Calreticulin as A Novel Potential Metastasis-Associated Protein in Myxoid Liposarcoma, as Revealed by Two-Dimensional Difference Gel Electrophoresis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Protein Expression Profiling
2.3. Protein Identification
2.4. Western Blotting
2.5. Immunohistochemistry
2.6. Gene Silencing Assay
2.7. Cell Proliferation Assay
2.8. Cell Invasion Assay
3. Results
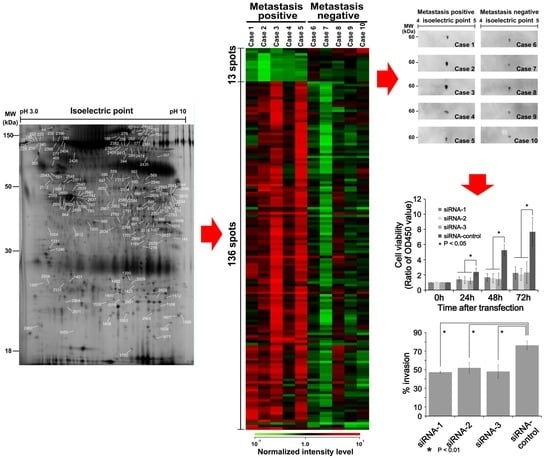
3.1. Protein Expression Profiles of MLS Cells Using 2D-DIGE
3.2. Comparative Study between Sample Groups with Different Metastatic Statuses
3.3. Calreticulin Expression was Associated with Metastasis
3.4. Immunohistochemical Localization of Calreticulin in Tumor Cells
3.5. In Vitro Functional Analysis of Calreticulin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fletcher, C.D.M.; Bridge, J.A.; Hogendoorn, P.; Mertens, F. WHO Classification of Tumours of Soft Tissue and Bone, 4th ed.; WHO Press: Geneva, Switzerland, 2013. [Google Scholar]
- Grosso, F.; Jones, R.L.; Demetri, G.D.; Judson, I.R.; Blay, J.Y.; Le Cesne, A.; Sanfilippo, R.; Casieri, P.; Collini, P.; Dileo, P.; et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: A retrospective study. Lancet Oncol. 2007, 8, 595–602. [Google Scholar] [CrossRef]
- Forni, C.; Minuzzo, M.; Virdis, E.; Tamborini, E.; Simone, M.; Tavecchio, M.; Erba, E.; Grosso, F.; Gronchi, A.; Aman, P.; et al. Trabectedin (ET-743) promotes differentiation in myxoid liposarcoma tumors. Mol. Cancer Ther. 2009, 8, 449–457. [Google Scholar] [CrossRef] [Green Version]
- Ueda, T.; Kakunaga, S.; Ando, M.; Yonemori, K.; Sugiura, H.; Yamada, K.; Kawai, A. Phase I and pharmacokinetic study of trabectedin, a DNA minor groove binder, administered as a 24-h continuous infusion in Japanese patients with soft tissue sarcoma. Investig. New Drugs 2014, 32, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Di Giandomenico, S.; Frapolli, R.; Bello, E.; Uboldi, S.; Licandro, S.A.; Marchini, S.; Beltrame, L.; Brich, S.; Mauro, V.; Tamborini, E.; et al. Mode of action of trabectedin in myxoid liposarcomas. Oncogene 2014, 33, 5201–5210. [Google Scholar] [CrossRef]
- Graadt van Roggen, J.F.; Hogendoorn, P.C.; Fletcher, C.D. Myxoid tumours of soft tissue. Histopathology 1999, 35, 291–312. [Google Scholar] [CrossRef]
- Demicco, E.G.; Torres, K.E.; Ghadimi, M.P.; Colombo, C.; Bolshakov, S.; Hoffman, A.; Peng, T.; Bovee, J.V.; Wang, W.L.; Lev, D.; et al. Involvement of the PI3K/Akt pathway in myxoid/round cell liposarcoma. Mod. Pathol. 2012, 25, 212–221. [Google Scholar] [CrossRef]
- Barretina, J.; Taylor, B.S.; Banerji, S.; Ramos, A.H.; Lagos-Quintana, M.; Decarolis, P.L.; Shah, K.; Socci, N.D.; Weir, B.A.; Ho, A.; et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010, 42, 715–721. [Google Scholar] [CrossRef]
- Negri, T.; Virdis, E.; Brich, S.; Bozzi, F.; Tamborini, E.; Tarantino, E.; Jocolle, G.; Cassinelli, G.; Grosso, F.; Sanfilippo, R.; et al. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clin. Cancer Res. 2010, 16, 3581–3593. [Google Scholar] [CrossRef]
- Petricoin, E.F.; Ardekani, A.M.; Hitt, B.A.; Levine, P.J.; Fusaro, V.A.; Steinberg, S.M.; Mills, G.B.; Simone, C.; Fishman, D.A.; Kohn, E.C.; et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002, 359, 572–577. [Google Scholar] [CrossRef]
- Kondo, T. Inconvenient truth: Cancer biomarker development by using proteomics. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 861–865. [Google Scholar] [CrossRef]
- Kondo, T.; Hirohashi, S. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat. Protoc. 2007, 1, 2940–2956. [Google Scholar] [CrossRef]
- Thelin-Jarnum, S.; Lassen, C.; Panagopoulos, I.; Mandahl, N.; Aman, P. Identification of genes differentially expressed in TLS-CHOP carrying myxoid liposarcomas. Int. J. Cancer 1999, 83, 30–33. [Google Scholar] [CrossRef] [Green Version]
- Michalak, M.; Corbett, E.F.; Mesaeli, N.; Nakamura, K.; Opas, M. Calreticulin: One protein, one gene, many functions. Biochem. J. 1999, 344, 281–292. [Google Scholar] [CrossRef]
- Zamanian, M.; Veerakumarasivam, A.; Abdullah, S.; Rosli, R. Calreticulin and cancer. Pathol. Oncol. Res. 2013, 19, 149–154. [Google Scholar] [CrossRef]
- Lwin, Z.M.; Guo, C.; Salim, A.; Yip, G.W.; Chew, F.T.; Nan, J.; Thike, A.A.; Tan, P.H.; Bay, B.H. Clinicopathological significance of calreticulin in breast invasive ductal carcinoma. Mod. Pathol. 2010, 23, 1559–1566. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.N.; Chang, C.C.; Su, T.E.; Hsu, W.M.; Jeng, Y.M.; Ho, M.C.; Hsieh, F.J.; Lee, P.H.; Kuo, M.L.; Lee, H.; et al. Identification of calreticulin as a prognosis marker and angiogenic regulator in human gastric cancer. Ann. Surg. Oncol. 2009, 16, 524–533. [Google Scholar] [CrossRef]
- Wu, M.; Bai, X.; Xu, G.; Wei, J.; Zhu, T.; Zhang, Y.; Li, Q.; Liu, P.; Song, A.; Zhao, L.; et al. Proteome analysis of human androgen-independent prostate cancer cell lines: Variable metastatic potentials correlated with vimentin expression. Proteomics 2007, 7, 1973–1983. [Google Scholar] [CrossRef]
- Vaksman, O.; Davidson, B.; Trope, C.; Reich, R. Calreticulin expression is reduced in high-grade ovarian serous carcinoma effusions compared with primary tumors and solid metastases. Hum. Pathol. 2013, 44, 2677–2683. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, C.; Dong, M.; Zhou, J.; Liu, Q.; Dong, Q.; Li, F. Overexpression of calreticulin contributes to the development and progression of pancreatic cancer. J. Cell. Physiol. 2014, 229, 887–897. [Google Scholar] [CrossRef]
- Du, X.L.; Hu, H.; Lin, D.C.; Xia, S.H.; Shen, X.M.; Zhang, Y.; Luo, M.L.; Feng, Y.B.; Cai, Y.; Xu, X.; et al. Proteomic profiling of proteins dysregulted in Chinese esophageal squamous cell carcinoma. J. Mol. Med. 2007, 85, 863–875. [Google Scholar] [CrossRef]
- Kageyama, S.; Isono, T.; Iwaki, H.; Wakabayashi, Y.; Okada, Y.; Kontani, K.; Yoshimura, K.; Terai, A.; Arai, Y.; Yoshiki, T. Identification by proteomic analysis of calreticulin as a marker for bladder cancer and evaluation of the diagnostic accuracy of its detection in urine. Clin. Chem. 2004, 50, 857–866. [Google Scholar] [CrossRef]
- Coppolino, M.; Leung-Hagesteijn, C.; Dedhar, S.; Wilkins, J. Inducible interaction of integrin alpha 2 beta 1 with calreticulin. Dependence on the activation state of the integrin. J. Biol. Chem. 1995, 270, 23132–23138. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chen, C.N.; Chu, C.Y.; Lu, J.; Wang, B.J.; Chen, C.H.; Huang, M.C.; Lin, T.H.; Pan, C.C.; Chen, S.S.; et al. Calreticulin activates beta1 integrin via fucosylation by fucosyltransferase 1 in J82 human bladder cancer cells. Biochem. J. 2014, 460, 69–78. [Google Scholar] [CrossRef]
- Papp, S.; Fadel, M.P.; Kim, H.; McCulloch, C.A.; Opas, M. Calreticulin affects fibronectin-based cell-substratum adhesion via the regulation of c-Src activity. J. Biol. Chem. 2007, 282, 16585–16598. [Google Scholar] [CrossRef]
- Szabo, E.; Papp, S.; Opas, M. Differential calreticulin expression affects focal contacts via the calmodulin/CaMK II pathway. J. Cell. Physiol. 2007, 213, 269–277. [Google Scholar] [CrossRef]
- Opas, M.; Szewczenko-Pawlikowski, M.; Jass, G.K.; Mesaeli, N.; Michalak, M. Calreticulin modulates cell adhesiveness via regulation of vinculin expression. J. Cell Biol. 1996, 135, 1913–1923. [Google Scholar] [CrossRef] [Green Version]
- Shu, Q.; Li, W.; Li, H.; Sun, G. Vasostatin inhibits VEGF-induced endothelial cell proliferation, tube formation and induces cell apoptosis under oxygen deprivation. Int. J. Mol. Sci. 2014, 15, 6019–6030. [Google Scholar] [CrossRef]
- Hisaoka, M.; Matsuyama, A.; Nakamoto, M. Aberrant calreticulin expression is involved in the dedifferentiation of dedifferentiated liposarcoma. Am. J. Pathol. 2012, 180, 2076–2083. [Google Scholar] [CrossRef]
- Chiang, W.F.; Hwang, T.Z.; Hour, T.C.; Wang, L.H.; Chiu, C.C.; Chen, H.R.; Wu, Y.J.; Wang, C.C.; Wang, L.F.; Chien, C.Y.; et al. Calreticulin, an endoplasmic reticulum-resident protein, is highly expressed and essential for cell proliferation and migration in oral squamous cell carcinoma. Oral Oncol. 2013, 49, 534–541. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chen, C.N.; Wang, B.; Hsu, W.M.; Chen, S.T.; Chang, K.J.; Chang, C.C.; Lee, H. Changes in tumor growth and metastatic capacities of J82 human bladder cancer cells suppressed by down-regulation of calreticulin expression. Am. J. Pathol. 2011, 179, 1425–1433. [Google Scholar] [CrossRef]
- Kondo, T. Cancer biomarker development and two-dimensional difference gel electrophoresis (2D-DIGE). Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2019, 1867, 2–8. [Google Scholar] [CrossRef]




| Case No. | Gender | Age | Location | Metastasis | Metastasis Duration (month) | Therapy for Advanced Disease | Follow-Up Duration (month) | Follow-Up Status | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | female | 51 | thigh | positive | 19 | CTx 1 and RTx 2 | 21 | DOD 3 | |
| 2 | female | 61 | thigh | positive | 17 | CTx and RTx | 75 | AWD 4 | |
| 3 | Male | 31 | forearm | positive | 65 | CTx | 84 | DOD | |
| 4 | Male | 69 | buttock | positive | 7 | CTx and RTx | 41 | DOD | |
| 5 | Male | 41 | foot | positive | 0 | CTx | 21 | DOD | |
| 6 | Male | 44 | thigh | negative | - | - | 169 | CDF 5 | |
| 7 | female | 52 | inguinal | negative | - | - | 150 | CDF | |
| 8 | female | 35 | lower leg | negative | - | - | 88 | CDF | |
| 9 | Male | 65 | thigh | negative | - | - | 77 | CDF | |
| 10 | female | 47 | thigh | negative | - | - | 68 | CDF | |
| Spots No. | Welch Test p-Value | Ratio of Means | Identified Protein |
|---|---|---|---|
| 44 | 1.31E-02 | 0.42 | Endoplasmin |
| 62 | 4.78E-02 | 0.38 | Keratin, type I cytoskeletal 10 |
| 74 | 3.15E-02 | 0.38 | Keratin, type II cytoskeletal 1 |
| 85 | 9.56E-03 | 0.28 | Trypsin-1 |
| 91 | 2.82E-02 | 0.36 | Trypsin-1 |
| 163 | 4.96E-02 | 0.39 | Keratin, type II cytoskeletal 1 |
| 177 | 4.58E-02 | 0.38 | Keratin, type II cytoskeletal 2 epidermal |
| 182 | 3.49E-02 | 0.36 | Trypsin-1 |
| 185 | 1.46E-02 | 0.36 | Aconitate hydratase, mitochondrial |
| 188 | 2.02E-02 | 0.76 | Ig mu chain C region |
| 216 | 6.69E-03 | 2.19 | Keratin, type II cytoskeletal 2 epidermal |
| 221 | 9.04E-03 | 2.38 | Serum albumin |
| 222 | 8.09E-03 | 2.11 | Serum albumin |
| 223 | 1.11E-02 | 2.01 | Cancer-associated gene 1 protein |
| 228 | 2.84E-02 | 2.03 | Serum albumin |
| 229 | 1.46E-02 | 2.30 | Serum albumin |
| 230 | 6.48E-03 | 2.38 | Serum albumin |
| 232 | 8.57E-03 | 2.02 | Serum albumin |
| 276 | 7.57E-03 | 0.40 | Keratin, type II cytoskeletal 1 |
| 277 | 4.07E-03 | 0.32 | 78 kDa glucose-regulated protein |
| 279 | 4.09E-03 | 0.35 | Trypsin-1 |
| 280 | 1.49E-02 | 0.49 | Glutamine-rich protein 2 |
| 281 | 3.24E-04 | 0.29 | 78 kDa glucose-regulated protein |
| 282 | 3.46E-03 | 0.42 | Keratin, type I cytoskeletal 10 |
| 286 | 2.75E-02 | 0.42 | Trypsin-1 |
| 340 | 2.29E-02 | 0.48 | Calreticulin |
| 344 | 3.01E-02 | 0.47 | Uncharacterized protein KIAA1529 |
| 365 | 1.63E-02 | 0.50 | Alpha-1-antichymotrypsin |
| 397 | 2.92E-02 | 0.40 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial |
| 469 | 2.12E-02 | 0.43 | Keratin, type II cytoskeletal 1 |
| 511 | 1.30E-02 | 0.44 | Vimentin |
| 559 | 2.28E-02 | 0.46 | Keratin, type II cytoskeletal 1 |
| 562 | 1.68E-02 | 0.49 | Trypsin-1 |
| 569 | 1.96E-02 | 0.46 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 590 | 1.48E-02 | 0.44 | Keratin, type I cytoskeletal 10 |
| 598 | 1.82E-02 | 0.44 | Keratin, type II cytoskeletal 1 |
| 647 | 2.21E-02 | 0.45 | Keratin, type I cytoskeletal 10 |
| 680 | 4.46E-02 | 0.47 | Keratin, type II cytoskeletal 1 |
| 694 | 2.21E-02 | 0.44 | Keratin, type II cytoskeletal 1 |
| 702 | 4.46E-02 | 0.45 | Fumarate hydratase, mitochondrial |
| 713 | 3.92E-02 | 0.35 | Serpin H1 |
| 742 | 2.25E-02 | 0.48 | Trypsin-1 |
| 744 | 4.98E-02 | 0.47 | Growth factor receptor-bound protein 1 |
| 762 | 3.23E-02 | 0.45 | Keratin, type II cytoskeletal 1 |
| 787 | 4.99E-02 | 0.48 | Protein Shroom3 |
| 788 | 2.14E-02 | 0.45 | Glutamate receptor, ionotropic kainate 3 |
| 790 | 5.59E-03 | 0.34 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
| 802 | 4.20E-02 | 0.42 | Keratin, type II cytoskeletal 1 |
| 809 | 2.25E-02 | 0.43 | 3-ketoacyl-CoA thiolase, mitochondrial |
| 810 | 1.19E-02 | 0.46 | Cytochrome b-c1 complex subunit 2, mitochondrial |
| 811 | 3.14E-02 | 0.37 | Trypsin-1 |
| 841 | 3.97E-02 | 0.47 | Heterogeneous nuclear ribonucleoprotein D0 |
| 848 | 2.15E-02 | 0.49 | Trypsin-2 |
| 864 | 6.12E-03 | 0.48 | Guanine nucleotide-binding protein G(i), alpha-2 subunit |
| 903 | 9.55E-03 | 0.46 | Keratin, type I cytoskeletal 9 |
| 986 | 3.96E-02 | 0.50 | Annexin A2 |
| 988 | 2.39E-02 | 0.45 | Keratin, type I cytoskeletal 10 |
| 995 | 2.85E-02 | 0.47 | Keratin, type I cytoskeletal 10 |
| 1000 | 4.91E-02 | 0.46 | L-lactate dehydrogenase B chain |
| 1007 | 3.03E-02 | 0.42 | Keratin, type II cytoskeletal 1 |
| 1008 | 3.87E-02 | 0.37 | Keratin, type II cytoskeletal 1 |
| 1015 | 4.07E-02 | 0.38 | Keratin, type I cytoskeletal 10 |
| 1054 | 7.46E-04 | 0.31 | Insulin receptor |
| 1103 | 2.42E-02 | 0.27 | Transmembrane protease, serine 13 |
| 1148 | 1.88E-02 | 0.48 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
| 1169 | 7.59E-03 | 0.36 | Glutamine-rich protein 2 |
| 1216 | 1.34E-02 | 0.44 | Probable ATP-dependent RNA helicase DDX56 |
| 1231 | 3.64E-02 | 0.50 | Tropomyosin alpha-3 chain |
| 1246 | 2.71E-02 | 0.39 | Keratin, type II cytoskeletal 1 |
| 1390 | 5.25E-03 | 0.43 | Zinc finger CCHC domain-containing protein 11 |
| 1397 | 4.35E-02 | 0.33 | Keratin, type II cytoskeletal 2 epidermal |
| 1401 | 4.83E-02 | 0.48 | Keratin, type I cytoskeletal 10 |
| 1421 | 1.16E-02 | 0.48 | Lethal(2) giant larvae protein homolog 2 |
| 1452 | 2.00E-02 | 0.48 | Uncharacterized protein KIAA1529 |
| 1480 | 4.67E-02 | 0.46 | Lethal(2) giant larvae protein homolog 2 |
| 1512 | 4.27E-02 | 0.48 | Uncharacterized protein KIAA1529 |
| 1515 | 9.61E-03 | 0.33 | CD83 antigen |
| 1526 | 5.63E-03 | 0.45 | Uncharacterized protein KIAA1529 |
| 1556 | 2.99E-02 | 0.45 | Glutamine-rich protein 2 |
| 1593 | 2.46E-02 | 0.45 | Zinc finger protein 616 |
| 1599 | 1.08E-02 | 0.39 | Keratin, type II cytoskeletal 1 |
| 1606 | 9.59E-03 | 0.48 | ADP-ribosylation factor 1 |
| 1607 | 2.00E-02 | 0.37 | Peptidyl-prolyl cis-trans isomerase B |
| 1628 | 2.29E-02 | 0.45 | Uncharacterized protein KIAA1529 |
| 1655 | 7.58E-03 | 0.48 | LIM and calponin homology domains-containing protein 1 |
| 1677 | 3.13E-02 | 0.50 | Echinoderm microtubule-associated protein-like 6 |
| 1700 | 3.14E-04 | 0.39 | Keratin, type I cytoskeletal 10 |
| 2381 | 2.87E-02 | 0.31 | Aconitate hydratase, mitochondrial |
| 2382 | 1.02E-02 | 2.18 | Uncharacterized protein KIAA1529 |
| 2396 | 7.10E-03 | 2.04 | RNA-binding protein NOB1 |
| 2398 | 2.15E-03 | 2.23 | Uncharacterized protein KIAA1529 |
| 2404 | 5.14E-03 | 2.54 | Keratin, type I cytoskeletal 10 |
| 2409 | 2.52E-02 | 0.45 | Teneurin-1 |
| 2413 | 3.00E-02 | 0.50 | Moesin |
| 2419 | 2.82E-02 | 0.42 | Trypsin-2 |
| 2426 | 3.59E-02 | 0.42 | Heat shock cognate 71 kDa protein |
| 2435 | 1.51E-02 | 0.40 | Growth arrest-specific protein 2 |
| 2512 | 3.66E-02 | 0.49 | Glutamine-rich protein 2 |
| 2513 | 1.88E-02 | 0.46 | Glutamine-rich protein 2 |
| 2534 | 3.22E-02 | 0.47 | Keratin, type I cytoskeletal 10 |
| 2543 | 4.61E-02 | 0.46 | Acetolactate synthase-like protein |
| 2546 | 1.80E-02 | 0.39 | Keratin, type I cytoskeletal 10 |
| 2548 | 1.13E-02 | 0.45 | Keratin, type I cytoskeletal 10 |
| 2550 | 1.86E-02 | 0.47 | Crumbs homolog 1 |
| 2551 | 7.32E-03 | 0.48 | Keratin, type I cytoskeletal 10 |
| 2559 | 1.31E-02 | 0.47 | Vimentin |
| 2584 | 3.45E-02 | 0.41 | Uncharacterized protein KIAA1529 |
| 2586 | 1.88E-02 | 0.41 | Transcriptional adapter 3-like |
| 2593 | 3.80E-02 | 0.47 | Complement C2 |
| 2616 | 2.05E-02 | 0.43 | Coiled-coil domain-containing protein 116 |
| 2633 | 3.94E-02 | 0.38 | Serpin H1 |
| 2634 | 7.83E-03 | 0.41 | Serpin H1 |
| 2637 | 7.27E-03 | 0.41 | Keratin, type II cytoskeletal 2 epidermal |
| 2639 | 2.95E-02 | 0.49 | Zinc finger protein 512 |
| 2640 | 3.84E-02 | 0.49 | Not identified |
| 2643 | 3.58E-02 | 0.40 | Serpin H1 |
| 2644 | 4.81E-02 | 0.50 | Serpin H1 |
| 2652 | 2.00E-02 | 0.47 | Glutamine-rich protein 2 |
| 2653 | 1.30E-02 | 0.43 | Actin, cytoplasmic 1 |
| 2663 | 2.23E-02 | 0.50 | Glutamine-rich protein 2 |
| 2667 | 4.24E-03 | 0.39 | Keratin, type II cytoskeletal 1 |
| 2674 | 1.93E-02 | 0.50 | Keratin, type I cytoskeletal 10 |
| 2692 | 1.38E-02 | 0.50 | Actin, cytoplasmic 1 |
| 2695 | 4.04E-02 | 0.37 | Mitogen-activated protein kinase kinase kinase 8 |
| 2699 | 1.11E-02 | 0.40 | Actin, cytoplasmic 1 |
| 2700 | 9.58E-03 | 0.39 | Actin, cytoplasmic 1 |
| 2701 | 2.18E-02 | 0.35 | Keratin, type II cytoskeletal 1 |
| 2709 | 3.71E-02 | 0.43 | Echinoderm microtubule-associated protein-like 6 |
| 2710 | 1.52E-02 | 0.38 | Keratin, type I cytoskeletal 10 |
| 2783 | 2.46E-02 | 0.39 | Stress-70 protein, mitochondrial |
| 2791 | 3.75E-02 | 0.46 | Uncharacterized protein KIAA1529 |
| 2792 | 1.02E-03 | 0.37 | Uncharacterized protein KIAA1529 |
| 2796 | 1.72E-02 | 0.49 | Poly(ADP-ribose) glycohydrolase ARH3 |
| 2812 | 1.24E-02 | 0.44 | Transmembrane protein 18 |
| 2819 | 1.33E-02 | 0.36 | Glutamine-rich protein 2 |
| 2834 | 3.37E-02 | 0.48 | Zinc finger protein 584 |
| 2837 | 3.88E-02 | 0.45 | Uncharacterized protein KIAA1529 |
| 2868 | 3.12E-02 | 0.46 | Uncharacterized protein KIAA1529 |
| 2878 | 1.36E-02 | 0.48 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial |
| 2929 | 2.47E-02 | 0.49 | Growth factor receptor-bound protein 10 |
| 2931 | 6.12E-03 | 0.47 | Apolipoprotein A-I |
| 2934 | 4.76E-02 | 0.49 | Apolipoprotein A-I |
| 2951 | 2.17E-02 | 0.50 | FAST kinase domain-containing protein 1 |
| 2959 | 1.16E-02 | 0.43 | Uncharacterized protein KIAA1529 |
| 2964 | 4.21E-02 | 0.45 | Kelch-like protein 38 |
| 2971 | 2.88E-02 | 0.47 | Keratin, type II cytoskeletal 1 |
| 2985 | 2.49E-02 | 0.29 | Keratin, type I cytoskeletal 10 |
| 2993 | 2.87E-02 | 0.47 | Glutamine-rich protein 2 |
| 2997 | 6.08E-03 | 0.48 | Keratin, type I cytoskeletal 10 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tajima, T.; Kito, F.; Yoshida, A.; Kawai, A.; Kondo, T. Calreticulin as A Novel Potential Metastasis-Associated Protein in Myxoid Liposarcoma, as Revealed by Two-Dimensional Difference Gel Electrophoresis. Proteomes 2019, 7, 13. https://doi.org/10.3390/proteomes7020013
Tajima T, Kito F, Yoshida A, Kawai A, Kondo T. Calreticulin as A Novel Potential Metastasis-Associated Protein in Myxoid Liposarcoma, as Revealed by Two-Dimensional Difference Gel Electrophoresis. Proteomes. 2019; 7(2):13. https://doi.org/10.3390/proteomes7020013
Chicago/Turabian StyleTajima, Takashi, Fusako Kito, Akihiko Yoshida, Akira Kawai, and Tadashi Kondo. 2019. "Calreticulin as A Novel Potential Metastasis-Associated Protein in Myxoid Liposarcoma, as Revealed by Two-Dimensional Difference Gel Electrophoresis" Proteomes 7, no. 2: 13. https://doi.org/10.3390/proteomes7020013