Identification of Chiral-Specific Carbon Nanotube Binding Peptides Using a Modified Biopanning Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phage Display with CNT Target
2.2. Phage Display Sans Target
2.3. Sanger Sequencing
2.4. High Throughput Sequencing
2.5. Dispersion Experiment
2.6. Gold Decoration Experiment
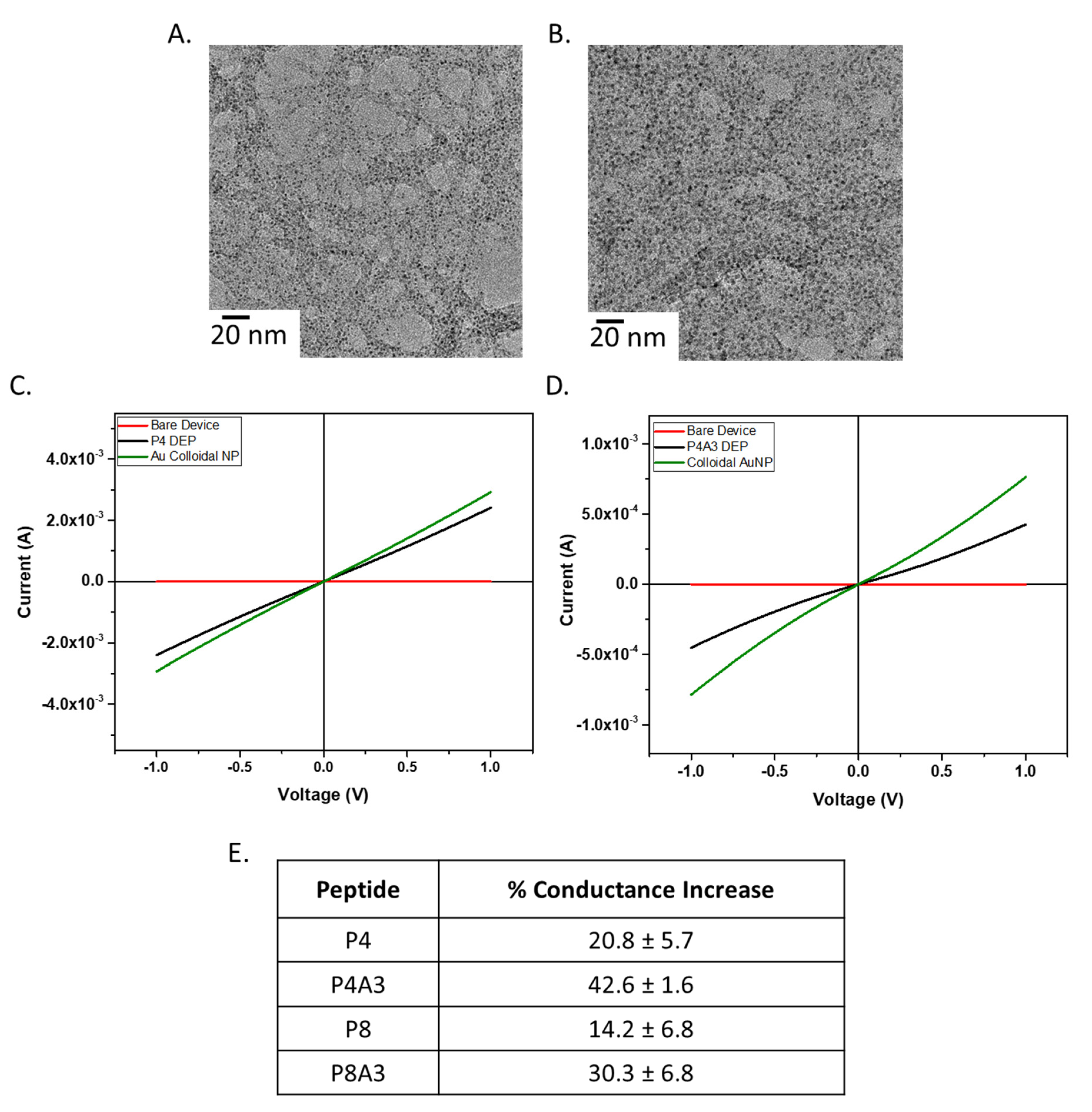
2.6.1. Characterization by Transmission Electron Microscopy (TEM)/Scanning Electron Microscopy (SEM)
2.6.2. Electrical Measurements by Dielectrophoresis (DEP)
3. Results and Discussion
3.1. Identification of Peptides
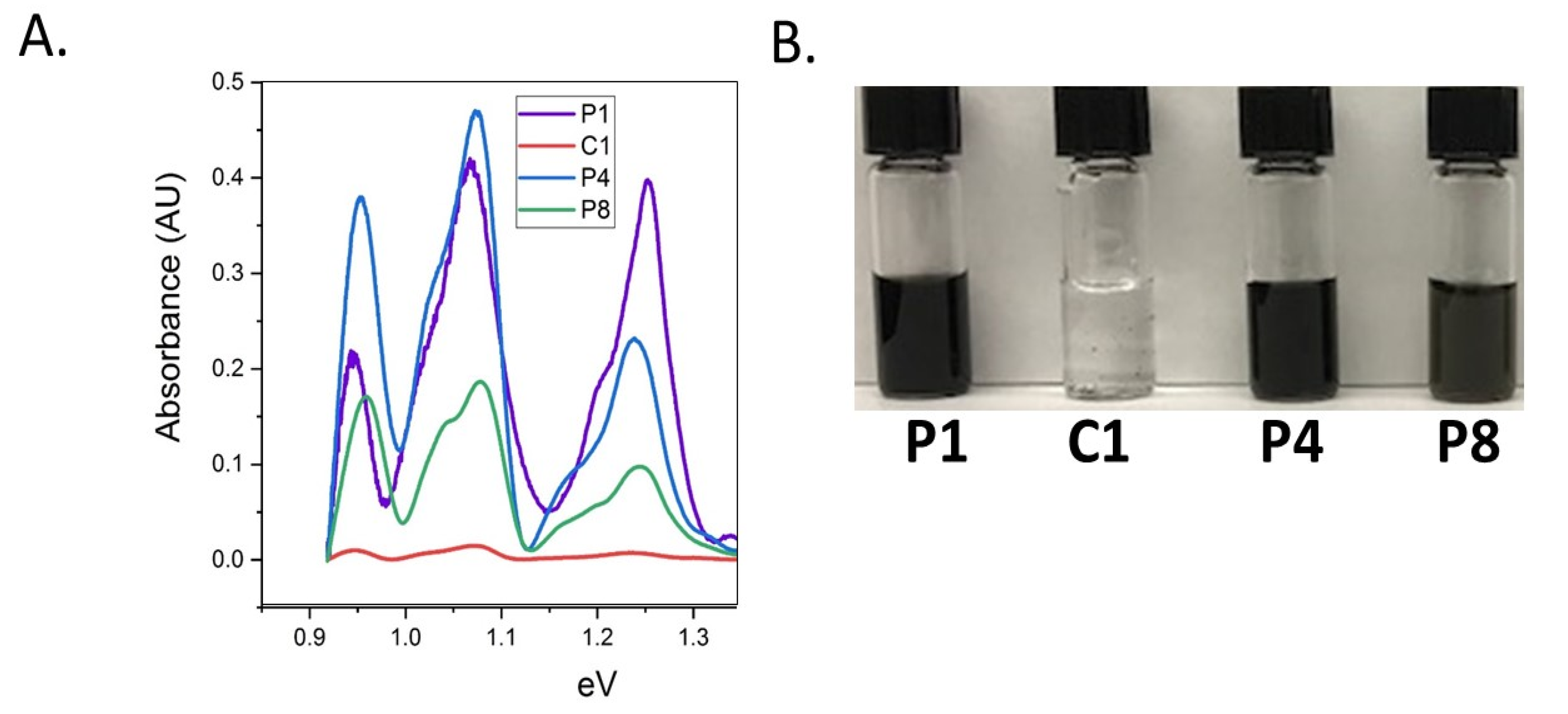
3.2. Peptide CNT Dispersion: Binding Characterization
3.3. SWNT Functionalized with Gold Nanoparticles (AuNP) Using Multifunctional Peptides
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Traditional Method: | Unique Method: | ||||
|---|---|---|---|---|---|
| Sanger | HTS | NHTS-PD | |||
| CoMoCAT | HiPCO | CoMoCAT | HiPCO | CoMoCAT | HiPCO |
| TAKYLPMRPGPL | TAKYLPMRPGPL | TAKYLPMRPGPL | TAKYLPMRPGPL | AVYQQSKVLTAR | GSVQKLSATPWV |
| QASDSLRSVGPG | QASDSLRSVGPG | AVYQQSKVLTAR | TANYLPMRPGPL | GSVQKLSATPWV | GLRETQVTALQK |
| TAKYLPMRPGPL | TFNTEAMWTSWP * | GSVQKLSATPWV | TAKYLPLRPGPL | SVDGWLEPPTST | LADNAFAHRQRC |
| SAKYLPMRPGPL * | WEGGGAGKLSAA * | SVDGWLEPPTST | TAQYLPMRPGPL | ALNWTELHGQAT | MGNVSHQPIPLP |
| DVAKSYQSFTNE * | AVYQQSKVLTAR* | TANYLPMRPGPL | TAKYLPMRPVPL | DAATQGVLARHY | SHVDQMHRPHRP |
| HLEALSDLVNRN * | FNDGSSKPLDDL * | ALNWTELHGQAT | TAKYLPMRPGLL | TPYVTHYSLNPF | YGHLEPRTPREL |
| DAATQGVLARHY * | ALNWTELHGQAT * | DAATQGVLARHY | TAKYLPMRPWPL | NSYASPLEPKVS | VPDRLVAYPIVL |
| GLGPALHHNIVA * | SPHSAASVLASL * | TAKYLPLRPGPL | TARYLPMRPGPL | FNDGSSKPLDDL | IANPSLFPPMSM |
| ID | Peptide Sequence | Identified |
|---|---|---|
| P1 | HSSYWYAFNNKT | HTS-PD Literature to CNT |
| C1 | TAKYLPMRPGPL | Traditional to CNT |
| P4 | TPYVTHYSLNPF | NHTS-PD to CoMoCAT |
| P8 | LADNAFAHRQRC | NHTS-PD to HiPCO |
| Peptide | Average Resistance (Ohms) |
|---|---|
| P4 | 342 ± 36.1 Ω |
| P4A3 | 2065 ± 367.8 Ω |
| P8 | 560 ± 200.1 Ω |
| P8A3 | 1128 ± 292.5 Ω |
References
- Zheng, M.; Semke, E.D. Enrichment of Single Chirality Carbon Nanotubes. Amer. Chem. Soc. 2007, 129, 6084–6085. [Google Scholar] [CrossRef]
- Zheng, M.; Jagota, A.; Semke, E.D.; Diner, B.A.; McLean, R.S.; Lustig, S.R.; Richardson, R.E.; Tassi, N.G. DNA-Assisted Dispersion and Separation of Carbon Nanotubes. Nat. Mater. 2003, 2, 338–342. [Google Scholar] [CrossRef]
- Tromp, R.M.; Afzali, A.; Freitag, M.; Mitzi, D.B.; Chen, Z. Novel Strategy for Diameter-Selective Separation and Functionali-zation of Single-Wall Carbon Nanotubes. Nano Lett. 2008, 8, 469–472. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.J.; Bachilo, S.M.; Huffman, C.B.; Moore, V.C.; Strano, M.S.; Haroz, E.H.; Rialon, K.L.; Boul, P.J.; Noon, W.H.; Kittrell, C.; et al. Band Gap Fluorescence From Individual Single-Walled Carbon Nanotubes. Science 2002, 297, 593–596. [Google Scholar] [CrossRef] [Green Version]
- Kuang, Z.; Kim, S.N.; Goodson, W.; Farmer, B.L.; Naik, R.R. Biomimetic Chemosensor: Designing Peptide Recognition Elements for Surface Functionalization of Carbon Nanotube Field Effect Transistors. ACS Nano 2010, 4, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Politi, J.; Zappavigna, S.; Rea, I.; Grieco, P.; Caliò, A.; Luce, A.; Caraglia, M.; De Stefano, L. Peptide Functionalization of Silicon for De-tection and Classification of Prostatic Cells. J. Sens. 2017, 2017, 6792396. [Google Scholar] [CrossRef] [Green Version]
- Ermini, M.L.; Song, X.C.; Špringer, T.; Homola, J. Peptide Functionalization of Gold Nanoparticles for the Detection of Carcinoembryonic Antigen in Blood Plasma via SPR-Based Biosensor. Front. Chem. 2019, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Sim, D.; Krabacher, R.; Chávez, J.L.; Martin, J.A.; Islam, A.E.; Kuang, Z.; Maruyama, B.; Naik, R.R.; Kim, S.S. Peptide-Functionalized Single-Walled Carbon Nanotube Field-Effect Transistors for Monitoring Volatile Organic Compounds in Breath. In Proceedings of the IEEE International Flexible Electronics Technology Conference, Vancouver, BC, Canada, 11–14 August 2019. [Google Scholar]
- Sapsford, K.E.; Algar, W.R.; Berti, L.; Gemmill, K.B.; Casey, B.J.; Oh, E.; Stewart, M.H.; Medintz, I.L. Functionalizing Nanoparticles with Biological Molecules: Developing Chemistries that Facilitate Nanotechnology. Chem. Rev. 2013, 113, 1904–2074. [Google Scholar] [CrossRef]
- Pender, M.J.; Sowards, L.A.; Hartgerink, J.D.; Stone, M.O.; Naik, R.R. Peptide-Mediated Formation of Single-Wall Carbon Nanotube Composites. Nano Lett. 2006, 6, 40–44. [Google Scholar] [CrossRef]
- Cui, Y.; Kim, S.N.; Naik, R.R.; McAlpine, M.C. Biomimetic Peptide Nanosensors. Acc. Chem. Res. 2011, 45, 696–704. [Google Scholar] [CrossRef]
- Naik, R.R.; Jones, S.E.; Murray, C.J.; McAuliffe, J.C.; Vaia, R.A.; Stone, M.O. Peptide Templates for Nanoparticle Synthesis Derived from Polymerase Chain Reaction-Driven Phage Display. Adv. Funct. Mater. 2004, 14, 25. [Google Scholar] [CrossRef]
- Van Dorst, B.; Mehta, J.; Rouah-Martin, E.; Blust, R.; Robbens, J. Phage Display as a Method for Discovering Cellular Targets of Small Molecules. Methods 2012, 58, 56–61. [Google Scholar] [CrossRef]
- Qianglong, Z.; Shi, L.; Peng, G.; Fei-Shi, L. High-throughput sequencing technology and its application. J. Northeast. Agric. Univ. 2014, 21, 84–96. [Google Scholar]
- Kukovecz, A.; Kramberger, C.; Georgakilas, V.; Prato, M.; Kuzmany, H. A detailed Raman study on thin single-wall carbon nanotubes prepared by the HiPCO process. Eur. Phys. J. B-Condens. Matter Complex Syst. 2002, 28, 223–230. [Google Scholar] [CrossRef]
- Bachilo, S.M.; Balzano, L.; Herrera, J.E.; Pompeo, F.; Resasco, D.E.; Weisman, R.B. Narrow (n,m)-Distribution of Single-Walled Carbon Nanotubes Grown Using a Solid Supported Catalyst. J. Am. Chem. Soc. 2003, 125, 11186–11187. [Google Scholar] [CrossRef]
- Chen, Y.; Ciuparu, D.; Lim, S.; Yang, Y.; Haller, G.L.; Pfefferle, L. Synthesis of Uniform Diameter Single-Wall Carbon Nanotubes in Co-MCM-41: Effects of the Catalyst Prereduction and Nanotube Growth Temperatures. J. Catal. 2004, 225, 453–465. [Google Scholar] [CrossRef]
- Luo, Z.; Pfefferle, L.D.; Haller, G.L.; Papadimitrakopoulos, F. (n, m) Abundance Evaluation of Single-Walled Carbon Nanotubes by Fluorescence and Absorption Spectroscopy. J. Am. Chem. Soc. 2006, 128, 15511–15516. [Google Scholar] [CrossRef]
- Kitiyanan, B.; Alvarez, W.; Harwell, J.; Resasco, D. Controlled production of single-wall carbon nanotubes by catalytic decomposition of CO on bimetallic Co-Mo catalysts. Chem. Phys. Let. 2000, 317, 497–503. [Google Scholar] [CrossRef]
- He, Z.; Zhou, J. Probing Carbon Nanotube-Amino Acid Interactions in Aqueous Solution with Molecular Dynamics Simulations. Carbon 2014, 78, 500–509. [Google Scholar] [CrossRef]
- LeMieux, M.C.; Roberts, M.; Barman, S.; Jin, Y.W.; Kim, J.M.; Bao, Z. Self-Sorted, Aligned Nanotube Networks for Thin-Film Transistors. Science 2008, 321, 101–104. [Google Scholar] [CrossRef]
- Brown, S.; Jespersen, T.; Nygård, J. A Genetic Analysis of Carbon-Nanotube-Binding Proteins. Small 2008, 4, 416–420. [Google Scholar] [CrossRef]
- Su, Z.; Leung, T.; Honek, J.F. Conformational Selectivity of Peptides for Single-Walled Carbon Nano-tubes. J. Phys. Chem. B 2006, 110, 23623–23627. [Google Scholar] [CrossRef]
- Kim, S.N.; Singh, K.M.; Ouchen, F.; Grote, J.G.; Naik, R.R. DNA Mediated Solubilization of Single Wall Carbon Nano-tubes. In Proceedings of the 2008 IEEE National Aerospace and Electronics Conference, Dayton, OH, USA, 16–18 July 2008; pp. 94–96. [Google Scholar]
- Rastogi, R.; Kaushal, R.; Tripathi, S.K.; Sharma, A.L.; Kaur, I.; Bharadwaj, L.M. Comparative Study of Carbon Nanotube Dispersion Using Surfactants. J. Colloid Interface Sci. 2008, 328, 421–428. [Google Scholar] [CrossRef]
- Li, Z.; Kameda, T.; Isoshima, T.; Kobatake, E.; Tanaka, T.; Ito, Y.; Kawamoto, M. Solubilization of Single-Walled Carbon Nanotubes Using a Peptide Aptamer in Water below the Critical Micelle Concentration. Langmuir 2015, 31, 3482–3488. [Google Scholar] [CrossRef] [PubMed]
- Pantarotto, D.; Partidos, C.D.; Hoebeke, J.; Brown, F.; Kramer, E.; Briand, J.-P.; Muller, S.; Prato, M.; Bianco, A. Immunization with Peptide-Functionalized Carbon Nanotubes Enhances Virus-Specific Neutralizing Antibody Responses. Chem. Biol. 2003, 10, 961–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinals, R.L.; Ledesma, F.; Yang, D.; Navarro, N.; Jeong, S.; Pak, J.E.; Kuo, L.; Chuang, Y.-C.; Cheng, Y.-W.; Sun, H.-Y.; et al. Rapid SARS-CoV-2 spike protein detection by carbon nanotube-based near-infrared nanosensors. Nano Lett. 2021, 21, 2272–2280. [Google Scholar] [CrossRef] [PubMed]
- Tohidifar, L.; Hadipour, N.L. Tracing chirality, diameter dependence, and temperature-controlling of single-walled carbon nanotube non-covalent functionalization by biologically compatible peptide: Insights from molecular dynamics simulations. J. Mol. Modeling 2019, 25, 274. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Dieckmann, G.R.; Nielsen, S.O. Role of Peptide-Peptide Interactions in Stabilizing Peptide-Wrapped Single-Walled Carbon Nanotubes: A Molecular Dynamics Study. Pept. Sci. Orig. Res. Biomol. 2009, 92, 156–163. [Google Scholar] [CrossRef]
- Kim, S.N.; Kuang, Z.; Slocik, J.M.; Jones, S.E.; Cui, Y.; Farmer, B.L.; McAlpine, M.C.; Naik, R.R. Preferential Binding of Peptides to Graphene Edges and Planes. J. Am. Chem. Soc. 2011, 133, 14480–14483. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.H.; Wang, Q.; Collins, F. Dispersion of Carbon Nanotubes with SDS Surfactants: A Study From a Binding Energy Perspective. Chem. Sci. 2011, 2, 1407–1413. [Google Scholar] [CrossRef]
- Kim, S.N.; Singh, K.; Ouchen, F.; Grote, J.G.; Naik, R.R. Solubilization of Single Wall Carbon Nanotubes with Salmon Sperm DNA. TechConnect Briefs. 2008, 1, 132–135. [Google Scholar]
- Miyata, Y.; Yanagi, K.; Maniwa, Y.; Kataura, H. Optical Evaluation of the Metal-to-Semiconductor Ratio of Single-Wall Carbon Nanotubes. J. Phys. Chem. 2008, 112, 13187–13191. [Google Scholar] [CrossRef]
- Gravely, M.; Safaee, M.M.; Roxbury, D. Oligonucleotide Length Determines Intracellular Stability of DNA-Wrapped Carbon Nanotubes. bioRxiv 2019, 642413. [Google Scholar] [CrossRef]
- Oyama, Y.; Saito, R.; Sato, K.; Jiang, J.; Samsonidze, G.; Grüneis, A.; Miyauchi, Y.; Maruyama, S.; Jorio, A.; Dresselhaus, G.; et al. Photoluminescence Intensity of Single Wall Carbon Nanotubes. Carbon 2006, 44, 873–879. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krabacher, R.; Kim, S.; Ngo, Y.; Slocik, J.; Harsch, C.; Naik, R. Identification of Chiral-Specific Carbon Nanotube Binding Peptides Using a Modified Biopanning Method. Chemosensors 2021, 9, 245. https://doi.org/10.3390/chemosensors9090245
Krabacher R, Kim S, Ngo Y, Slocik J, Harsch C, Naik R. Identification of Chiral-Specific Carbon Nanotube Binding Peptides Using a Modified Biopanning Method. Chemosensors. 2021; 9(9):245. https://doi.org/10.3390/chemosensors9090245
Chicago/Turabian StyleKrabacher, Rachel, Steve Kim, Yen Ngo, Joseph Slocik, Christina Harsch, and Rajesh Naik. 2021. "Identification of Chiral-Specific Carbon Nanotube Binding Peptides Using a Modified Biopanning Method" Chemosensors 9, no. 9: 245. https://doi.org/10.3390/chemosensors9090245






