, B.; Godin, B.; Granier, F.; Boutin, J.-P.; Job, D.; Jullien, M.; Grappin, P. Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 2008, 20, 3022–3037. [Google Scholar] [CrossRef] [PubMed]
- Shen-Miller, J.; Mudgett, M.B.; Schopf, J.W.; Clarke, S.; Berger, R. Exceptional seed longevity and robust growth: Ancient sacred lotus from China. Am. J. Bot. 1995, 82, 1367–1380. [Google Scholar] [CrossRef]
- Shen-Miller, J. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res. 2002, 12, 131–143. [Google Scholar] [CrossRef]
- Shen-Miller, J.; Aung, L.H.; Turek, J.; Schopf, J.W.; Tholandi, M.; Yang, M.; Czaja, A. Centuries-old viable fruit of sacred lotus Nelumbo nucifera Gaertn var. China Antique. Trop. Plant Biol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- Job, C.; Rajjou, L.; Lovigny, Y.; Belghazi, M.; Job, D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Kishor, P.B.K.; Sangam, S.; Amrutha, R.N.; Laxmi, P.S.; Naidu, K.R.; Rao, K.R.S.S.; Rao, S.; Reddy, K.J.; Theriappan, P.; Sreenivasulu, N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr. Sci. 2005, 88, 424–438. [Google Scholar]
- Miller, G.; Honig, A.; Stein, H.; Suzuki, N.; Mittler, R.; Zilberstein, A. Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 2009, 284, 26482–26492. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Nakamura, T.; Han, S.Y.; Takabe, T. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol. Biol. 1995, 27, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.M.; Andrade, M.O.; Gomes, A.P.S.; DaMatta, F.M.; Baracat-Pereirea, M.C.; Fontes, E.P.B. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 2006, 57, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Romao, M.J.; Turk, D.; Gomis-Ruth, F.X.; Huber, R.; Schumacher, G.; Mollering, H.; Russmann, L. Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 Å resolution. J. Mol. Biol. 1992, 226, 1111–1130. [Google Scholar] [CrossRef]
- Shintani, D.; DellaPenna, D. Elevating the vitamin E content of plants through metabolic engineering. Science 1998, 282, 2098–2100. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 2004, 14, 93–107. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Huizinga, D.H.; Denton, R.; Koehler, K.G.; Tomasello, A.; Wood, L.; Sen, S.E.; Crowell, D.N. Farnesylcysteine lyase is involved in negative regulation of abscisic acid signaling in Arabidopsis. Mol. Plant 2010, 3, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Galau, G.A.; Hughes, W.D.; Dure, L. Abscisic acid induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol. 1986, 7, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Gaubier, P.; Raynal, M.; Hull, G.; Huestis, G.M.; Grellet, F.; Arenas, C.; Pagès, M.; Delseny, M. Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol. Gen. Genet. 1993, 238, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 1993, 101, 1119–1120. [Google Scholar] [CrossRef] [PubMed]
- Manfre, A.J.; Lanni, L.M.; Marcotte, W.R.J. The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 Is required for normal seed development. Plant Physiol. 2006, 140, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Kalemba, E.M.; Pukacka, S. Possible roles of LEA proteins and sHSPs in seed protection: A short review. Biol. Lett. 2007, 44, 3–16. [Google Scholar]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, K.L.; Giese, K.C.; Buan, N.R.; Vierling, E. Interactions between small heat shock protein subunits and substrate in small heat shock protein-substrate complexes. J. Biol. Chem. 2004, 279, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Dapena, P.; Castano, R.; Almoguera, C.; Jordano, J. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol. 2006, 142, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Su, P.-H.; Li, H.-M. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ.-Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Berjak, P.; Pammenter, N.W. From Avicennia to Zizania: Seed recalcitrance in perspective. Ann. Bot. 2008, 101, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Von Teichman, I.; van Wyk, A.E. Structural aspects and trends in the evolution of recalcitrant seeds in dicotyledons. Seed Sci. Res. 1994, 4, 225–239. [Google Scholar] [CrossRef]
- Farnsworth, E. The ecology and physiology of viviparous and recalcitrant seeds. Annu. Rev. Ecol. Evol. Syst. 2000, 31, 107–138. [Google Scholar] [CrossRef]
- Dickie, J.B.; Pritchard, H.W. Systematic and evolutionary aspects of desiccation tolerance in seeds. In Desiccation and Plant Survival; Black, M., Pritchard, H.W., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 239–262. [Google Scholar]
- Tweddle, J.C.; Dickie, J.B.; Baskin, C.C.; Baskin, J.M. Ecological aspects of seed desiccation sensitivity. J. Ecol. 2003, 91, 294–304. [Google Scholar] [CrossRef]
- González-Morales, S.A.; Chávez-Montes, R.A.; Hayano-Kanashiro, C.; Alejo-Jacuinde, G.; Rico-Cambron, T.Y.; de Folter, S.; Herrera-Estrella, L. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, E5232–E5241. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.M.; Moore, J.P. Programming desiccation-tolerance: From plants to seeds to resurrection plants. Curr. Opin. Plant Biol 2011, 14, 340–345. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villegente, M.; Marmey, P.; Job, C.; Galland, M.; Cueff, G.; Godin, B.; Rajjou, L.; Balliau, T.; Zivy, M.; Fogliani, B.; et al. A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda. Proteomes 2017, 5, 19. https://doi.org/10.3390/proteomes5030019
Villegente M, Marmey P, Job C, Galland M, Cueff G, Godin B, Rajjou L, Balliau T, Zivy M, Fogliani B, et al. A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda. Proteomes. 2017; 5(3):19. https://doi.org/10.3390/proteomes5030019
Chicago/Turabian StyleVillegente, Matthieu, Philippe Marmey, Claudette Job, Marc Galland, Gwendal Cueff, Béatrice Godin, Loïc Rajjou, Thierry Balliau, Michel Zivy, Bruno Fogliani, and et al. 2017. "A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda" Proteomes 5, no. 3: 19. https://doi.org/10.3390/proteomes5030019
APA StyleVillegente, M., Marmey, P., Job, C., Galland, M., Cueff, G., Godin, B., Rajjou, L., Balliau, T., Zivy, M., Fogliani, B., Sarramegna-Burtet, V., & Job, D. (2017). A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda. Proteomes, 5(3), 19. https://doi.org/10.3390/proteomes5030019






