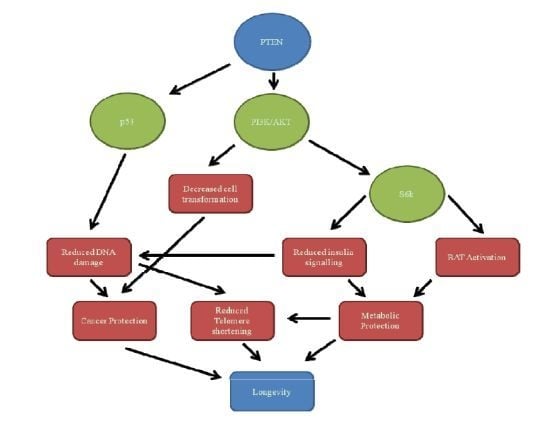
PTEN, Longevity and Age-Related Diseases
Abstract
:1. Introduction
2. PTEN Regulations
2.1. Regulates PTEN
2.1.1. Mutations
2.1.2. Transcriptional Regulation
2.1.3. Post-Translational Regulation
| PTEN regulators | Effects |
|---|---|
| Haploinsuffiency | Reduced PTEN function, higher susceptibility to tumours [16] |
| C2-domain mutations | Negatively affect PTEN stability, phosphorylation, protein interactions and proper localization [17,18,19] |
| N-terminal mutations | Negatively affect protein stability [17] |
| PPARγ | Up-regulation of PTEN transcription [25] |
| EGR-1 | Up-regulation of PTEN mRNA leading to apoptosis [28] |
| Ras | Down-regulates PTEn via c-Jun, induces PI3k-AKT pathway [33] |
| Promoter hypermethylation | Possible inhibition of PTEN expression [35] |
| STT phosphorylation | Significantly negatively affect PTEN stability while positively affecting phosphatase activity [21,22] |
| mir-21 | Down-regulates PTEN transcription [41,42,43,44,45] |
| PICT1 | Phosphorylates PTEN C-terminal, increases stability [21,47,48] |
| Phosphorylation of T366, S370, S380, T382-3, S385 | Affects protein stability and decreases PTEN’s ability to interact with membranes [49] |
| Chk1 | Induces CK2-mediated phosphorylation of PTEN, recovers stalled cell cycle [48,50] |
| ROCK | Phosphorylates PTEN to increase PTEN localization to membrane [51] |
| PEST-sequences | Destabilizes PTEN [48] |
| NEDD4-1 | Ubiquitinates PTEN via L13, L289 [19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56] |
| PCAF | Promotes PTEN acetylation to decrease PTEN catalytic activity [57] |
| ROS | Decreases PTEN catalytic activity [58] |
| NHERF & MAGI-2 | Regulates PTEN localization and recruitment to the membrane [47,61] |
| NFκB | Down-regulates PTEN transcription [63] |
2.2. Regulated by PTEN
2.2.1. In the Cytoplasm
2.2.2. In the Nucleus
| Regulated by PTEN | Effects |
|---|---|
| PIP3/AKT pathway | Increased longevity [73,74,75,76], reduced insulin signalling [77], tumour suppression [78], reduced DNA damage [79] |
| JNK pathway | Downregulated via phosphorylation PI3K [65] |
| p53 | Increased stability and transcription of p53 [67] |
| CENP-C | Binds with PTEN, maintains chromosomal integrity [72] |
| Rad51 | Positively regulated via E2F-1, repairs DSB [72] |
3. Cell Functions

3.1. PI3K
3.2. Membrane
3.3. Nucleus
3.3.1. Localization
3.3.2. Nuclear Interactions
3.4. DNA Damage
3.5. Stem Cells
3.6. Senescence/Apoptosis
3.7. Caloric Restriction
4. Cancer Properties
4.1. Mutations
4.1.1. Complete Loss/Haploinsufficiency
4.1.2. Germline Mutations
4.1.3. Somatic Mutations
4.2. Regulatory Effects Causing Cancer
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Li, J.; Yen, C.; Liaw, D.; Podsypanina, K.; Bose, S.; Wang, S.I.; Puc, J.; Miliaresis, C.; Rodgers, L.; McCombie, R.; et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Steck, P.A.; Pershouse, M.A.; Jasser, S.A.; Yung, W.K.; Lin, H.; Ligon, A.H.; Langford, L.A.; Baumgard, M.L.; Hattier, T.; Davis, T.; et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997, 15, 356–362. [Google Scholar] [CrossRef]
- Ali, I.U.; Schriml, L.M.; Dean, M. Mutational spectra of PTEN/MMAC1 gene: A tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 1999, 91, 1922–1932. [Google Scholar] [CrossRef]
- Ortega-Molina, A.; Efeyan, A.; Lopez-Guadamillas, E.; Munoz-Martin, M.; Gomez-Lopez, G.; Canamero, M.; Mulero, F.; Pastor, J.; Martinez, S.; Romanos, E.; et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell. Metab. 2012, 15, 382–394. [Google Scholar] [CrossRef]
- Sulis, M.L.; Parsons, R. PTEN: From pathology to biology. Trends Cell Biol. 2003, 13, 478–483. [Google Scholar] [CrossRef]
- Chen, H.; Rossier, C.; Morris, M.A.; Scott, H.S.; Gos, A.; Bairoch, A.; Antonarakis, S.E. A testis-specific gene, TPTE, encodes a putative transmembrane tyrosine phosphatase and maps to the pericentromeric region of human chromosomes 21 and 13, and to chromosomes 15, 22, and Y. Hum. Genet. 1999, 105, 399–409. [Google Scholar] [CrossRef]
- Wu, Y.; Dowbenko, D.; Pisabarro, M.T.; Dillard-Telm, L.; Koeppen, H.; Lasky, L.A. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J. Biol. Chem. 2001, 276, 21745–21753. [Google Scholar] [CrossRef]
- Walker, S.M.; Downes, C.P.; Leslie, N.R. TPIP: A novel phosphoinositide 3-phosphatase. Biochem. J. 2001, 360, 277–283. [Google Scholar] [CrossRef]
- Lee, J.O.; Yang, H.; Georgescu, M.M.; di Cristofano, A.; Maehama, T.; Shi, Y.; Dixon, J.E.; Pandolfi, P.; Pavletich, N.P. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999, 99, 323–334. [Google Scholar] [CrossRef]
- Maehama, T.; Dixon, J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef]
- Ashcroft, M.; Ludwig, R.L.; Woods, D.B.; Copeland, T.D.; Weber, H.O.; MacRae, E.J.; Vousden, K.H. Phosphorylation of HDM2 by Akt. Oncogene 2002, 21, 1955–1962. [Google Scholar] [CrossRef]
- Chang, C.J.; Mulholland, D.J.; Valamehr, B.; Mosessian, S.; Sellers, W.R.; Wu, H. PTEN nuclear localization is regulated by oxidative stress and mediates p53-dependent tumor suppression. Mol. Cell. Biol. 2008, 28, 3281–3289. [Google Scholar] [CrossRef]
- Ming, M.; Feng, L.; Shea, C.R.; Soltani, K.; Zhao, B.; Han, W.; Smart, R.C.; Trempus, C.S.; He, Y.Y. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 2011, 71, 5287–5295. [Google Scholar] [CrossRef]
- Jaskelioff, M.; Muller, F.L.; Paik, J.H.; Thomas, E.; Jiang, S.; Adams, A.C.; Sahin, E.; Kost-Alimova, M.; Protopopov, A.; Cadinanos, J.; et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 2011, 469, 102–106. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef]
- Kwabi-Addo, B.; Giri, D.; Schmidt, K.; Podsypanina, K.; Parsons, R.; Greenberg, N.; Ittmann, M. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. USA 2001, 98, 11563–11568. [Google Scholar] [CrossRef]
- Han, S.Y.; Kato, H.; Kato, S.; Suzuki, T.; Shibata, H.; Ishii, S.; Shiiba, K.; Matsuno, S.; Kanamaru, R.; Ishioka, C. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000, 60, 3147–3151. [Google Scholar]
- Waite, K.A.; Eng, C. Protean PTEN: Form and function. Am. J. Hum. Genet. 2002, 70, 829–844. [Google Scholar] [CrossRef]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef]
- Salmena, L.; Carracedo, A.; Pandolfi, P.P. Tenets of PTEN tumor suppression. Cell 2008, 133, 403–414. [Google Scholar] [CrossRef]
- Vazquez, F.; Ramaswamy, S.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000, 20, 5010–5018. [Google Scholar] [CrossRef]
- Leslie, N.R.; Downes, C.P. PTEN function: How normal cells control it and tumour cells lose it. Biochem. J. 2004, 382, 1–11. [Google Scholar] [CrossRef]
- Vazquez, F.; Grossman, S.R.; Takahashi, Y.; Rokas, M.V.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001, 276, 48627–48630. [Google Scholar] [CrossRef]
- Pilarski, R.; Eng, C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J. Med. Genet. 2004, 41, 323–326. [Google Scholar] [CrossRef]
- Patel, L.; Pass, I.; Coxon, P.; Downes, C.P.; Smith, S.A.; Macphee, C.H. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr. Biol. 2001, 11, 764–768. [Google Scholar] [CrossRef]
- Dionisi, M.; Alexander, S.P.; Bennett, A.J. Oleamide activates peroxisome proliferator-activated receptor gamma (PPARgamma) in vitro. Lipids Health Dis. 2012, 11, 51. [Google Scholar] [CrossRef]
- Kim, H.J.; Ham, S.A.; Kim, M.Y.; Hwang, J.S.; Lee, H.; Kang, E.S.; Yoo, T.; Woo, I.S.; Yabe-Nishimura, C.; Paek, K.S.; et al. PPARdelta coordinates angiotensin II-induced senescence in vascular smooth muscle cells through PTEN-mediated inhibition of superoxide generation. J. Biol. Chem. 2011, 286, 44585–44593. [Google Scholar] [CrossRef]
- Virolle, T.; Adamson, E.D.; Baron, V.; Birle, D.; Mercola, D.; Mustelin, T.; de Belle, I. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat. Cell Biol. 2001, 3, 1124–1128. [Google Scholar] [CrossRef]
- Sperandio, S.; Fortin, J.; Sasik, R.; Robitaille, L.; Corbeil, J.; de Belle, I. The transcription factor Egr1 regulates the HIF-1alpha gene during hypoxia. Mol. Carcinog. 2009, 48, 38–44. [Google Scholar] [CrossRef]
- Moorehead, R.A.; Hojilla, C.V.; de Belle, I.; Wood, G.A.; Fata, J.E.; Adamson, E.D.; Watson, K.L.; Edwards, D.R.; Khokha, R. Insulin-like growth factor-II regulates PTEN expression in the mammary gland. J. Biol. Chem. 2003, 278, 50422–50427. [Google Scholar] [CrossRef]
- Codina, M.; Garcia de la serrana, D.; Sanchez-Gurmaches, J.; Montserrat, N.; Chistyakova, O.; Navarro, I.; Gutierrez, J. Metabolic and mitogenic effects of IGF-II in rainbow trout (Oncorhynchus mykiss) myocytes in culture and the role of IGF-II in the PI3K/Akt and MAPK signalling pathways. Gen. Comp. Endocrinol. 2008, 157, 116–124. [Google Scholar] [CrossRef]
- Marshall, C.J. Ras effectors. Curr. Opin. Cell Biol. 1996, 8, 197–204. [Google Scholar] [CrossRef]
- Hettinger, K.; Vikhanskaya, F.; Poh, M.K.; Lee, M.K.; de Belle, I.; Zhang, J.T.; Reddy, S.A.; Sabapathy, K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007, 14, 218–229. [Google Scholar] [CrossRef]
- Vasudevan, K.M.; Gurumurthy, S.; Rangnekar, V.M. Suppression of PTEN expression by NF-kappa B prevents apoptosis. Mol. Cell. Biol. 2004, 24, 1007–1021. [Google Scholar] [CrossRef]
- Garcia, J.M.; Silva, J.; Pena, C.; Garcia, V.; Rodriguez, R.; Cruz, M.A.; Cantos, B.; Provencio, M.; Espana, P.; Bonilla, F. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer 2004, 41, 117–124. [Google Scholar] [CrossRef]
- Goel, A.; Arnold, C.N.; Niedzwiecki, D.; Carethers, J.M.; Dowell, J.M.; Wasserman, L.; Compton, C.; Mayer, R.J.; Bertagnolli, M.M.; Boland, C.R. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004, 64, 3014–3021. [Google Scholar] [CrossRef]
- Kang, Y.H.; Lee, H.S.; Kim, W.H. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab. Invest. 2002, 82, 285–291. [Google Scholar]
- Mirmohammadsadegh, A.; Marini, A.; Nambiar, S.; Hassan, M.; Tannapfel, A.; Ruzicka, T.; Hengge, U.R. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006, 66, 6546–6552. [Google Scholar] [CrossRef]
- Zysman, M.A.; Chapman, W.B.; Bapat, B. Considerations when analyzing the methylation status of PTEN tumor suppressor gene. Am. J. Pathol. 2002, 160, 795–800. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Stewart, L.M.; Ajayi, L.; Gray, I.C.; Gray, N.E.; Roberts, K.G.; Watson, G.J.; Kaisary, A.V.; Snary, D. The expression profile for the tumour suppressor gene PTEN and associated polymorphic markers. Br. J. Cancer 2000, 82, 1671–1676. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Lang, M.; Wehbe, H.; Maheshwari, S.; Mendell, J.T.; Jiang, J.; Schmittgen, T.D.; Patel, T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006, 130, 2113–2129. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Okahara, F.; Ikawa, H.; Kanaho, Y.; Maehama, T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004, 279, 45300–45303. [Google Scholar] [CrossRef]
- Georgescu, M.M.; Kirsch, K.H.; Akagi, T.; Shishido, T.; Hanafusa, H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl. Acad. Sci. USA 1999, 96, 10182–10187. [Google Scholar] [CrossRef]
- Torres, J.; Pulido, R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001, 276, 993–998. [Google Scholar] [CrossRef]
- Maccario, H.; Perera, N.M.; Davidson, L.; Downes, C.P.; Leslie, N.R. PTEN is destabilized by phosphorylation on Thr366. Biochem. J. 2007, 405, 439–444. [Google Scholar] [CrossRef]
- Martin, S.A.; Ouchi, T. Cellular commitment to reentry into the cell cycle after stalled DNA is determined by site-specific phosphorylation of Chk1 and PTEN. Mol. Cancer Ther. 2008, 7, 2509–2516. [Google Scholar] [CrossRef]
- Li, Z.; Dong, X.; Wang, Z.; Liu, W.; Deng, N.; Ding, Y.; Tang, L.; Hla, T.; Zeng, R.; Li, L.; et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 2005, 7, 399–404. [Google Scholar] [CrossRef]
- Al-Khouri, A.M.; Ma, Y.; Togo, S.H.; Williams, S.; Mustelin, T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 2005, 280, 35195–35202. [Google Scholar] [CrossRef]
- Miller, S.J.; Lou, D.Y.; Seldin, D.C.; Lane, W.S.; Neel, B.G. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002, 528, 145–153. [Google Scholar] [CrossRef]
- Wu, W.; Wang, X.; Zhang, W.; Reed, W.; Samet, J.M.; Whang, Y.E.; Ghio, A.J. Zinc-induced PTEN protein degradation through the proteasome pathway in human airway epithelial cells. J. Biol. Chem. 2003, 278, 28258–28263. [Google Scholar] [CrossRef]
- Tang, Y.; Eng, C. p53 Down-regulates phosphatase and tensin homologue deleted on chromosome 10 protein stability partially through caspase-mediated degradation in cells with proteasome dysfunction. Cancer Res. 2006, 66, 6139–6148. [Google Scholar] [CrossRef]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef]
- Okumura, K.; Mendoza, M.; Bachoo, R.M.; DePinho, R.A.; Cavenee, W.K.; Furnari, F.B. PCAF modulates PTEN activity. J. Biol. Chem. 2006, 281, 26562–26568. [Google Scholar] [CrossRef]
- Lee, S.R.; Yang, K.S.; Kwon, J.; Lee, C.; Jeong, W.; Rhee, S.G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002, 277, 20336–20342. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, S.R.; Yang, K.S.; Ahn, Y.; Kim, Y.J.; Stadtman, E.R.; Rhee, S.G. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. USA 2004, 101, 16419–16424. [Google Scholar] [CrossRef]
- Leslie, N.R.; Bennett, D.; Lindsay, Y.E.; Stewart, H.; Gray, A.; Downes, C.P. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003, 22, 5501–5510. [Google Scholar] [CrossRef]
- Wu, X.; Hepner, K.; Castelino-Prabhu, S.; Do, D.; Kaye, M.B.; Yuan, X.J.; Wood, J.; Ross, C.; Sawyers, C.L.; Whang, Y.E. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc. Natl. Acad. Sci. USA 2000, 97, 4233–4238. [Google Scholar] [CrossRef]
- Takahashi, Y.; Morales, F.C.; Kreimann, E.L.; Georgescu, M.M. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006, 25, 910–920. [Google Scholar] [CrossRef]
- Xia, D.; Srinivas, H.; Ahn, Y.H.; Sethi, G.; Sheng, X.; Yung, W.K.; Xia, Q.; Chiao, P.J.; Kim, H.; Brown, P.H.; et al. Mitogen-activated protein kinase kinase-4 promotes cell survival by decreasing PTEN expression through an NF kappa B-dependent pathway. J. Biol. Chem. 2007, 282, 3507–3519. [Google Scholar]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef]
- Vivanco, I.; Palaskas, N.; Tran, C.; Finn, S.P.; Getz, G.; Kennedy, N.J.; Jiao, J.; Rose, J.; Xie, W.; Loda, M.; et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell 2007, 11, 555–569. [Google Scholar] [CrossRef]
- Stambolic, V.; MacPherson, D.; Sas, D.; Lin, Y.; Snow, B.; Jang, Y.; Benchimol, S.; Mak, T.W. Regulation of PTEN transcription by p53. Mol. Cell 2001, 8, 317–325. [Google Scholar] [CrossRef]
- Freeman, D.J.; Li, A.G.; Wei, G.; Li, H.H.; Kertesz, N.; Lesche, R.; Whale, A.D.; Martinez-Diaz, H.; Rozengurt, N.; Cardiff, R.D.; et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 2003, 3, 117–130. [Google Scholar] [CrossRef]
- Chen, Z.; Trotman, L.C.; Shaffer, D.; Lin, H.K.; Dotan, Z.A.; Niki, M.; Koutcher, J.A.; Scher, H.I.; Ludwig, T.; Gerald, W.; et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005, 436, 725–730. [Google Scholar] [CrossRef]
- Alimonti, A.; Nardella, C.; Chen, Z.; Clohessy, J.G.; Carracedo, A.; Trotman, L.C.; Cheng, K.; Varmeh, S.; Kozma, S.C.; Thomas, G.; et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J. Clin. Investig. 2010, 120, 681–693. [Google Scholar] [CrossRef]
- Li, A.G.; Piluso, L.G.; Cai, X.; Wei, G.; Sellers, W.R.; Liu, X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell 2006, 23, 575–587. [Google Scholar] [CrossRef]
- Puc, J.; Parsons, R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle 2005, 4, 927–929. [Google Scholar] [CrossRef]
- Shen, W.H.; Balajee, A.S.; Wang, J.; Wu, H.; Eng, C.; Pandolfi, P.P.; Yin, Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007, 128, 157–170. [Google Scholar] [CrossRef]
- Partridge, L.; Bruning, J.C. Forkhead transcription factors and ageing. Oncogene 2008, 27, 2351–2363. [Google Scholar] [CrossRef]
- Adler, A.S.; Sinha, S.; Kawahara, T.L.; Zhang, J.Y.; Segal, E.; Chang, H.Y. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 2007, 21, 3244–3257. [Google Scholar] [CrossRef]
- Salminen, A.; Ojala, J.; Huuskonen, J.; Kauppinen, A.; Suuronen, T.; Kaarniranta, K. Interaction of aging-associated signaling cascades: Inhibition of NF-kappaB signaling by longevity factors FoxOs and SIRT1. Cell. Mol. Life Sci. 2008, 65, 1049–1058. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K. NF-kappaB signaling in the aging process. J. Clin. Immunol. 2009, 29, 397–405. [Google Scholar] [CrossRef]
- Carracedo, A.; Pandolfi, P.P. The PTEN-PI3K pathway: Of feedbacks and cross-talks. Oncogene 2008, 27, 5527–5541. [Google Scholar] [CrossRef]
- Scanga, S.E.; Ruel, L.; Binari, R.C.; Snow, B.; Stambolic, V.; Bouchard, D.; Peters, M.; Calvieri, B.; Mak, T.W.; Woodgett, J.R.; et al. The conserved PI3'K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 2000, 19, 3971–3977. [Google Scholar] [CrossRef]
- Wang, Y. Bulky DNA lesions induced by reactive oxygen species. Chem. Res. Toxicol. 2008, 21, 276–281. [Google Scholar] [CrossRef]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009, 4, 127–150. [Google Scholar] [CrossRef]
- Backman, S.A.; Stambolic, V.; Suzuki, A.; Haight, J.; Elia, A.; Pretorius, J.; Tsao, M.S.; Shannon, P.; Bolon, B.; Ivy, G.O.; et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 2001, 29, 396–403. [Google Scholar]
- Cho, K.S.; Lee, J.H.; Kim, S.; Kim, D.; Koh, H.; Lee, J.; Kim, C.; Kim, J.; Chung, J. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 6144–6149. [Google Scholar] [CrossRef]
- Alessi, D.R.; Andjelkovic, M.; Caudwell, B.; Cron, P.; Morrice, N.; Cohen, P.; Hemmings, B.A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996, 15, 6541–6551. [Google Scholar]
- Brognard, J.; Sierecki, E.; Gao, T.; Newton, A.C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 2007, 25, 917–931. [Google Scholar] [CrossRef]
- Gao, T.; Furnari, F.; Newton, A.C. PHLPP: A phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 2005, 18, 13–24. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef]
- Guertin, D.A.; Sabatini, D.M. Defining the role of mTOR in cancer. Cancer Cell 2007, 12, 9–22. [Google Scholar] [CrossRef]
- Manning, B.D. Balancing Akt with S6K: Implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 2004, 167, 399–403. [Google Scholar] [CrossRef]
- Puig, O.; Tjian, R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005, 19, 2435–2446. [Google Scholar] [CrossRef]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef]
- Cardone, M.H.; Roy, N.; Stennicke, H.R.; Salvesen, G.S.; Franke, T.F.; Stanbridge, E.; Frisch, S.; Reed, J.C. Regulation of cell death protease caspase-9 by phosphorylation. Science 1998, 282, 1318–1321. [Google Scholar] [CrossRef]
- Zhou, B.P.; Liao, Y.; Xia, W.; Zou, Y.; Spohn, B.; Hung, M.C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001, 3, 973–982. [Google Scholar] [CrossRef]
- Mayo, L.D.; Donner, D.B. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA 2001, 98, 11598–11603. [Google Scholar] [CrossRef]
- Calnan, D.R.; Brunet, A. The FoxO code. Oncogene 2008, 27, 2276–2288. [Google Scholar] [CrossRef]
- Van Der Heide, L.P.; Hoekman, M.F.; Smidt, M.P. The ins and outs of FoxO shuttling: Mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 2004, 380, 297–309. [Google Scholar] [CrossRef]
- Huang, H.; Tindall, D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120, 2479–2487. [Google Scholar] [CrossRef]
- Van der Horst, A.; Burgering, B.M. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 440–450. [Google Scholar] [CrossRef]
- Peng, S.L. Foxo in the immune system. Oncogene 2008, 27, 2337–2344. [Google Scholar] [CrossRef]
- Das, S.; Dixon, J.E.; Cho, W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA 2003, 100, 7491–7496. [Google Scholar] [CrossRef]
- Deichmann, M.; Thome, M.; Benner, A.; Egner, U.; Hartschuh, W.; Naher, H. PTEN/MMAC1 expression in melanoma resection specimens. Br. J. Cancer 2002, 87, 1431–1436. [Google Scholar] [CrossRef]
- McMenamin, M.E.; Soung, P.; Perera, S.; Kaplan, I.; Loda, M.; Sellers, W.R. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999, 59, 4291–4296. [Google Scholar]
- Lachyankar, M.B.; Condon, P.J.; Daou, M.C.; de, A.K.; Levine, J.B.; Obermeier, A.; Ross, A.H. Novel functional interactions between Trk kinase and p75 neurotrophin receptor in neuroblastoma cells. J. Neurosci. Res. 2003, 71, 157–172. [Google Scholar] [CrossRef]
- Sano, T.; Lin, H.; Chen, X.; Langford, L.A.; Koul, D.; Bondy, M.L.; Hess, K.R.; Myers, J.N.; Hong, Y.K.; Yung, W.K.; et al. Differential expression of MMAC/PTEN in glioblastoma multiforme: Relationship to localization and prognosis. Cancer Res. 1999, 59, 1820–1824. [Google Scholar]
- Gimm, O.; Perren, A.; Weng, L.P.; Marsh, D.J.; Yeh, J.J.; Ziebold, U.; Gil, E.; Hinze, R.; Delbridge, L.; Lees, J.A.; et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 2000, 156, 1693–1700. [Google Scholar] [CrossRef]
- Tamura, M.; Gu, J.; Matsumoto, K.; Aota, S.; Parsons, R.; Yamada, K.M. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998, 280, 1614–1617. [Google Scholar] [CrossRef]
- Liliental, J.; Moon, S.Y.; Lesche, R.; Mamillapalli, R.; Li, D.; Zheng, Y.; Sun, H.; Wu, H. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 2000, 10, 401–404. [Google Scholar] [CrossRef]
- Raftopoulou, M.; Etienne-Manneville, S.; Self, A.; Nicholls, S.; Hall, A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science 2004, 303, 1179–1181. [Google Scholar] [CrossRef]
- Li, D.M.; Sun, H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997, 57, 2124–2129. [Google Scholar]
- Whang, Y.E.; Wu, X.; Suzuki, H.; Reiter, R.E.; Tran, C.; Vessella, R.L.; Said, J.W.; Isaacs, W.B.; Sawyers, C.L. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc. Natl. Acad. Sci. USA 1998, 95, 5246–5250. [Google Scholar] [CrossRef]
- Lian, Z.; di Cristofano, A. Class reunion: PTEN joins the nuclear crew. Oncogene 2005, 24, 7394–7400. [Google Scholar] [CrossRef]
- Lachyankar, M.B.; Sultana, N.; Schonhoff, C.M.; Mitra, P.; Poluha, W.; Lambert, S.; Quesenberry, P.J.; Litofsky, N.S.; Recht, L.D.; Nabi, R.; et al. A role for nuclear PTEN in neuronal differentiation. J. Neurosci. 2000, 20, 1404–1413. [Google Scholar]
- Perren, A.; Komminoth, P.; Saremaslani, P.; Matter, C.; Feurer, S.; Lees, J.A.; Heitz, P.U.; Eng, C. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 2000, 157, 1097–1103. [Google Scholar]
- Deleris, P.; Bacqueville, D.; Gayral, S.; Carrez, L.; Salles, J.P.; Perret, B.; Breton-Douillon, M. SHIP-2 and PTEN are expressed and active in vascular smooth muscle cell nuclei, but only SHIP-2 is associated with nuclear speckles. J. Biol. Chem. 2003, 278, 38884–38891. [Google Scholar] [CrossRef]
- Ginn-Pease, M.E.; Eng, C. Increased nuclear phosphatase and tensin homologue deleted on chromosome 10 is associated with G0–G1 in MCF-7 cells. Cancer Res. 2003, 63, 282–286. [Google Scholar]
- Perren, A.; Weng, L.P.; Boag, A.H.; Ziebold, U.; Thakore, K.; Dahia, P.L.; Komminoth, P.; Lees, J.A.; Mulligan, L.M.; Mutter, G.L.; et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am. J. Pathol. 1999, 155, 1253–1260. [Google Scholar]
- Planchon, S.M.; Waite, K.A.; Eng, C. The nuclear affairs of PTEN. J. Cell Sci. 2008, 121, 249–253. [Google Scholar] [CrossRef]
- Chung, J.H.; Ginn-Pease, M.E.; Eng, C. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res. 2005, 65, 4108–4116. [Google Scholar] [CrossRef]
- Liu, J.L.; Mao, Z.; LaFortune, T.A.; Alonso, M.M.; Gallick, G.E.; Fueyo, J.; Yung, W.K. Cell cycle-dependent nuclear export of phosphatase and tensin homologue tumor suppressor is regulated by the phosphoinositide-3-kinase signaling cascade. Cancer Res. 2007, 67, 11054–11063. [Google Scholar] [CrossRef]
- Gil, A.; Andres-Pons, A.; Fernandez, E.; Valiente, M.; Torres, J.; Cervera, J.; Pulido, R. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol. Biol. Cell 2006, 17, 4002–4013. [Google Scholar] [CrossRef]
- Radu, A.; Neubauer, V.; Akagi, T.; Hanafusa, H.; Georgescu, M.M. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol. Cell. Biol. 2003, 23, 6139–6149. [Google Scholar] [CrossRef]
- Van Gent, D.C.; Hoeijmakers, J.H.; Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar]
- Cleveland, D.W.; Mao, Y.; Sullivan, K.F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 2003, 112, 407–421. [Google Scholar] [CrossRef]
- Lindsay, Y.; McCoull, D.; Davidson, L.; Leslie, N.R.; Fairservice, A.; Gray, A.; Lucocq, J.; Downes, C.P. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 2006, 119, 5160–5168. [Google Scholar] [CrossRef]
- Matheu, A.; Maraver, A.; Klatt, P.; Flores, I.; Garcia-Cao, I.; Borras, C.; Flores, J.M.; Vina, J.; Blasco, M.A.; Serrano, M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 2007, 448, 375–379. [Google Scholar] [CrossRef]
- Wang, C.; Jurk, D.; Maddick, M.; Nelson, G.; Martin-Ruiz, C.; von Zglinicki, T. DNA damage response and cellular senescence in tissues of aging mice. Aging cell 2009, 8, 311–323. [Google Scholar] [CrossRef]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef]
- Friedberg, E.C. DNA damage and repair. Nature 2003, 421, 436–440. [Google Scholar] [CrossRef]
- Cline, S.D.; Hanawalt, P.C. Who’s on first in the cellular response to DNA damage? Nat. Rev. Mol. Cell Biol. 2003, 4, 361–372. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef]
- Seviour, E.G.; Lin, S.Y. The DNA damage response: Balancing the scale between cancer and ageing. Aging 2010, 2, 900–907. [Google Scholar]
- Cleaver, J.E. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 2005, 5, 564–573. [Google Scholar] [CrossRef]
- Cleaver, J.E.; Lam, E.T.; Revet, I. Disorders of nucleotide excision repair: The genetic and molecular basis of heterogeneity. Nat. Revs. Genet. 2009, 10, 756–768. [Google Scholar] [CrossRef]
- Sugasawa, K. UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. J. Mol. Histol. 2006, 37, 189–202. [Google Scholar] [CrossRef]
- Sugasawa, K.; Ng, J.M.; Masutani, C.; Iwai, S.; van der Spek, P.J.; Eker, A.P.; Hanaoka, F.; Bootsma, D.; Hoeijmakers, J.H. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 1998, 2, 223–232. [Google Scholar] [CrossRef]
- Niggli, H.J.; Rothlisberger, R. Cyclobutane-type pyrimidine photodimer formation and induction of ornithine decarboxylase in human skin fibroblasts after UV irradiation. J. Investig. Dermatol. 1988, 91, 579–584. [Google Scholar]
- Vink, A.A.; Berg, R.J.; de Gruijl, F.R.; Roza, L.; Baan, R.A. Induction, repair and accumulation of thymine dimers in the skin of UV-B-irradiated hairless mice. Carcinogenesis 1991, 12, 861–864. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Ming, M.; Shea, C.R.; Guo, X.; Li, X.; Soltani, K.; Han, W.; He, Y.Y. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc. Natl. Acad. Sci. USA 2010, 107, 22623–22628. [Google Scholar]
- Gupta, A.; Yang, Q.; Pandita, R.K.; Hunt, C.R.; Xiang, T.; Misri, S.; Zeng, S.; Pagan, J.; Jeffery, J.; Puc, J.; et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle 2009, 8, 2198–2210. [Google Scholar] [CrossRef]
- Brown, E.J.; Baltimore, D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003, 17, 615–628. [Google Scholar] [CrossRef]
- Chehab, N.H.; Malikzay, A.; Appel, M.; Halazonetis, T.D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000, 14, 278–288. [Google Scholar]
- Yoon, K.; Smart, R.C. C/EBPalpha is a DNA damage-inducible p53-regulated mediator of the G1 checkpoint in keratinocytes. Mol. Cell. Biol. 2004, 24, 10650–10660. [Google Scholar] [CrossRef]
- Groszer, M.; Erickson, R.; Scripture-Adams, D.D.; Lesche, R.; Trumpp, A.; Zack, J.A.; Kornblum, H.I.; Liu, X.; Wu, H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 2001, 294, 2186–2189. [Google Scholar] [CrossRef]
- Kwon, C.H.; Zhu, X.; Zhang, J.; Knoop, L.L.; Tharp, R.; Smeyne, R.J.; Eberhart, C.G.; Burger, P.C.; Baker, S.J. Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat. Genet. 2001, 29, 404–411. [Google Scholar] [CrossRef]
- Zhang, J.; Grindley, J.C.; Yin, T.; Jayasinghe, S.; He, X.C.; Ross, J.T.; Haug, J.S.; Rupp, D.; Porter-Westpfahl, K.S.; Wiedemann, L.M.; et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 2006, 441, 518–522. [Google Scholar] [CrossRef]
- Yilmaz, O.H.; Valdez, R.; Theisen, B.K.; Guo, W.; Ferguson, D.O.; Wu, H.; Morrison, S.J. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006, 441, 475–482. [Google Scholar] [CrossRef]
- Wan, X.; Li, J.; Xie, X.; Lu, W. PTEN augments doxorubicin-induced apoptosis in PTEN-null Ishikawa cells. Int. J. Gynecol. Cancer 2007, 17, 808–812. [Google Scholar] [CrossRef]
- Flores, E.R.; Tsai, K.Y.; Crowley, D.; Sengupta, S.; Yang, A.; McKeon, F.; Jacks, T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002, 416, 560–564. [Google Scholar] [CrossRef]
- Lehman, J.A.; Waning, D.L.; Batuello, C.N.; Cipriano, R.; Kadakia, M.P.; Mayo, L.D. Induction of apoptotic genes by a p73-phosphatase and tensin homolog (p73-PTEN) protein complex in response to genotoxic stress. J. Biol. Chem. 2011, 286, 36631–36640. [Google Scholar] [CrossRef]
- Liu, J.L.; Sheng, X.; Hortobagyi, Z.K.; Mao, Z.; Gallick, G.E.; Yung, W.K. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell. Biol. 2005, 25, 6211–6224. [Google Scholar] [CrossRef]
- Ming, M.; He, Y.Y. PTEN in DNA damage repair. Cancer Lett. 2012, 319, 125–129. [Google Scholar] [CrossRef]
- Matheu, A.; Maraver, A.; Collado, M.; Garcia-Cao, I.; Canamero, M.; Borras, C.; Flores, J.M.; Klatt, P.; Vina, J.; Serrano, M. Anti-aging activity of the Ink4/Arf locus. Aging Cell 2009, 8, 152–161. [Google Scholar] [CrossRef]
- Bartke, A. Insulin and aging. Cell Cycle 2008, 7, 3338–3343. [Google Scholar] [CrossRef]
- Bartke, A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: Novel findings. Aging Cell 2008, 7, 285–290. [Google Scholar] [CrossRef]
- Kamagate, A.; Kim, D.H.; Zhang, T.; Slusher, S.; Gramignoli, R.; Strom, S.C.; Bertera, S.; Ringquist, S.; Dong, H.H. FoxO1 links hepatic insulin action to endoplasmic reticulum stress. Endocrinology 2010, 151, 3521–3535. [Google Scholar] [CrossRef]
- Um, S.H.; Frigerio, F.; Watanabe, M.; Picard, F.; Joaquin, M.; Sticker, M.; Fumagalli, S.; Allegrini, P.R.; Kozma, S.C.; Auwerx, J.; et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004, 431, 200–205. [Google Scholar] [CrossRef]
- Um, S.H.; D’Alessio, D.; Thomas, G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006, 3, 393–402. [Google Scholar] [CrossRef]
- Mehta, L.H.; Roth, G.S. Caloric restriction and longevity: The science and the ascetic experience. Ann. N. Y. Acad. Sci. 2009, 1172, 28–33. [Google Scholar]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span—From yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Dorman, J.B.; Albinder, B.; Shroyer, T.; Kenyon, C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 1995, 141, 1399–1406. [Google Scholar]
- Masse, I.; Molin, L.; Billaud, M.; Solari, F. Lifespan and dauer regulation by tissue-specific activities of Caenorhabditis elegans DAF-18. Dev. Biol. 2005, 286, 91–101. [Google Scholar] [CrossRef]
- Morris, J.Z.; Tissenbaum, H.A.; Ruvkun, G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 1996, 382, 536–539. [Google Scholar] [CrossRef]
- Hempenstall, S.; Picchio, L.; Mitchell, S.E.; Speakman, J.R.; Selman, C. The impact of acute caloric restriction on the metabolic phenotype in male C57BL/6 and DBA/2 mice. Mech. Ageing Dev. 2010, 131, 111–118. [Google Scholar] [CrossRef]
- Jiang, H.; Schiffer, E.; Song, Z.; Wang, J.; Zurbig, P.; Thedieck, K.; Moes, S.; Bantel, H.; Saal, N.; Jantos, J.; et al. Proteins induced by telomere dysfunction and DNA damage represent biomarkers of human aging and disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11299–11304. [Google Scholar] [CrossRef]
- Moore, T.; Beltran, L.; Carbajal, S.; Strom, S.; Traag, J.; Hursting, S.D.; di Giovanni, J. Dietary energy balance modulates signaling through the Akt/mammalian target of rapamycin pathways in multiple epithelial tissues. Cancer Prev. Res. 2008, 1, 65–76. [Google Scholar] [CrossRef]
- Kozak, L.P.; Anunciado-Koza, R. UCP1: Its involvement and utility in obesity. Int. J. Obes. 2008, 32, S32–S38. [Google Scholar] [CrossRef]
- Nedergaard, J.; Cannon, B. The changed metabolic world with human brown adipose tissue: Therapeutic visions. Cell Metab. 2010, 11, 268–272. [Google Scholar] [CrossRef]
- Risinger, J.I.; Hayes, A.K.; Berchuck, A.; Barrett, J.C. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997, 57, 4736–4738. [Google Scholar]
- Tashiro, H.; Blazes, M.S.; Wu, R.; Cho, K.R.; Bose, S.; Wang, S.I.; Li, J.; Parsons, R.; Ellenson, L.H. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997, 57, 3935–3940. [Google Scholar]
- Saal, L.H.; Gruvberger-Saal, S.K.; Persson, C.; Lovgren, K.; Jumppanen, M.; Staaf, J.; Jonsson, G.; Pires, M.M.; Maurer, M.; Holm, K.; et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008, 40, 102–107. [Google Scholar] [CrossRef]
- Cantley, L.C.; Neel, B.G. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 4240–4245. [Google Scholar] [CrossRef]
- Suzuki, A.; Yamaguchi, M.T.; Ohteki, T.; Sasaki, T.; Kaisho, T.; Kimura, Y.; Yoshida, R.; Wakeham, A.; Higuchi, T.; Fukumoto, M.; et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 2001, 14, 523–534. [Google Scholar] [CrossRef]
- Crackower, M.A.; Oudit, G.Y.; Kozieradzki, I.; Sarao, R.; Sun, H.; Sasaki, T.; Hirsch, E.; Suzuki, A.; Shioi, T.; Irie-Sasaki, J.; et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell 2002, 110, 737–749. [Google Scholar] [CrossRef]
- Kimura, T.; Suzuki, A.; Fujita, Y.; Yomogida, K.; Lomeli, H.; Asada, N.; Ikeuchi, M.; Nagy, A.; Mak, T.W.; Nakano, T. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development 2003, 130, 1691–1700. [Google Scholar] [CrossRef] [Green Version]
- Podsypanina, K.; Ellenson, L.H.; Nemes, A.; Gu, J.; Tamura, M.; Yamada, K.M.; Cordon-Cardo, C.; Catoretti, G.; Fisher, P.E.; Parsons, R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA 1999, 96, 1563–1568. [Google Scholar] [CrossRef]
- Di Cristofano, A.; Kotsi, P.; Peng, Y.F.; Cordon-Cardo, C.; Elkon, K.B.; Pandolfi, P.P. Impaired Fas response and autoimmunity in Pten+/− mice. Science 1999, 285, 2122–2125. [Google Scholar] [CrossRef]
- Di Cristofano, A.; de Acetis, M.; Koff, A.; Cordon-Cardo, C.; Pandolfi, P.P. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 2001, 27, 222–224. [Google Scholar] [CrossRef]
- Dahia, P.L. PTEN, a unique tumor suppressor gene. Endocr. Relat. Cancer 2000, 7, 115–129. [Google Scholar] [CrossRef]
- Marsh, D.J.; Kum, J.B.; Lunetta, K.L.; Bennett, M.J.; Gorlin, R.J.; Ahmed, S.F.; Bodurtha, J.; Crowe, C.; Curtis, M.A.; Dasouki, M.; et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999, 8, 1461–1472. [Google Scholar] [CrossRef]
- Rustad, C.F.; Bjornslett, M.; Heimdal, K.R.; Maehle, L.; Apold, J.; Moller, P. Germline PTEN mutations are rare and highly penetrant. Hered. Cancer Clin. Pract. 2006, 4, 177–185. [Google Scholar] [CrossRef]
- Liaw, D.; Marsh, D.J.; Li, J.; Dahia, P.L.; Wang, S.I.; Zheng, Z.; Bose, S.; Call, K.M.; Tsou, H.C.; Peacocke, M.; et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997, 16, 64–67. [Google Scholar] [CrossRef]
- Marsh, D.J.; Dahia, P.L.; Zheng, Z.; Liaw, D.; Parsons, R.; Gorlin, R.J.; Eng, C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997, 16, 333–334. [Google Scholar] [CrossRef]
- Suzuki, A.; de la Pompa, J.L.; Stambolic, V.; Elia, A.J.; Sasaki, T.; del Barco Barrantes, I.; Ho, A.; Wakeham, A.; Itie, A.; Khoo, W.; et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 1998, 8, 1169–1178. [Google Scholar] [CrossRef]
- Blumenthal, G.M.; Dennis, P.A. PTEN hamartoma tumor syndromes. Eur. J. Human Genet. 2008, 16, 1289–1300. [Google Scholar] [CrossRef]
- Backman, S.A.; Ghazarian, D.; So, K.; Sanchez, O.; Wagner, K.U.; Hennighausen, L.; Suzuki, A.; Tsao, M.S.; Chapman, W.B.; Stambolic, V.; et al. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc. Natl. Acad. Sci. USA 2004, 101, 1725–1730. [Google Scholar] [CrossRef]
- Li, G.; Robinson, G.W.; Lesche, R.; Martinez-Diaz, H.; Jiang, Z.; Rozengurt, N.; Wagner, K.U.; Wu, D.C.; Lane, T.F.; Liu, X.; et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development 2002, 129, 4159–4170. [Google Scholar]
- Suzuki, G. Characterization of TP53, APC, and PTEN mutational statuses of 120 human cancer cell lines by using yeast-based analyses. Hokkaido Igaku Zasshi 2003, 78, 437–449. [Google Scholar]
- Yanagi, S.; Kishimoto, H.; Kawahara, K.; Sasaki, T.; Sasaki, M.; Nishio, M.; Yajima, N.; Hamada, K.; Horie, Y.; Kubo, H.; et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J. Clin. Investig. 2007, 117, 2929–2940. [Google Scholar] [CrossRef]
- Busa, T.; Chabrol, B.; Perret, O.; Longy, M.; Philip, N. Novel PTEN germline mutation in a family with mild phenotype: Difficulties in genetic counseling. Gene 2013, 512, 194–197. [Google Scholar] [CrossRef]
- McBride, K.L.; Varga, E.A.; Pastore, M.T.; Prior, T.W.; Manickam, K.; Atkin, J.F.; Herman, G.E. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010, 3, 137–141. [Google Scholar] [CrossRef]
- Simpson, L.; Parsons, R. PTEN: Life as a tumor suppressor. Exp. Cell Res. 2001, 264, 29–41. [Google Scholar] [CrossRef]
- Oldham, S.; Stocker, H.; Laffargue, M.; Wittwer, F.; Wymann, M.; Hafen, E. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development 2002, 129, 4103–4109. [Google Scholar]
- Gao, X.; Neufeld, T.P.; Pan, D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 2000, 221, 404–418. [Google Scholar] [CrossRef]
- Whiteman, D.C.; Zhou, X.P.; Cummings, M.C.; Pavey, S.; Hayward, N.K.; Eng, C. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int. J. Cancer 2002, 99, 63–67. [Google Scholar] [CrossRef]
- Zhou, X.P.; Gimm, O.; Hampel, H.; Niemann, T.; Walker, M.J.; Eng, C. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am. J. Pathol. 2000, 157, 1123–1128. [Google Scholar] [CrossRef]
- Fridberg, M.; Servin, A.; Anagnostaki, L.; Linderoth, J.; Berglund, M.; Soderberg, O.; Enblad, G.; Rosen, A.; Mustelin, T.; Jerkeman, M.; et al. Protein expression and cellular localization in two prognostic subgroups of diffuse large B-cell lymphoma: Higher expression of ZAP70 and PKC-beta II in the non-germinal center group and poor survival in patients deficient in nuclear PTEN. Leuk. Lymphoma 2007, 48, 2221–2232. [Google Scholar] [CrossRef]
- Zhou, X.P.; Loukola, A.; Salovaara, R.; Nystrom-Lahti, M.; Peltomaki, P.; de la Chapelle, A.; Aaltonen, L.A.; Eng, C. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002, 161, 439–447. [Google Scholar] [CrossRef]
- Tachibana, M.; Shibakita, M.; Ohno, S.; Kinugasa, S.; Yoshimura, H.; Ueda, S.; Fujii, T.; Rahman, M.A.; Dhar, D.K.; Nagasue, N. Expression and prognostic significance of PTEN product protein in patients with esophageal squamous cell carcinoma. Cancer 2002, 94, 1955–1960. [Google Scholar] [CrossRef]
- Liu, Y.; Majumder, S.; McCall, W.; Sartor, C.I.; Mohler, J.L.; Gregory, C.W.; Earp, H.S.; Whang, Y.E. Inhibition of HER-2/neu kinase impairs androgen receptor recruitment to the androgen responsive enhancer. Cancer Res. 2005, 65, 3404–3409. [Google Scholar]
- Okumura, K.; Zhao, M.; DePinho, R.A.; Furnari, F.B.; Cavenee, W.K. PTEN: A novel anti-oncogenic function independent of phosphatase activity. Cell Cycle 2005, 4, 540–542. [Google Scholar] [CrossRef]
- Okumura, K.; Zhao, M.; Depinho, R.A.; Furnari, F.B.; Cavenee, W.K. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc. Natl. Acad. Sci. USA 2005, 102, 2703–2706. [Google Scholar] [CrossRef]
- Soria, J.C.; Lee, H.Y.; Lee, J.I.; Wang, L.; Issa, J.P.; Kemp, B.L.; Liu, D.D.; Kurie, J.M.; Mao, L.; Khuri, F.R. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res. 2002, 8, 1178–1184. [Google Scholar]
- Vasudevan, K.M.; Burikhanov, R.; Goswami, A.; Rangnekar, V.M. Suppression of PTEN expression is essential for antiapoptosis and cellular transformation by oncogenic Ras. Cancer Res. 2007, 67, 10343–10350. [Google Scholar] [CrossRef]
- Chung, J.H.; Ostrowski, M.C.; Romigh, T.; Minaguchi, T.; Waite, K.A.; Eng, C. The ERK1/2 pathway modulates nuclear PTEN-mediated cell cycle arrest by cyclin D1 transcriptional regulation. Hum. Mol. Genet. 2006, 15, 2553–2559. [Google Scholar] [CrossRef]
© 2013 by the authors. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tait, I.S.; Li, Y.; Lu, J. PTEN, Longevity and Age-Related Diseases. Biomedicines 2013, 1, 17-48. https://doi.org/10.3390/biomedicines1010017
Tait IS, Li Y, Lu J. PTEN, Longevity and Age-Related Diseases. Biomedicines. 2013; 1(1):17-48. https://doi.org/10.3390/biomedicines1010017
Chicago/Turabian StyleTait, Izak S., Yan Li, and Jun Lu. 2013. "PTEN, Longevity and Age-Related Diseases" Biomedicines 1, no. 1: 17-48. https://doi.org/10.3390/biomedicines1010017