Microdevices for Non-Invasive Detection of Bladder Cancer
Abstract
:1. Introduction
2. Standards of Detection and Evaluation
2.1. Invasive Techniques
2.2. Non-Invasive Techniques
3. Types of Urine Markers
3.1. Invasive Urine Markers
3.2. Non-Invasive Urine Markers
4. Increasing the Detection Sensitivity of Non-Invasive Markers Using Microdevices
4.1. Cell-Based Detection
4.1.1. Affinity-Based Detection
4.1.2. Detection by Membrane Capacitance
4.1.3. Impedance-Based Detection
4.2. Cell-Free Detection
4.2.1. DNA-Bonded Substrates
4.2.2. Antibody-Bonded Substrates
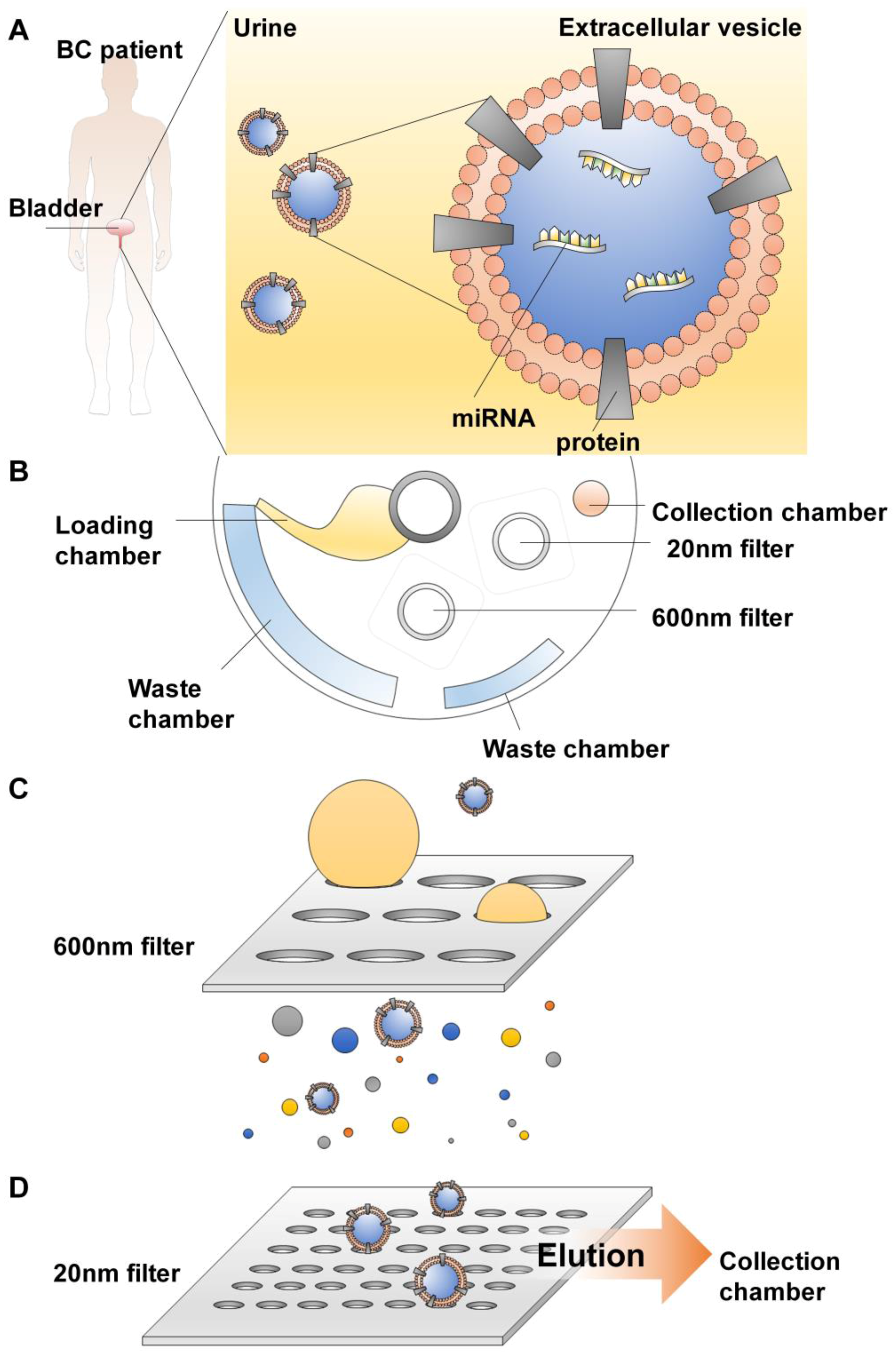
4.2.3. Extracellular Vesicles (EV) Isolation
5. Future Perspectives
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| APOA1 | Apolipoprotein A1 |
| BTA | Bladder tumor antigen |
| CDC | Cell division cycle |
| CDH1 | Epithelial cadherin |
| cx | Connexin |
| CXCL | Chemokine |
| CXCR | CXC chemokine receptor |
| DAPK | Death-associated protein kinase |
| DEP | Dielectrophoresis/dielectrophoretic |
| Drg-1 | Developmentally regulated GTP binding protein 1 |
| EDIL-3 | EGF-like Repeats and Discoidin Domains 3 |
| ELISA | Enzyme-linked immunosorbent assay |
| EpCAM | Epithelial cell adhesion molecule |
| EV | Extracellular vesicles |
| FDA | Food and Drug Administration |
| FISH | Fluorescence in situ hybridization |
| Gal-1 | Galectin-1IGF: Insulin-like growth factor |
| IHC | Immunohistochemistry |
| IL8R | Interleukin 8 receptor |
| HOXA | Homeobox A |
| MDK | Midkine |
| NMP22 | Nuclear matrix protein-22 |
| pAkt | Phosphorylated protein kinase B |
| PDMS | Polydimethylsiloxane |
| pRB | Retinoblastoma protein |
| PTEN | Phosphatase and tensin homolog |
| RARβ | Retinoic acid receptor beta |
| RC | Radical cystectomy |
| SLD5 | Synthetic Lethality with Dpb11-1 |
| TACSTD2 | Tumor-Associated Calcium Signal Transducer 2 |
| TURBT | Transurethral resection of bladder tumor |
References
- van Osch, F.H.M.; Jochems, S.H.J.; van Schooten, F.J.; Bryan, R.T.; Zeegers, M.P. Significant role of lifetime cigarette smoking in worsening bladder cancer and upper tract urothelial carcinoma prognosis: A meta-analysis. J. Urol. 2016, 195, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Wyszynski, A.; Tanyos, S.A.; Rees, J.R.; Marsit, C.J.; Kelsey, K.T.; Schned, A.R.; Pendleton, E.M.; Celaya, M.O.; Zens, M.S.; Karagas, M.R.; et al. Body mass and smoking are modifiable risk factors for recurrent bladder cancer. Cancer 2014, 120, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Koebnick, C.; Michaud, D.; Moore, S.C.; Park, Y.; Hollenbeck, A.; Ballard-Barbash, R.; Schatzkin, A.; Leitzmann, M.F. Body mass index, physical activity, and bladder cancer in a large prospective study. Cancer Epidemiol. Prev. Biomark. 2008, 17, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Ploeg, M.; Aben, K.K.H.; Kiemeney, L.A. The present and future burden of urinary bladder cancer in the world. World J. Urol. 2009, 27, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Krapcho, M.; Miller, D.; Bishop, K.; Kosary, C.L.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. Contents of the seer cancer statistics review (csr), 1975–2014. In SEER Cancer Statistics Review, 1975–2014; National Cancer Institute: Bethesda, MD, USA, 2016. [Google Scholar]
- Kaufman, D.S.; Shipley, W.U.; Feldman, A.S. Bladder cancer. Lancet 2009, 374, 239–249. [Google Scholar] [CrossRef]
- Bellmunt, J.; Orsola, A.; Leow, J.J.; Wiegel, T.; De Santis, M.; Horwich, A. Bladder cancer: Esmo practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii40–iii48. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Goodison, S.; Rosser, C.J.; Urquidi, V. Bladder cancer detection and monitoring: Assessment of urine- and blood-based marker tests. Mol. Diagn. Ther. 2013, 17, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Kassouf, W.; Kamat, A.M.; Zlotta, A.; Bochner, B.H.; Moore, R.; So, A.; Izawa, J.; Rendon, R.A.; Lacombe, L.; Aprikian, A.G. Canadian guidelines for treatment of non-muscle invasive bladder cancer: A focus on intravesical therapy. Can. Urol. Assoc. J. 2010, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Sievert, K.D.; Amend, B.; Nagele, U.; Schilling, D.; Bedke, J.; Horstmann, M.; Hennenlotter, J.; Kruck, S.; Stenzl, A. Economic aspects of bladder cancer: What are the benefits and costs? World J. Urol. 2009, 27, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Botteman, M.F.; Pashos, C.L.; Redaelli, A.; Laskin, B.; Hauser, R. The health economics of bladder cancer: A comprehensive review of the published literature. Pharmacoeconomics 2003, 21, 1315–1330. [Google Scholar] [CrossRef] [PubMed]
- Herr, H.W. The risk of urinary tract infection after flexible cystoscopy in patients with bladder tumor who did not receive prophylactic antibiotics. J. Urol. 2015, 193, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Barbadoro, P.; Labricciosa, F.M.; Recanatini, C.; Gori, G.; Tirabassi, F.; Martini, E.; Gioia, M.G.; D’Errico, M.M.; Prospero, E. Catheter-associated urinary tract infection: Role of the setting of catheter insertion. Am. J. Infect. Control 2015, 43, 707–710. [Google Scholar] [CrossRef] [PubMed]
- van der Aa, M.N.M.; Steyerberg, E.W.; Sen, E.F.; Zwarthoff, E.C.; Kirkels, W.J.; van der Kwast, T.H.; Essink-Bot, M.-L. Patients’ perceived burden of cystoscopic and urinary surveillance of bladder cancer: A randomized comparison. BJU Int. 2008, 101, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Witjes, J.A.; Lebret, T.; Compérat, E.M.; Cowan, N.C.; De Santis, M.; Bruins, H.M.; Hernández, V.; Espinós, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 eau guidelines on muscle-invasive and metastatic bladder cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Milowsky, M.I.; Rumble, R.B.; Booth, C.M.; Gilligan, T.; Eapen, L.J.; Hauke, R.J.; Boumansour, P.; Lee, C.T. Guideline on muscle-invasive and metastatic bladder cancer (european association of urology guideline): American society of clinical oncology clinical practice guideline endorsement. J. Clin. Oncol. 2016, 34, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Hennenlotter, J.; Huber, S.; Todenhofer, T.; Kuehs, U.; Schilling, D.; Aufderklamm, S.; Gakis, G.; Schwentner, C.; Stenzl, A. Point-of-care tests for bladder cancer: The influencing role of hematuria. Adv. Urol. 2011, 2011, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Oge, O.; Kozaci, D.; Gemalmaz, H. The bta stat test is nonspecific for hematuria: An experimental hematuria model. J. Urol. 2002, 167, 1318–1320. [Google Scholar] [CrossRef]
- Hautmann, S.; Toma, M.; Gomez, M.F.L.; Friedrich, M.G.; Jaekel, T.; Michl, U.; Schroeder, G.L.; Huland, H.; Juenemann, K.-P.; Lokeshwar, V.B. Immunocyt and the ha-haase urine tests for the detection of bladder cancer: A side-by-side comparison. Eur. Urol. 2004, 46, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zippe, C.D.; Pandrangi, L.; Nelson, D.; Agarwal, A. Exclusion criteria enhance the specificity and positive predictive value of nmp22 and bta stat. J. Urol. 1999, 162, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Dimashkieh, H.; Wolff, D.J.; Smith, T.M.; Houser, P.M.; Nietert, P.J.; Yang, J. Evaluation of urovysion and cytology for bladder cancer detection: A study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol. 2013, 121, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Lavery, H.; Zaharieva, B.; McFaddin, A.; Heerema, N.; Pohar, K. A prospective comparison of urovysion fish and urine cytology in bladder cancer detection. BMC Cancer 2017, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.; Dana, T. Screening adults for bladder cancer: A review of the evidence for the U.S. Preventive services task force. Ann. Intern. Med. 2010, 153, 461–468. [Google Scholar] [CrossRef] [PubMed]
- de Bekker-Grob, E.W.; van der Aa, M.N.M.; Zwarthoff, E.C.; Eijkemans, M.J.C.; van Rhijn, B.W.; van der Kwast, T.H.; Steyerberg, E.W. Non-muscle-invasive bladder cancer surveillance for which cystoscopy is partly replaced by microsatellite analysis of urine: A cost-effective alternative? BJU Int. 2009, 104, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Svatek, R.S.; Sagalowsky, A.I.; Lotan, Y. Economic impact of screening for bladder cancer using bladder tumor markers: A decision analysis. Urol. Oncol. 2006, 24, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Elvira, K.; i Solvas, X.; Wootton, R.; deMello, A. The past, present and potential for microfluidic reactor technology in chemical synthesis. Nat. Chem. 2013, 5, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gomez-Sjoberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Prakadan, S.; Shalek, A.; Weitz, D. Scaling by shrinking: Empowering single-cell ‘omics’ with microfluidic devices. Nat. Rev. Genet. 2017, 18, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi Warkiani, M.; Luan Khoo, B.; Wu, L.; Kah Ping Tay, A.; Asgar Bhagat, A.S.; Han, J.; Teck Lim, C. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2015, 11, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, W.; Wang, Z.; Tang, Y.; Deng, Y.; Xu, L.; Tian, J.; Shi, Q. Ex vivo expansion of circulating lung tumor cells based on one-step microfluidics-based immunomagnetic isolation. Analyst 2016, 141, 3621–3625. [Google Scholar] [CrossRef] [PubMed]
- Ohnaga, T.; Shimada, Y.; Takata, K.; Obata, T.; Okumura, T.; Nagata, T.; Kishi, H.; Muraguchi, A.; Tsukada, K. Capture of esophageal and breast cancer cells with polymeric microfluidic devices for ctc isolation. Mol. Clin. Oncol. 2016, 4, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed]
- Renier, C.; Pao, E.; Che, J.; Liu, H.E.; Lemaire, C.A.; Matsumoto, M.; Triboulet, M.; Srivinas, S.; Jeffrey, S.S.; Rettig, M.; et al. Label-free isolation of prostate circulating tumor cells using vortex microfluidic technology. NPJ Precis. Oncol. 2017, 15. [Google Scholar] [CrossRef]
- Ghodbane, M.; Stucky, E.C.; Maguire, T.J.; Schloss, R.S.; Shreiber, D.I.; Zahn, J.D.; Yarmush, M.L. Development and validation of a microfluidic immunoassay capable of multiplexing parallel samples in microliter volumes. Lab Chip 2015, 15, 3211–3221. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Ren, K.; Shu, Y.; Chen, Y.; Shen, B.; Wu, H. Recent developments in microfluidics for cell studies. Adv. Mater. 2014, 26, 5525–5532. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Fang, Q.; den Toonder, J.M.J. Microfluidics for cell-based high throughput screening platforms—A review. Anal. Chim. Acta 2016, 903, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.; Buckley, D.; Fu, R.; Gore, J.L.; Gustafson, K.; Griffin, J.; Grusing, S.; Selph, S. Emerging Approaches to Diagnosis and Treatment of Non-Muscle-Invasive Bladder Cancer; MD: Rockville, MA, USA, 2015. [Google Scholar]
- Lotan, Y.; O’Sullivan, P.; Raman, J.D.; Shariat, S.F.; Kavalieris, L.; Frampton, C.; Guilford, P.; Luxmanan, C.; Suttie, J.; Crist, H.; et al. Clinical comparison of noninvasive urine tests for ruling out recurrent urothelial carcinoma. Urol. Oncol. 2017, 35. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Steven, K.; Guldberg, P. Size-based enrichment of exfoliated tumor cells in urine increases the sensitivity for DNA-based detection of bladder cancer. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Yi, L.; Lin, X.; Lin, L.; Li, H.; Lin, J.M. A non-invasive genomic diagnostic method for bladder cancer using size-based filtration and microchip electrophoresis. Talanta 2015, 144, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Rajesh, A.; Prasad, S.R.; Gaitonde, K.; Lall, C.G.; Mouraviev, V.; Aeron, G.; Bracken, R.B.; Sandrasegaran, K. Urinary bladder cancer: Role of mr imaging. Radiographics 2012, 32, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Cha, E.K.; Matsumoto, K.; Baba, S.; Chromecki, T.F.; Fajkovic, H.; Sun, M.; Karakiewicz, P.I.; Scherr, D.S.; Shariat, S.F. Immunohistochemical biomarkers for bladder cancer prognosis. Int. J. Urol. 2011, 18, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Naito, H.; Wakabayashi, T.; Yoshida, H.; Muramatsu, F.; Iba, T.; Kidoya, H.; Takakura, N. Regulation of sld5 gene expression by mir-370 during acute growth of cancer cells. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.D.; De La Cerda, J.; Tuziak, T.; Rosen, D.; Xiao, L.; Shen, Y.; Sabichi, A.L.; Czerniak, B.; Grossman, B.H. Analysis of the expression of biomarkers in urinary bladder cancer using a tissue microarray. Mol. Carcinog. 2008, 47, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Lee, S.D.; Lee, J.Z.; Chung, M.K.; Ha, H.K. Expression analysis and clinical significance of cxcl16/cxcr6 in patients with bladder cancer. Oncol. Lett. 2012, 5, 229–235. [Google Scholar] [PubMed]
- Nishizawa, K.; Nishiyama, H.; Oishi, S.; Tanahara, N.; Kotani, H.; Mikami, Y.; Toda, Y.; Evans, B.J.; Peiper, S.C.; Saito, R.; et al. Fluorescent imaging of high-grade bladder cancer using a specific antagonist for chemokine receptor cxcr4. Int. J. Cancer 2010, 127, 1180–1187. [Google Scholar] [CrossRef] [PubMed]
- Darling, D.; Luxmanan, C.; O’Sullivan, P.; Lough, T.; Suttie, J. Clinical utility of cxbladder for the diagnosis of urothelial carcinoma. Adv. Ther. 2017, 34, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Reid-nicholson, M.D.; Ramalingam, P.; Adeagbo, B.; Cheng, N.; Peiper, S.C.; Terris, M.K. The use of urovysion t fluorescence in situ hybridization in the diagnosis and surveillance of non-urothelial carcinoma of the bladder. Mod. Pathol. 2009, 22, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez Baños, J.L.; del Henar Rebollo Rodrigo, M.; Antolín Juárez, F.M.; García, B.M. Usefulness of the bta stat test for the diagnosis of bladder cancer. Urology 2001, 57, 685–689. [Google Scholar] [CrossRef]
- Van Rhijn, B.W.G.; Van Der Poel, H.G.; Van Der Kwast, T.H. Urine markers for bladder cancer surveillance: A systematic review. Eur. Urol. 2005, 47, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Ponsky, L.E.; Sharma, S.; Pandrangi, L.; Kedia, S.; Nelson, D.; Agarwal, A.; Zippe, C.D. Screening and monitoring for bladder cancer: Refining the use of nmp22. J. Urol. 2001, 166, 75–78. [Google Scholar] [CrossRef]
- Moonen, P.M.J.; Kiemeney, L.A.L.M.; Witjes, J.A. Urinary nmp22® bladderchek® test in the diagnosis of superficial bladder cancer. Eur. Urol. 2005, 48, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Natalin, R.A.; Landman, J. Where next for the endoscope? Nat. Rev. Urol. 2009, 6, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Kamat, A.M.; Bagcioglu, M.; Huri, E. What is new in non-muscle-invasive bladder cancer in 2016? Turk. J. Urol. 2017, 43, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Han, A.L.; Veeneman, B.A.; El-Sawy, L.; Day, K.C.; Day, M.L.; Tomlins, S.A.; Keller, E.T. Fibulin-3 promotes muscle-invasive bladder cancer. Oncogene 2017, 36, 5243–5251. [Google Scholar] [CrossRef] [PubMed]
- Tetu, B. Diagnosis of urothelial carcinoma from urine. Mod. Pathol. 2009, 22 (Suppl. 2), S53–S59. [Google Scholar] [CrossRef] [PubMed]
- Young, R.H. Tumor-like lesions of the urinary bladder. Mod. Pathol. 2009, 22 (Suppl. 2), S37–S52. [Google Scholar] [CrossRef] [PubMed]
- Abrol, S.; Jairath, A.; Ganpule, S.; Ganpule, A.; Mishra, S.; Sabnis, R.; Desai, M. Can ct virtual cystoscopy replace conventional cystoscopy in early detection of bladder cancer? Adv. Urol. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczyk, M.A.; Nyk, L.; Szostek, P.; Szemplinski, S.; Borowka, A.; Dobruch, J. The role of endoscopic bladder tumour assessment in the management of patients subjected to transurethral bladder tumour resection. Eur. J. Cancer Care (Engl.) 2017, 26. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Boorjian, S.A.; Chou, R.; Clark, P.E.; Daneshmand, S.; Konety, B.R.; Pruthi, R.; Quale, D.Z.; Ritch, C.R.; Seigne, J.D.; et al. Diagnosis and treatment of non-muscle invasive bladder cancer: Aua/suo guideline. J. Urol. 2016, 196, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.A.; Zanganeh, S.; Akbarnejad, E.; Salehi, F.; Abdolahad, M. Microfluidic device for label-free quantitation and distinction of bladder cancer cells from the blood cells using micro machined silicon based electrical approach; suitable in urinalysis assays. J. Pharm. Biomed. Anal. 2017, 134, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Zigeuner, R.; Shariat, S.F.; van Rhijn, B.W.; Comperat, E.; Sylvester, R.J.; Kaasinen, E.; Bohle, A.; Palou Redorta, J.; et al. Eau guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2013. Eur. Urol. 2013, 64, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Owens, C.L.; Vandenbussche, C.J.; Burroughs, F.H.; Rosenthal, D.L. A review of reporting systems and terminology for urine cytology. Cancer Cytopathol. 2013, 121, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.D.; Yafi, F.A.; Brimo, F.; Steinberg, J.; Aprikian, A.G.; Tanguay, S.; Kassouf, W. Prognostic value of urinary cytology and other biomarkers for recurrence and progression in bladder cancer: A prospective study. World J. Urol. 2016, 34, 1405–1409. [Google Scholar] [CrossRef] [PubMed]
- Turco, P.; Houssami, N.; Bulgaresi, P.; Troni, G.M.; Galanti, L.; Cariaggi, M.P.; Cifarelli, P.; Crocetti, E.; Ciatto, S. Is conventional urinary cytology still reliable for diagnosis of primary bladder carcinoma? Accuracy based on data linkage of a consecutive clinical series and cancer registry. Acta Cytol. 2011, 55, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Yager, P.; Edwards, T.; Fu, E.; Helton, K.; Nelson, K.; Tam, M.R.; Weigl, B.H. Microfluidic diagnostic technologies for global public health. Nature 2006, 442, 412–418. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, P.; Sharples, K.; Dalphin, M.; Davidson, P.; Gilling, P.; Cambridge, L.; Harvey, J.; Toro, T.; Giles, N.; Luxmanan, C.; et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J. Urol. 2012, 188, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Greene, K.L.; Berry, A.; Konety, B.R. Diagnostic utility of the immunocyt/ucyt+ test in bladder cancer. Rev. Urol. 2006, 8, 190–197. [Google Scholar] [PubMed]
- Oge, O.; Atsü, N.; Sahin, A.; Ozen, H. Comparison of bta stat and nmp22 tests in the detection of bladder cancer. Scand. J. Urol. Nephrol. 2000, 34, 349–351. [Google Scholar] [PubMed]
- Thomas, L.; Leyh, H.; Marberger, M.; Bombardieri, E.; Bassi, P.; Pagano, F.; Pansadoro, V.; Sternberg, C.N.; Boccon-Gibod, L.; Ravery, V.; et al. Multicenter trial of the quantitative bta trak assay in the detection of bladder cancer. Clin. Chem. 1999, 45, 472–477. [Google Scholar] [PubMed]
- Bibbo, M.; Kern, W.H. Urinary tract. In Comprehensive Cytopathology, 3rd ed.; Elsevier: Philadelphia, PA, USA, 2008. [Google Scholar]
- Kundal, V.K.; Pandith, A.A.; Hamid, A.; Shah, A.; Kundal, R.; Wani, S.M. Role of nmp22 bladder check test in early detection of bladder cancer with recurrence. Asian Pac. J. Cancer Prev. 2010, 141, 1279–1282. [Google Scholar]
- Pichler, R.; Tulchiner, G.; Fritz, J.; Schaefer, G.; Horninger, W.; Heidegger, I. Urinary ubc rapid and nmp22 test for bladder cancer surveillance in comparison to urinary cytology: Results from a prospective single-center study. Int. J. Med. Sci. 2017, 14, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nandi, S.; Tan, T.Z.; Ler, S.G.; Chia, K.S.; Lim, W.Y.; Butow, Z.; Vordos, D.; De la Taille, A.; Al-Haddawi, M.; et al. Highly sensitive and specific novel biomarkers for the diagnosis of transitional bladder carcinoma. Oncotarget 2015, 6, 13539–13549. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Chen, Y.-J.; Lai, C.-S.; Chen, Y.-T.; Chen, C.-L.; Yu, J.-S.; Chang, Y.-S. A negative-pressure-driven microfluidic chip for the rapid detection of a bladder cancer biomarker in urine using bead-based enzyme-linked immunosorbent assay. Biomicrofluidics 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Pursey, J.P.; Chen, Y.; Stulz, E.; Park, M.K.; Kongsuphol, P. Microfluidic electrochemical multiplex detection of bladder cancer DNA markers. Sens. Actuators B 2017, 251, 34–39. [Google Scholar] [CrossRef]
- Chuang, C.H.; Du, Y.C.; Wu, T.F.; Chen, C.H.; Lee, D.H.; Chen, S.M.; Huang, T.C.; Wu, H.P.; Shaikh, M.O. Immunosensor for the ultrasensitive and quantitative detection of bladder cancer in point of care testing. Biosens. Bioelectron. 2016, 84, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Macgregor-Ramiasa, M.; McNicholas, K.; Ostrikov, K.; Li, J.; Michael, M.; Gleadle, J.M.; Vasilev, K. A platform for selective immuno-capture of cancer cells from urine. Biosens. Bioelectron. 2017, 96, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.-K.; Sunkara, V.; Park, J.; Kim, T.-H.; Han, J.-R.; Kim, C.-J.; Choi, H.-I.; Kim, Y.-K.; Cho, Y.-K. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano 2017, 11, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Chaudhuri, P.K.; Lim, C.T.; Warkiani, M.E. Advancing techniques and insights in circulating tumor cell (ctc) research. In Ex Vivo Engineering of the Tumor Microenvironment; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Imrich, S.; Hachmeister, M.; Gires, O. Epcam and its potential role in tumor-initiating cells. Cell Adhes. Migr. 2012, 6, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.T.; Shimwell, N.J.; Wei, W.; Devall, A.J.; Pirrie, S.J.; James, N.D.; Zeegers, M.P.; Cheng, K.K.; Martin, A.; Ward, D.G. Urinary epcam in urothelial bladder cancer patients: Characterisation and evaluation of biomarker potential. Br. J. Cancer 2014, 110, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005, 2, S4–S11. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.W.Y.; Chan, L.W.; Tang, N.L.S.; Tong, J.H.M.; Lo, K.W.; Lee, T.L.; Cheung, H.Y.; Wong, W.S.; Chan, P.S.F.; Lai, F.M.M.; et al. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin. Cancer Res. 2002, 8, 464–470. [Google Scholar] [PubMed]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific pcr: A novel pcr assay for methylation status of cpg islands (DNA methylation/tumor suppressor genes/pl6/p15). Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed]
- Wojdacz, T.K.; Hansen, L.L.; Dobrovic, A. A new approach to primer design for the control of pcr bias in methylation studies. BMC Res. Notes 2008, 1. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.P. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Ke, H.L.; Huang, S.P.; Wu, W.J.; Chen, Y.K.; Chang, L.L. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of e-cadherin, p16, p14, rassf1a genes in urine. Urol. Oncol. 2010, 28, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Singleton, D.G.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecka, H.; Stulz, E.; Radecki, J. A highly sensitive electrochemical genosensor based on co-porphyrin-labelled DNA. Chem. Commun. 2014, 50, 4196–4199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-free DNA (cfdna): Clinical significance and utility in cancer shaped by emerging technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-H.; Wu, C.-H.; Lee, Y.-L.; Huang, P.-H.; Kao, Y.-L.; Shiau, M.-Y. Evaluation of nuclear matrix protein-22 as a clinical diagnostic marker for bladder cancer. Urology 2004, 64, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.T.; Zhang, Y. Bead-based microfluidic immunoassays: The next generation. Biosens. Bioelectron. 2007, 22, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.H.C.; Uddayasankar, U.; Wheeler, A.R. Immunoassays in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Chen, C.-L.; Chen, H.-W.; Chung, T.; Wu, C.-C.; Chen, C.-D.; Hsu, C.-W.; Chen, M.-C.; Tsui, K.-H.; Chang, P.-L.; et al. Discovery of novel bladder cancer biomarkers by comparative urine proteomics using itraq technology. J. Proteom. Res. 2010, 9, 5803–5815. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Chen, H.-W.; Domanski, D.; Smith, D.S.; Liang, K.-H.; Wu, C.-C.; Chen, C.-L.; Chung, T.; Chen, M.-C.; Chang, Y.-S.; et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J. Proteom. 2012, 75, 3529–3545. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, C.; Wu, H.; Zhang, T.; Wang, J.; Wang, S.; Chang, J. Identification of apo-a1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteom. Sci. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Extracellular vesicles isolation and their biomarker potential: Are we ready for testing? Ann. Transl. Med. 2017, 5, 3–6. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Boing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed]
- Wendler, F.; Favicchio, R.; Simon, T.; Alifrangis, C.; Stebbing, J.; Giamas, G. Extracellular vesicles swarm the cancer microenvironment: From tumor—Stroma communication to drug intervention. Nat. Publ. Group 2016, 36, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, T.; Kosaka, N.; Ochiya, T. The roles of extracellular vesicles in cancer biology: Toward the development of novel cancer biomarkers. Proteomics 2014, 14, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Zaripov, M.M.; Skvortsova, T.E.; Lekchnov, E.A. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Lai, Y.F.; Tang, P.; Chien, K.Y.; Yu, J.S.; Tsai, C.H.; Chen, H.W.; Wu, C.C.; Chung, T.; Hsu, C.W.; et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J. Proteom. Res. 2012, 11, 5611–5629. [Google Scholar] [CrossRef] [PubMed]
- Beckham, C.J.; Olsen, J.; Yin, P.-N.; WU, C.-H.; TING, H.-J.; Hagen, F.K.; Scosyrev, E.; Messing, E.M.; Lee, Y.-F. Bladder cancer exosomes contain edil-3/del1 and facilitate cancer progression. J. Urol. 2014, 192, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Camussi, G.; Valadi, H.; Nazarenko, I.; Ekström, K.; Wang, X.; Principe, S.; Shah, N.; Ashraf, N.M.; Fatima, F.; et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014, 11, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Yancik, R.; Ries, L.A.G. Cancer in older persons: An international issue in an aging world. Semin. Oncol. 2004, 31, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Rabinovitch, P.S.; Martin, G.M. Healthy aging: The ultimate preventative medicine. Science 2015, 350, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Islami, F.; Torre, L.A.; Jemal, A. Global trends of lung cancer mortality and smoking prevalence. Transl. Lung Cancer Res. 2015, 4, 327–338. [Google Scholar] [PubMed]
- Sullivan, P.S.; Nooraie, F.; Sanchez, H.; Hirschowitz, S.; Levin, M.; Rao, P.N.; Rao, J. Comparison of immunocyt, urovysion, and urine cytology in detection of recurrent urothelial carcinoma. Cancer Cytopathol. 2009, 117, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Brausi, M.A. Primary prevention and early detection of bladder cancer: Two main goals for urologists. Eur. Urol. 2013, 63, 242–243. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Marberger, M.J.; Lotan, Y.; Sanchez-Carbayo, M.; Zippe, C.; Ludecke, G.; Boman, H.; Sawczuk, I.; Friedrich, M.G.; Casella, R.; et al. Variability in the performance of nuclear matrix protein 22 for the detection of bladder cancer. J. Urol. 2006, 176, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Lotan, Y.; Shariat, S.F.; Schmitz-Drager, B.J.; Sanchez-Carbayo, M.; Jankevicius, F.; Racioppi, M.; Minner, S.J.; Stohr, B.; Bassi, P.F.; Grossman, H.B. Considerations on implementing diagnostic markers into clinical decision making in bladder cancer. Urol. Oncol. 2010, 28, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Vrooman, O.P.; Witjes, J.A. Urinary markers in bladder cancer. Eur. Urol. 2008, 53, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Budman, L.I.; Kassouf, W.; Steinberg, J.R. Biomarkers for detection and surveillance of bladder cancer. Can. Urol. Assoc. J. 2008, 2, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, E.; Miyake, S.; Yogo, Y. Review of enzyme-linked immunosorbent assays (elisas) for analyses of neonicotinoid insecticides in agro-environments. J. Agric. Food Chem. 2013, 61, 12459–12472. [Google Scholar] [CrossRef] [PubMed]
- Andreu-Perez, J.; Leff, D.R.; Ip, H.M.; Yang, G.Z. From wearable sensors to smart implants—Toward pervasive and personalized healthcare. IEEE Trans. Bio-Med. Eng. 2015, 62, 2750–2762. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, X.; Khimji, I.; Akbas, R.; Qiu, W.; Edwards, D.; Cramer, D.W.; Ye, B.; Demirci, U. Integration of cell phone imaging with microchip elisa to detect ovarian cancer he4 biomarker in urine at the point-of-care. Lab. Chip. 2011, 11, 3411–3418. [Google Scholar] [CrossRef] [PubMed]
- Kuhnemund, M.; Wei, Q.; Darai, E.; Wang, Y.; Hernandez-Neuta, I.; Yang, Z.; Tseng, D.; Ahlford, A.; Mathot, L.; Sjoblom, T.; et al. Targeted DNA sequencing and in situ mutation analysis using mobile phone microscopy. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nole, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, J.; Park, J. Methods to isolate extracellular vesicles for diagnosis. In Micro and Nano Systems Letters; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Ruppen, J.; Wildhaber, F.D.; Strub, C.; Hall, S.R.R.; Schmid, R.A.; Geiser, T.; Guenat, O.T. Towards personalized medicine: Chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip 2015, 15, 3076–3085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3d vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Genshaft, A.S.; Li, S.; Gallant, C.J.; Darmanis, S.; Prakadan, S.M.; Ziegler, C.G.K.; Lundberg, M.; Fredriksson, S.; Hong, J.; Regev, A.; et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Nature 2016, 188. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Chaudhuri, P.K.; Ramalingam, N.; Tan, D.S.; Lim, C.T.; Warkiani, M.E. Single-cell profiling approaches to probing tumor heterogeneity. Int. J. Cancer 2016, 139, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Gierahn, T.M.; Wadsworth, M.H.; Hughes, T.K.; Bryson, B.D.; Butler, A.; Satija, R.; Fortune, S.; Love, J.C.; Shalek, A.K. Seq-well: Portable, low-cost rna sequencing of single cells at high throughput. Nat. Methods 2017, 14, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W.; Siuzdak, G.; et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Grün, D.; Lyubimova, A.; Kester, L.; Wiebrands, K.; Basak, O.; Sasaki, N.; Clevers, H.; van Oudenaarden, A. Single-cell messenger rna sequencing reveals rare intestinal cell types. Nature 2015, 525, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Giustacchini, A.; Thongjuea, S.; Barkas, N.; Woll, P.S.; Povinelli, B.J.; Booth, C.A.G.; Sopp, P.; Norfo, R.; Rodriguez-Meira, A.; Ashley, N.; et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017, 23, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Proserpio, V.; Lönnberg, T. Single-cell technologies are revolutionizing the approach to rare cells. Immun. Cell Biol. 2016, 94, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Navin, N.E. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015, 25, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Saadatpour, A.; Lai, S.; Guo, G.; Yuan, G.-C. Single-cell analysis in cancer genomics. Trends Genet. 2015, 31, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Courtois, E.T.; Sengupta, D.; Tan, Y.; Chen, K.H.; Goh, J.J.L.; Kong, S.L.; Chua, C.; Hon, L.K.; Tan, W.S.; et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017, 49, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. Atac-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 109. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Grenci, G.; Jing, T.; Lim, Y.B.; Lee, S.C.; Thiery, J.P.; Han, J.; Lim, C.T. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
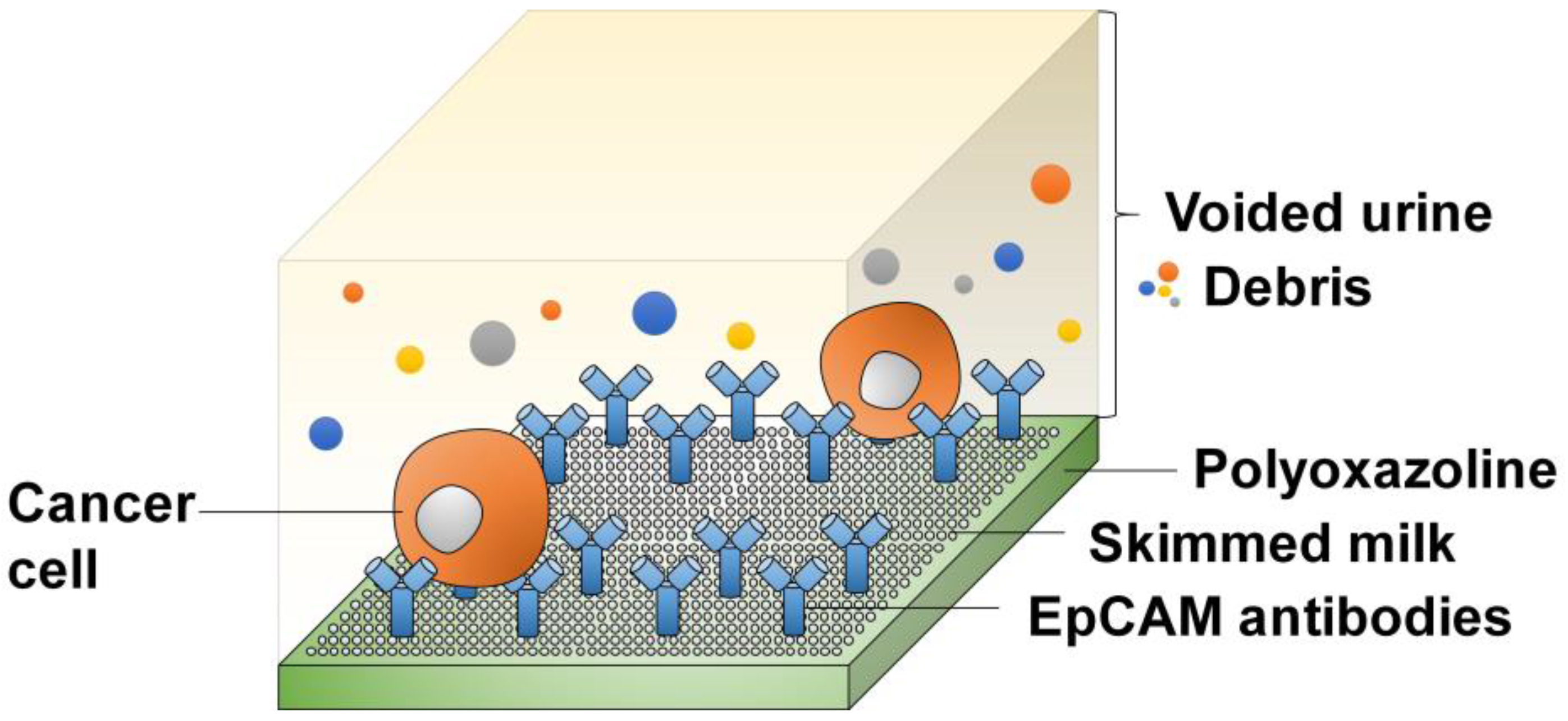
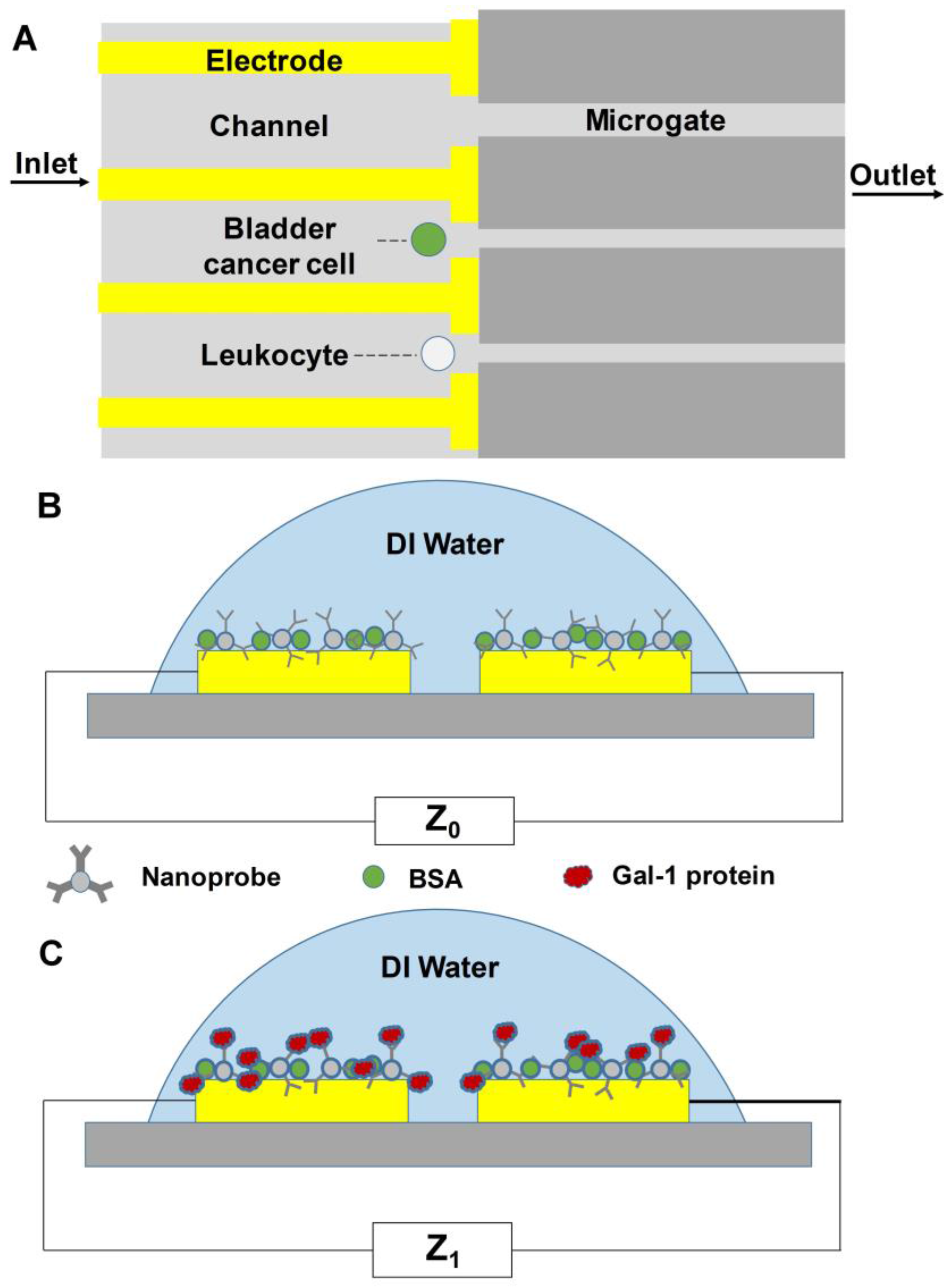
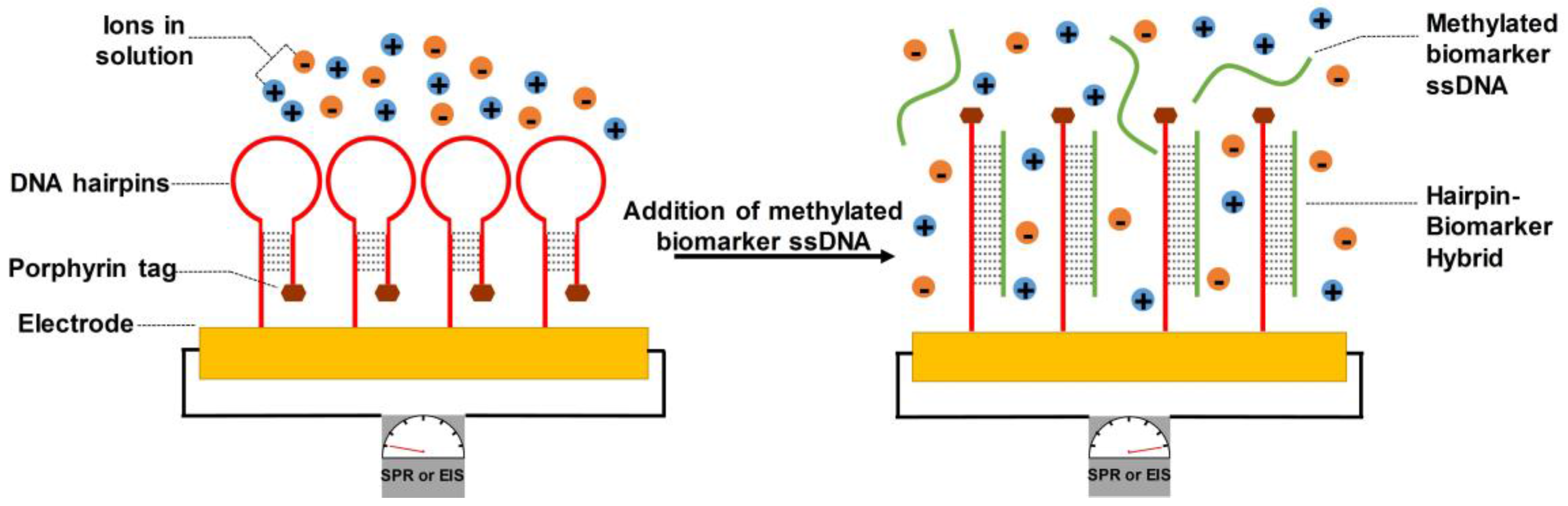
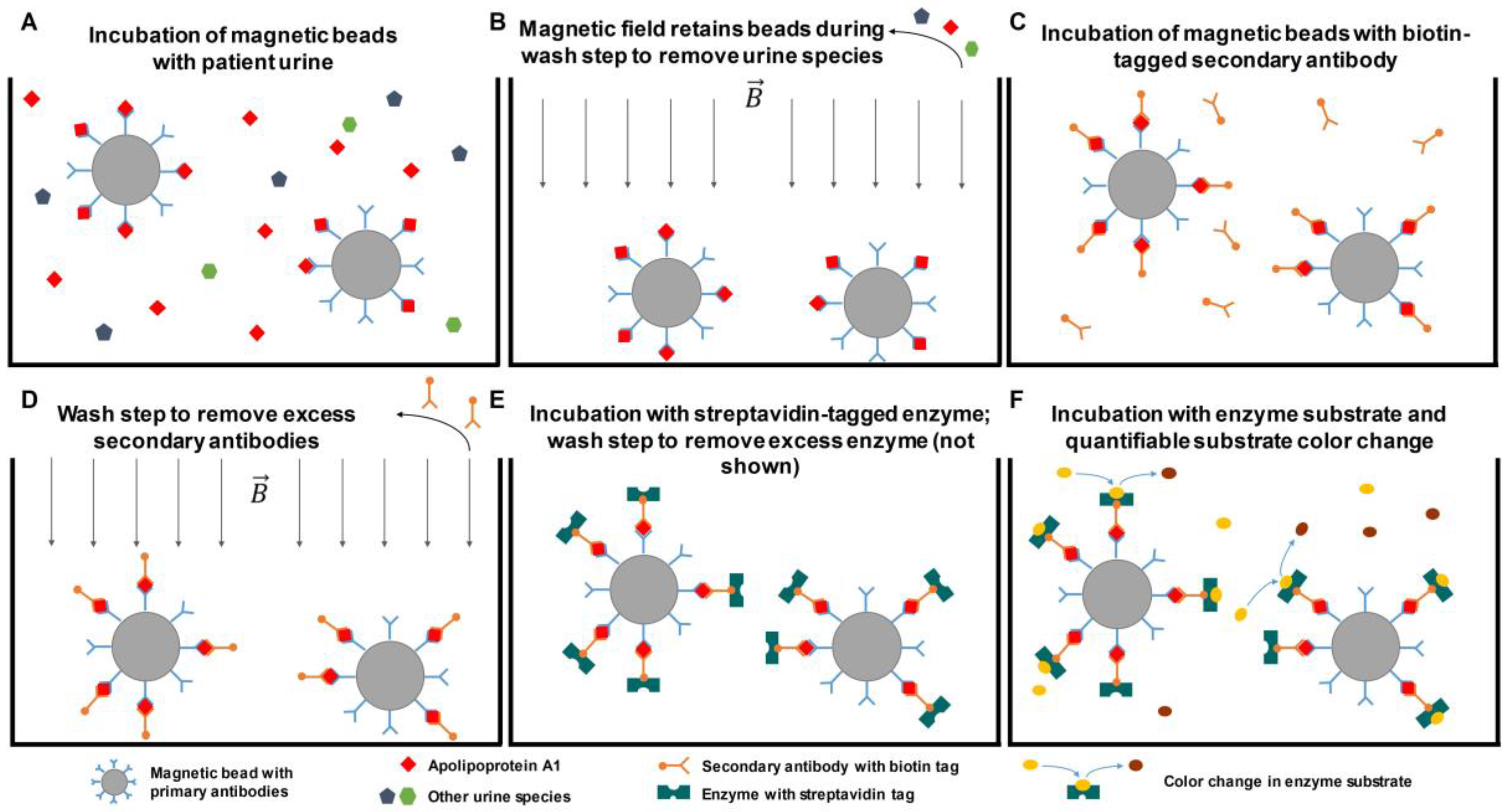
| Methods | Description of Diagnosis Method | Targeted Biomarkers/Genes |
|---|---|---|
| Urine cytology | Examine microscopically urinary sediment for the presence of tumor cells [41,42]. | N/A |
| Cystoscopy | Detect growths in the bladder and determine the need for a biopsy or surgery with the use of a cystoscope [42]. | N/A |
| Computed tomography (CT) | Show abnormalities or tumors in a detailed, cross-sectional view using X-ray. Measure the tumor size [43]. | N/A |
| Magnetic resonance imaging (MRI) | Produce detailed images of the tumor using a magnetic field. Measure the tumor size [43]. | N/A |
| Immunohistochemical Staining | Visualize protein presence in excised tissue via primary antibodies and secondary antibodies with fluorescent tags. | p21, p53, pRB, and p27 [44] Bcl-2 and caspase-3 [45] pAkt, PTEN, Drg-1, Cx-26, and L-plastin [46] CXCL16 and CXCR6 [47] CXCR4 [48] |
| CxBladder Detect | Test for the presence of mRNA associated with cancer-linked genes | IGF, HOXA, MDK, CDC, and IL8R [49] |
| UroVysion | Detect aneuploidy and loss of loci from patient’s urine sample via fluorescence in situ hybridization (FISH). | 9p21 loci, amplification of chromosomes 3, 7 and 17 [50] |
| BTA Trak and BTA Stat | Measure protein in the urine, quantitatively via ELISA (BTA Trak) or qualitatively via inspection for immobilized antibodies (BTA Stat). | Human complement factor H-related protein [51,52] |
| BladderChek | Measure protein in urine, quantitatively via ELISA or qualitatively via immunochromatography (BladderChek). | Nuclear matrix protein-22 [53,54] |
| ImmunoCyt/uCyt+ | Identify presence of high-molecular-weight glycosylated carcinoembryonic antigens and mucins via fluorescent antibody binding. | 19A211, LDQ10, and M344 [20] |
| Ref | [77] | [78] | [63] | [79] | [80] | [81] |
|---|---|---|---|---|---|---|
| Detection principle | Antibody capture on magnetic microbead | DNA hairpins bound to electrode | Membrane capacitance difference | Galectin-1 protein via immunosensor | Antibody capture | Size filtration of EVs |
| Urine samples | Yes | No | Yes | Yes | Yes | Yes |
| Processing rate/sample | 14.5 μL/40 min | 5 μL/20 min | 5 mL/60 min | 100 μL, 120 min | 1 mL, 15 min | 2 samples per run, 1 mL, 30 min |
| Sensitivity/% | ND | ND | ND | ND | >88% | ND |
| Lower Detection Limit | 10 ng antibody/mL urine | 250 fM biomarker DNA | ND | 0.0078 mg/mL of T24 cell lysate | ND | ND |
| Upper Detection Limit | 2000 ng antibody/mL urine | 100 nM biomarker DNA | ND | ND | ND | ND |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tzouanas, C.; Lim, J.S.Y.; Wen, Y.; Thiery, J.P.; Khoo, B.L. Microdevices for Non-Invasive Detection of Bladder Cancer. Chemosensors 2017, 5, 30. https://doi.org/10.3390/chemosensors5040030
Tzouanas C, Lim JSY, Wen Y, Thiery JP, Khoo BL. Microdevices for Non-Invasive Detection of Bladder Cancer. Chemosensors. 2017; 5(4):30. https://doi.org/10.3390/chemosensors5040030
Chicago/Turabian StyleTzouanas, Constantine, Joey Sze Yun Lim, Ya Wen, Jean Paul Thiery, and Bee Luan Khoo. 2017. "Microdevices for Non-Invasive Detection of Bladder Cancer" Chemosensors 5, no. 4: 30. https://doi.org/10.3390/chemosensors5040030





