Ribosome-Directed Therapies in Cancer
Abstract
:1. Introduction
2. Ribosome Biogenesis and Signal Transduction Pathways
3. Protein Synthesis by Ribosomes
4. Ribosome Biogenesis and Cancer Pathogenesis
5. Identification of Inhibitors That Have Target rRNA
- (a)
- rRNA transcription: the drugs that participate in this step, for example, are oxaliplatin, doxorubicin, and methotrexate,
- (b)
- Early rRNA processing: the drugs that act in this step, for example, are berberine HCl, negestrol acetate, and tanshinone IIA,
- (c)
- Late rRNA processing: the drugs that participate in this step, for example, are 5-fluorouracil and homoharringtonine.
6. Targeting Pol I Transcription for Therapeutic Effect
7. Identification of Inhibitors That Have a Target Translation Process
8. Drugs That Have a Ribosome as a Target in Cancer Diseases
9. rRNA and Ribosomal Protein Modification in Cancer Diseases
10. Other Roles of Ribosomal Proteins: Modulation of the Immune System
11. The Ribosome Inactivation Proteins as Anticancer Therapy
12. Future Perspective and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orelle, C.; Carlson, E.D.; Szal, T.; Florin, T.; Jewett, M.; Mankin, A.S. Protein synthesis by ribosomes with tethered subunits. Nature 2015, 524, 119–124. [Google Scholar] [CrossRef]
- Aleksashin, N.A.; Leppik, M.; Hockenberry, A.J.; Klepacki, D.; Vázquez-Laslop, N.; Jewett, M.C.; Remme, J.; Mankin, A.S. Assembly and functionality of the ribosome with tethered subunits. Nat. Commun. 2019, 10, 930. [Google Scholar] [CrossRef] [PubMed]
- Fabret, C.; Namy, O. Translational accuracy of a tethered ribosome. Nucleic Acids Res. 2021, 49, 5308–5318. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.G.; Wilusz, J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017, 31, 1717–1731. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Anger, A.M.; Armache, J.-P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 2017, 67, 71–83.e7. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, N.; Pusic, A.; Stumpf, C.R.; Shimizu, K.; Hsieh, A.C.; Xue, S.; Ishijima, J.; Shiroishi, T.; Barna, M. Ribosome-Mediated Specificity in Hox mRNA Translation and Vertebrate Tissue Patterning. Cell 2011, 145, 383–397. [Google Scholar] [CrossRef]
- Preiss, T. All Ribosomes Are Created Equal. Really? Trends Biochem. Sci. 2016, 41, 121–123. [Google Scholar] [CrossRef]
- Sulima, S.O.; Dinman, J.D. The Expanding Riboverse. Cells 2019, 8, 1205. [Google Scholar] [CrossRef] [Green Version]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- De la Cruz, J.; Karbstein, K.; Woolford, J.L., Jr. Functions of Ribosomal Proteins in Assembly of Eukaryotic Ribosomes In Vivo. Annu. Rev. Biochem. 2015, 84, 93–129. [Google Scholar] [CrossRef]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef]
- Espinar-Marchena, F.J.; Babiano, R.; Cruz, J. Placeholder factors in ribosome biogenesis: Please, pave my way. Microb. Cell 2017, 4, 144–168. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Bohnsack, M.T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019, 38, e100278. [Google Scholar] [CrossRef]
- Schmickel, R.D. Quantitation of Human Ribosomal DNA: Hybridization of Human DNA with Ribosomal RNA for Quantitation and Fractionation. Pediatr. Res. 1973, 7, 5–12. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008, 18, 13–18. [Google Scholar] [CrossRef]
- Gibbons, J.G.; Branco, A.T.; Godinho, S.A.; Yu, S.; Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 2485–2490. [Google Scholar] [CrossRef]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of Ribosomal DNA in the Human Chromosome Complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef] [Green Version]
- Scherl, A.; Couté, Y.; Déon, C.; Callé, A.; Kindbeiter, K.; Sanchez, J.-C.; Greco, A.; Hochstrasser, D.; Diaz, J.-J. Functional Proteomic Analysis of Human Nucleolus. Mol. Biol. Cell 2002, 13, 4100–4109. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed Proteomic Analysis of the Human Nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 2002, 13, 4100–4109. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lam, Y.W.; Leung, A.K.; Ong, S.-E.; Lyon, C.E.; Lamond, A.; Mann, M. Nucleolar proteome dynamics. Nature 2005, 433, 77–83. [Google Scholar] [CrossRef]
- Ahmad, Y.; Boisvert, F.-M.; Gregor, P.; Cobley, A.; Lamond, A.I. NOPdb: Nucleolar Proteome Database--2008 update. Nucleic Acids Res. 2009, 37, D181–D184. [Google Scholar] [CrossRef]
- Tafforeau, L.; Zorbas, C.; Langhendries, J.-L.; Mullineux, S.-T.; Stamatopoulou, V.; Mullier, R.; Wacheul, L.; Lafontaine, D.L. The Complexity of Human Ribosome Biogenesis Revealed by Systematic Nucleolar Screening of Pre-rRNA Processing Factors. Mol. Cell 2013, 51, 539–551. [Google Scholar] [CrossRef]
- Lempiäinen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef]
- Mahoney, S.J.; Dempsey, J.M.; Blenis, J. Cell Signaling in Protein Synthesis Ribosome Biogenesis and Translation Initiation and Elongation. Prog. Mol. Biol. Transl. Sci. 2009, 90, 53–107. [Google Scholar] [CrossRef]
- Brighenti, E.; Calabrese, C.; Liguori, G.; Giannone, F.A.; Trere, D.; Montanaro, L.; Derenzini, M. Interleukin 6 downregulates p53 expression and activity by stimulating ribosome biogenesis: A new pathway connecting inflammation to cancer. Oncogene 2014, 33, 4396–4406. [Google Scholar] [CrossRef] [Green Version]
- Van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Wu, C.; You, J.; Fu, J.; Wang, X.; Zhang, Y. Phosphatidylinositol 3-Kinase/Akt Mediates Integrin Signaling to Control RNA Polymerase I Transcriptional Activity. Mol. Cell. Biol. 2016, 36, 1555–1568. [Google Scholar] [CrossRef]
- Iadevaia, V.; Liu, R.; Proud, C.G. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 2014, 36, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Omlin, A.; De Bono, J.S. Development of Therapeutic Combinations Targeting Major Cancer Signaling Pathways. J. Clin. Oncol. 2013, 31, 1592–1605. [Google Scholar] [CrossRef]
- Yip, H.Y.K.; Papa, A. Signaling Pathways in Cancer: Therapeutic Targets, Combinatorial Treatments, and New Developments. Cells 2021, 10, 659. [Google Scholar] [CrossRef]
- Pardee, A.B. G 1 Events and Regulation of Cell Proliferation. Science 1989, 246, 603–608. [Google Scholar] [CrossRef]
- Pyronnet, S.; Sonenberg, N. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 2001, 11, 13–18. [Google Scholar] [CrossRef]
- Gilles, A.; Frechin, L.; Natchiar, K.; Biondani, G.; von Loeffelholz, O.; Holvec, S.; Malaval, J.-L.; Winum, J.-Y.; Klaholz, B.P.; Peyron, J.-F. Targeting the Human 80S Ribosome in Cancer: From Structure to Function and Drug Design for Innovative Adjuvant Therapeutic Strategies. Cells 2020, 9, 629. [Google Scholar] [CrossRef]
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11, a032896. [Google Scholar] [CrossRef]
- Silvera, D.; Formenti, S.C.; Schneider, R.J. Translational control in cancer. Nat. Rev. Cancer 2010, 10, 254–266. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D. Translational Control in Cancer Etiology. Cold Spring Harb. Perspect. Biol. 2013, 5, a012336. [Google Scholar] [CrossRef]
- Ruggero, D. Revisiting the Nucleolus: From Marker to Dynamic Integrator of Cancer Signaling. Sci. Signal. 2012, 5, pe38. [Google Scholar] [CrossRef]
- Montanaro, L.; Treré, D.; Derenzini, M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim. Biophys. Acta 2012, 1825, 101–110. [Google Scholar] [CrossRef]
- Pelletier, J.; Riaño-Canalias, F.; Almacellas, E.; Mauvezin, C.; Samino, S.; Feu, S.; Menoyo, S.; Domostegui, A.; Garcia-Cajide, M.; Salazar, R.; et al. Nucleotide depletion reveals the impaired ribosome biogenesis checkpoint as a barrier against DNA damage. EMBO J. 2020, 39, e103838. [Google Scholar] [CrossRef]
- Xu, X.; Xiong, X.; Sun, Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci. China Life Sci. 2016, 59, 656–672. [Google Scholar] [CrossRef]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA Polymerase I Transcription Machinery: An Emerging Target for the Treatment of Cancer. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef]
- Whittaker, S.; Martin, M.; Marais, R. All Roads Lead to the Ribosome. Cancer Cell 2010, 18, 5–6. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes, P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Klein, J.; Grummt, I. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 1999, 96, 6096–6101. [Google Scholar] [CrossRef] [Green Version]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef]
- Barna, M.; Pusic, A.; Zollo, O.; Costa, M.; Kondrashov, N.; Rego, E.; Rao, P.H.; Ruggero, D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 2008, 456, 971–975. [Google Scholar] [CrossRef]
- Staley, J.P.; Woolford, J.L., Jr. Assembly of ribosomes and spliceosomes: Complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009, 21, 109–118. [Google Scholar] [CrossRef]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The nucleolus: An emerging target for cancer therapy. Trends Mol. Med. 2013, 19, 643–654. [Google Scholar] [CrossRef]
- Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Nieto, B.; Gaspar, S.G.; Moriggi, G.; Pestov, D.G.; Bustelo, X.R.; Dosil, M. Identification of distinct maturation steps involved in human 40S ribosomal subunit biosynthesis. Nat. Commun. 2020, 11, 156. [Google Scholar] [CrossRef]
- Hein, N.; Cameron, D.P.; Hannan, K.M.; Nguyen, N.-Y.N.; Fong, C.Y.; Sornkom, J.; Wall, M.; Pavy, M.; Cullinane, C.; Diesch, J.; et al. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood 2017, 129, 2882–2895. [Google Scholar] [CrossRef]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, D.; Prattes, M.; Kofler, L.; Rössler, I.; Loibl, M.; Pertl, M.; Zisser, G.; Wolinski, H.; Pertschy, B.; Bergler, H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Klein, I.; Prattes, M.; Schmidt, C.; Kappel, L.; Zisser, G.; Gungl, A.; Krieger, E.; Pertschy, B.; Bergler, H. The Drug Diazaborine Blocks Ribosome Biogenesis by Inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 2014, 289, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Prattes, M.; Grishkovskaya, I.; Hodirnau, V.-V.; Rössler, I.; Klein, I.; Hetzmannseder, C.; Zisser, G.; Gruber, C.C.; Gruber, K.; Haselbach, D.; et al. Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine. Nat. Commun. 2021, 12, 3483. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Zeng, H.W.; Wang, J.X.; Yuan, X.; Zhang, C.; Fang, T.; Yang, P.M.; Wu, T.; Zhou, Y.D.; Nagle, D.G.; et al. The antitumor natural product tanshinone IIA inhibits protein kinase C and acts synergistically with 17-AAG. Cell Death Dis. 2018, 9, 165. [Google Scholar] [CrossRef]
- Lin, C.Y.; Chang, T.W.; Hsieh, W.H.; Hung, M.C.; Lin, I.H.; Lai, S.C.; Tzeng, Y.J. Simultaneous induction of apoptosis and necroptosis by Tanshinone IIA in human hepatocellular carcinoma HepG2 cells. Cell Death Discov. 2016, 2, 16065. [Google Scholar] [CrossRef]
- Chiu, S.C.; Huang, S.Y.; Chen, S.P.; Su, C.C.; Chiu, T.L.; Pang, C.-Y. Tanshinone IIA inhibits human prostate cancer cells growth by induction of endoplasmic reticulum stress in vitro and in vivo. Prostate Cancer Prostatic Dis. 2013, 16, 315–322. [Google Scholar] [CrossRef]
- Gregory, E.J.; Cohen, S.C.; Oines, D.W.; Mims, C.H. Megestrol acetate therapy for advanced breast cancer. J. Clin. Oncol. 1985, 3, 155–160. [Google Scholar] [CrossRef]
- Johnson, P.A.; Bonomi, P.D.; Anderson, K.M.; Wolter, J.M.; Economou, S.G. Megestrol acetate: First-line therapy for advanced breast cancer. Semin. Oncol. 1986, 13 (Suppl. 4), 15–19. [Google Scholar]
- Pronzato, P.; Brema, F.; Amoroso, D.; Bertelli, G.; Conte, P.; Martini, M.C.; Pastorino, G.; Rosso, R. Megestrol acetate: Phase II study of a single daily administration in advanced breast cancer. Breast Cancer Res. Treat. 1990, 17, 51–54. [Google Scholar] [CrossRef]
- Diogo, C.V.; Machado, N.G.; Barbosa, I.A.; Serafim, T.; Burgeiro, A.A.C.; Oliveira, P. Berberine as a Promising Safe Anti-Cancer Agent- Is there a Role for Mitochondria? Curr. Drug Targets 2011, 12, 850–859. [Google Scholar] [CrossRef]
- Luzina, O.A.; Salakhutdinov, N.F. Biological activity of usnic acid and its derivatives: Part 2. effects on higher organisms. Molecular and physicochemical aspects. Russ. J. Bioorg. Chem. 2016, 42, 249–268. [Google Scholar] [CrossRef]
- Luzina, O.A.; Salakhutdinov, N.F. Usnic acid and its derivatives for pharmaceutical use: A patent review (2000–2017). Expert Opin. Ther. Pat. 2018, 28, 477–491. [Google Scholar] [CrossRef]
- Kashyap, D.; Sharma, A.; Tuli, H.S.; Sak, K.; Mukherjee, T.; Bishayee, A. Molecular targets of celastrol in cancer: Recent trends and advancements. Crit. Rev. Oncol. Hematol. 2018, 128, 70–81. [Google Scholar] [CrossRef]
- Xiong, W.; Li, W.-H.; Jiang, Y.-X.; Liu, S.; Ai, Y.-Q.; Liu, R.; Chang, L.; Zhang, M.; Wang, X.-L.; Bai, H.; et al. Parecoxib: An Enhancer of Radiation Therapy for Colorectal Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 627–633. [Google Scholar] [CrossRef]
- Li, L.-Y.; Xiao, J.; Liu, Q.; Xia, K. Parecoxib inhibits glioblastoma cell proliferation, migration and invasion by up-regulating miRNA-29c. Biol. Open 2017, 6, 311–316. [Google Scholar] [CrossRef]
- Watanabe, M.; Kodaira, S.; Takahashi, T.; Tominaga, T.; Hojo, K.; Kato, T.; Kunitomo, K.; Isomoto, H.; Ohashi, Y.; Yasutomi, M. Randomized trial of the efficacy of adjuvant chemotherapy for colon cancer with combination therapy incorporating the oral pyrimidine 1-hexylcarbamoyl-5-fluorouracil. Langenbeck Arch. Surg. 2006, 391, 330–337. [Google Scholar] [CrossRef]
- Kubota, T.; Fujita, S.; Kodaira, S.; Yamamoto, T.; Josui, K.; Arisawa, Y.; Suto, A.; Ishibiki, K.; Abe, O.; Mabuchi, K.; et al. Antitumor Activity of Fluoropyrimidines and Thymidylate Synthetase Inhibition. Jpn. J. Cancer Res. 1991, 82, 476–482. [Google Scholar] [CrossRef]
- Shelton, J.; Lu, X.; Hollenbaugh, J.A.; Cho, J.H.; Amblard, F.; Schinazi, R.F. Metabolism, Biochemical Actions, and Chemical Synthesis of Anticancer Nucleosides, Nucleotides, and Base Analogs. Chem. Rev. 2016, 116, 14379–14455. [Google Scholar] [CrossRef]
- Fang, F.; Hoskins, J.; Butler, J.S. 5-Fluorouracil Enhances Exosome-Dependent Accumulation of Polyadenylated rRNAs. Mol. Cell. Biol. 2004, 24, 10766–10776. [Google Scholar] [CrossRef]
- Greenhalgh, D.A.; Parish, J.H. Effect of 5-fluorouracil combination therapy on RNA processing in human colonic carcinoma cells. Br. J. Cancer 1990, 61, 415–419. [Google Scholar] [CrossRef]
- Hoskins, J.; Butler, J.S. RNA-Based 5-Fluorouracil Toxicity Requires the Pseudouridylation Activity of Cbf5p. Genetics 2008, 179, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Kammler, S.; Lykke-Andersen, S.; Jensen, T.H. The RNA Exosome Component hRrp6 Is a Target for 5-Fluorouracil in Human Cells. Mol. Cancer Res. 2008, 6, 990–995. [Google Scholar] [CrossRef]
- Kilic, N.; Aras, S.; Cansaran-Duman, D. Determination of Vulpinic Acid Effect on Apoptosis and mRNA Expression Levels in Breast Cancer Cell Lines. Anti-Cancer Agents Med. Chem. 2018, 18, 2032–2041. [Google Scholar] [CrossRef]
- Zisser, G.; Ohmayer, U.; Mauerhofer, C.; Mitterer, V.; Klein, I.; Rechberger, G.N.; Wolinski, H.; Prattes, M.; Pertschy, B.; Milkereit, P.; et al. Viewing pre-60S maturation at a minute’s timescale. Nucleic Acids Res. 2018, 46, 3140–3151. [Google Scholar] [CrossRef]
- Burger, K.; Mühl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Hölzel, M.; et al. Chemotherapeutic Drugs Inhibit Ribosome Biogenesis at Various Levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; Karabekmez, M.E.; Eraslan, S.; Kırdar, B. Doxorubicin induces an extensive transcriptional and metabolic rewiring in yeast cells. Sci. Rep. 2018, 8, 13672. [Google Scholar] [CrossRef]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 2009, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arnaiz, C.; Busto, N.; Leal, J.M.; Garcia, B. New Insights into the Mechanism of the DNA/Doxorubicin Interaction. J. Phys. Chem. B 2014, 118, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D.; Bianchi, M.S. Genotoxicity of streptonigrin: A review. Mutat. Res. Mutat. Res. 2001, 488, 25–37. [Google Scholar] [CrossRef]
- Shen, M.; Wu, M.-Y.; Chen, L.-P.; Zhi, Q.; Gong, F.-R.; Chen, K.; Li, D.-M.; Wu, Y.; Tao, M.; Li, W. Cantharidin represses invasion of pancreatic cancer cells through accelerated degradation of MMP2 mRNA. Sci. Rep. 2015, 5, 11836. [Google Scholar] [CrossRef] [PubMed]
- Horigome, C.; Okada, T.; Matsuki, K.; Mizuta, K. A Ribosome Assembly Factor Ebp2p, the Yeast Homolog of EBNA1-Binding Protein 2, Is Involved in the Secretory Response. Biosci. Biotechnol. Biochem. 2008, 72, 1080–1086. [Google Scholar] [CrossRef] [Green Version]
- Wimalasena, T.T.; Enjalbert, B.; Guillemette, T.; Plumridge, A.; Budge, S.; Yin, Z.; Brown, A.J.; Archer, D.B. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 2008, 45, 1235–1247. [Google Scholar] [CrossRef]
- Yabuki, Y.; Katayama, M.; Kodama, Y.; Sakamoto, A.; Yatsuhashi, A.; Funato, K.; Mizuta, K. Arp2/3 complex and Mps3 are required for regulation of ribosome biosynthesis in the secretory stress response. Yeast 2017, 34, 155–163. [Google Scholar] [CrossRef]
- Rajagopalan, P.T.R.; Zhang, Z.; McCourt, L.; Dwyer, M.; Benkovic, S.J.; Hammes, G.G. Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics. Proc. Natl. Acad. Sci. USA 2002, 99, 13481–13486. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Chiang, E.-P.I. Low-Dose Methotrexate Inhibits Methionine S-Adenosyltransferase In Vitro and In Vivo. Mol. Med. 2011, 18, 423–432. [Google Scholar] [CrossRef]
- Albrecht, L.V.; Bui, M.H.; De Robertis, E.M. Canonical Wnt is inhibited by targeting one-carbon metabolism through methotrexate or methionine deprivation. Proc. Natl. Acad. Sci. USA 2019, 116, 2987–2995. [Google Scholar] [CrossRef]
- Liu, P.Y.; Tee, A.E.; Milazzo, G.; Hannan, K.; Maag, J.; Mondal, S.; Atmadibrata, B.; Bartonicek, N.; Peng, H.; Ho, N.; et al. The long noncoding RNA lncNB1 promotes tumorigenesis by interacting with ribosomal protein RPL35. Nat. Commun. 2019, 10, 5026. [Google Scholar] [CrossRef]
- Weissmiller, A.; Wang, J.; Lorey, S.L.; Howard, G.; Martinez, E.; Liu, Q.; Tansey, W.P. Inhibition of MYC by the SMARCB1 tumor suppressor. Nat. Commun. 2019, 10, 2014. [Google Scholar] [CrossRef]
- Mei, Y.; Wu, M. Noncoding RNAs Regulating p53 and c-Myc Signaling. In The Long and Short Non-Coding RNAs in Cancer Biology; Advances in Experimental Medicine and Biology; Springer: Singapore, 2016; Volume 927, pp. 337–365. [Google Scholar] [CrossRef]
- Dai, M.S.; Arnold, H.; Sun, X.X.; Sears, R.; Lu, H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 2007, 26, 3332–3345. [Google Scholar] [CrossRef]
- Liao, J.-M.; Zhou, X.; Gatignol, A.; Lu, H. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 2014, 33, 4916–4923. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Hao, Q.; Liao, J.-M.; Liao, P.; Lu, H. Ribosomal Protein S14 Negatively Regulates c-Myc Activity. J. Biol. Chem. 2013, 288, 21793–21801. [Google Scholar] [CrossRef]
- Challagundla, K.B.; Sun, X.-X.; Zhang, X.; DeVine, T.; Zhang, Q.; Sears, R.C.; Dai, M.-S. Ribosomal Protein L11 Recruits miR-24/miRISC To Repress c-Myc Expression in Response to Ribosomal Stress. Mol. Cell. Biol. 2011, 31, 4007–4021. [Google Scholar] [CrossRef]
- Wang, M.; Hu, Y.; Stearns, M.E. RPS2: A novel therapeutic target in prostate cancer. J. Exp. Clin. Cancer Res. 2009, 28, 6. [Google Scholar] [CrossRef]
- McCool, M.A.; Bryant, C.J.; Baserga, S.J. MicroRNAs and long non-coding RNAs as novel regulators of ribosome biogenesis. Biochem. Soc. Trans. 2020, 48, 595–612. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.-J.; Yuan, Y.-G.; Das, J.; Yasuda, H.; Kim, J.-H. MicroRNA-7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Sci. Rep. 2017, 7, 8365. [Google Scholar] [CrossRef]
- Ferreira, R.; Schneekloth, J.J.S., Jr.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [PubMed]
- Cong, R.; Das, S.; Ugrinova, I.; Kumar, S.; Mongelard, F.; Wong, J.; Bouvet, P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012, 40, 9441–9454. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, K.; Colis, L.; Liu, H.; Jäämaa, S.; Zhang, Z.; Hällström, T.A.; Moore, H.M.; Sirajuddin, P.; Laiho, M. Small Molecule BMH-Compounds That Inhibit RNA Polymerase I and Cause Nucleolar Stress. Mol. Cancer Ther. 2014, 13, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Durut, N.; Sáez-Vásquez, J. Nucleolin: Dual roles in rDNA chromatin transcription. Gene 2015, 556, 7–12. [Google Scholar] [CrossRef]
- Papadopoulos, K.; Mita, A.; Ricart, A.; Hufnagel, D.; Northfelt, D.; Von Hoff, D.; Darjania, L.; Lim, J.; Padgett, C.; Marschke, R. Pharmacokinetic findings from the phase I study of Quarfloxin (CX-3543): A protein-rDNA quadruplex inhibitor, in patients with advanced solid tumors. In Proceedings of the AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics, San Francisco, CA, USA, 22–26 October 2007. [Google Scholar]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA Polymerase I with an Oral Small Molecule CX-5461 Inhibits Ribosomal RNA Synthesis and Solid Tumor Growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Mars, J.-C.; Tremblay, M.G.; Valere, M.; Sibai, D.S.; Sabourin-Felix, M.; Lessard, F.; Moss, T. The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. NAR Cancer 2020, 2, zcaa032. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef]
- Negi, S.S.; Brown, P. rRNA synthesis inhibitor, CX-5461, activates ATM/ATR pathway in acute lymphoblastic leukemia, arrests cells in G2 phase and induces apoptosis. Oncotarget 2015, 6, 18094–18104. [Google Scholar] [CrossRef]
- Lee, H.C.; Wang, H.; Baladandayuthapani, V.; Lin, H.; He, J.; Jones, R.J.; Kuiatse, I.; Gu, D.; Wang, Z.; Ma, W.; et al. RNA Polymerase I Inhibition with CX-5461 as a Novel Therapeutic Strategy to TargetMYCin Multiple Myeloma. Br. J. Haematol. 2017, 177, 80–94. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Obinata, D.; Sandhu, S.; Selth, L.; Wong, S.Q.; Porter, L.H.; Lister, N.; Pook, D.; Pezaro, C.J.; Goode, D.L.; et al. Patient-derived Models of Abiraterone- and Enzalutamide-resistant Prostate Cancer Reveal Sensitivity to Ribosome-directed Therapy. Eur. Urol. 2018, 74, 562–572. [Google Scholar] [CrossRef]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.; Dos Santos, N.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Kim, D.-W.; Wu, N.; Kim, Y.-C.; Cheng, P.F.; Basom, R.; Kim, D.; Dunn, C.T.; Lee, A.Y.; Kim, K.; Lee, C.S.; et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev. 2016, 30, 1289–1299. [Google Scholar] [CrossRef]
- Cornelison, R.; Dobbin, Z.C.; Katre, A.A.; Jeong, D.H.; Zhang, Y.; Chen, D.; Petrova, Y.; Llaneza, D.C.; Steg, A.D.; Parsons, L.; et al. Targeting RNA-Polymerase I in Both Chemosensitive and Chemoresistant Populations in Epithelial Ovarian Cancer. Clin. Cancer Res. 2017, 23, 6529–6540. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.; Wright, W.C.; Chapple, R.H.; Zubair, A.F.; Sandhu, M.; Batchelder, J.E.; Huddle, B.C.; Low, J.; Blankenship, K.B.; Wang, Y.; et al. The chemotherapeutic CX-5461 primarily targets TOP2B and exhibits selective activity in high-risk neuroblastoma. Nat. Commun. 2021, 12, 6468. [Google Scholar] [CrossRef]
- Low, J.Y.; Sirajuddin, P.; Moubarek, M.; Agarwal, S.; Rege, A.; Guner, G.; Liu, H.; Yang, Z.; De Marzo, A.M.; Bieberich, C.; et al. Effective targeting of RNA polymerase I in treatment-resistant prostate cancer. Prostate 2019, 79, 1837–1851. [Google Scholar] [CrossRef]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef]
- Gupta, V.; Warner, J.R. Ribosome-omics of the human ribosome. RNA 2014, 20, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Abeysirigunawarden, S.C.; Chen, K.; Mayerle, M.; Ragunathan, K.; Luthey-Schulten, Z.; Ha, T.; Woodson, S.A. Protein-guided RNA dynamics during early ribosome assembly. Nature 2014, 506, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.; Rupeš, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [PubMed]
- Von der Haar, T. Mathematical and computational modelling of ribosomal movement and protein synthesis: An overview. Comput. Struct. Biotechnol. J. 2012, 1, e201204002. [Google Scholar] [CrossRef] [PubMed]
- Bielczyk-Maczyńska, E.; Hung, L.L.; Ferreira, L.; Fleischmann, T.; Weis, F.; Fernández-Pevida, A.; Harvey, S.A.; Wali, N.; Warren, A.J.; Barroso, I.; et al. The Ribosome Biogenesis Protein Nol9 Is Essential for Definitive Hematopoiesis and Pancreas Morphogenesis in Zebrafish. PLoS Genet. 2015, 11, e1005677. [Google Scholar] [CrossRef]
- Chung, J.; Bauer, D.E.; Ghamari, A.; Nizzi, C.P.; Deck, K.M.; Kingsley, P.D.; Yien, Y.Y.; Huston, N.C.; Chen, C.; Schultz, I.J.; et al. The mTORC1/4E-BP pathway coordinates hemoglobin production with L -leucine availability. Sci. Signal. 2015, 8, ra34. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [Green Version]
- De Loubresse, N.G.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef]
- Polacek, N.; Mankin, A.S. The Ribosomal Peptidyl Transferase Center: Structure, Function, Evolution, Inhibition. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 285–311. [Google Scholar] [CrossRef]
- Wilson, D.N. The A–Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 393–433. [Google Scholar] [CrossRef]
- Vázquez, D. Inhibitors of Protein Biosynthesis; Springer: Berlin/Heidelberg, Germany, 1979. [Google Scholar] [CrossRef]
- Schluenzen, F.; Zarivach, R.; Harms, J.; Bashan, A.; Tocilj, A.; Albrecht, R.; Yonath, A.; Franceschi, F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001, 413, 814–821. [Google Scholar] [CrossRef]
- Bulkley, D.; Innis, C.A.; Blaha, G.; Steitz, T.A. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. USA 2010, 107, 17158–17163. [Google Scholar] [CrossRef]
- Dunkle, J.A.; Xiong, L.; Mankin, A.S.; Cate, J.H.D. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA 2010, 107, 17152–17157. [Google Scholar] [CrossRef]
- Moellering, R.C. Linezolid: The First Oxazolidinone Antimicrobial. Ann. Intern. Med. 2003, 138, 135–142. [Google Scholar] [CrossRef]
- Patel, U.; Yan, Y.P.; Hobbs, F.W., Jr.; Kaczmarczyk, J.; Slee, A.M.; Pompliano, D.L.; Kurilla, M.G.; Bobkova, E.V. Oxazolidinones Mechanism of Action: Inhibition of the First Peptide Bond Formation. J. Biol. Chem. 2001, 276, 37199–37205. [Google Scholar] [CrossRef]
- Alvarez-Elcoro, S.; Enzler, M.J. The Macrolides: Erythromycin, Clarithromycin, and Azithromycin. Mayo Clin. Proc. 1999, 74, 613–634. [Google Scholar] [CrossRef]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef]
- Fernandes, P. Use of Antibiotic Core Structures to Generate New and Useful Macrolide Antibiotics. In Antibiotics: Current Innovations and Future Trends; Sánchez, S., Demain, A.L., Eds.; Caister Academic Press: Norfolk, UK, 2015; pp. 375–394. [Google Scholar] [CrossRef]
- Vázquez-Laslop, N.; Mankin, A.S. Context-Specific Action of Ribosomal Antibiotics. Annu. Rev. Microbiol. 2018, 72, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, A.; Machiyama, N.; Kinoshita, T.; Tanaka, N. Inhibition by kasugamycin of initiation complex formation on 30S ribosomes. Biochem. Biophys. Res. Commun. 1971, 43, 196–199. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Osterman, I.A.; Szal, T.; Tashlitsky, V.N.; Serebryakova, M.V.; Kusochek, P.; Bulkley, D.; Malanicheva, I.A.; Efimenko, T.A.; Efremenkova, O.V.; et al. Amicoumacin A Inhibits Translation by Stabilizing mRNA Interaction with the Ribosome. Mol. Cell 2014, 56, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Schuwirth, B.S.; Day, J.M.; Hau, C.W.; Janssen, G.R.; Dahlberg, A.E.; Cate, J.H.D.; Vila-Sanjurjo, A. Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 2006, 13, 879–886. [Google Scholar] [CrossRef]
- Dabbs, E.R.; Poldermans, B.; Bakker, H.; Van Knippenberg, P.H. Biochemical characterization of ribosomes of kasugamycin-dependent mutants ofEscherichia coli. FEBS Lett. 1980, 117, 164–166. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Clemons, W.M.; Carter, A.P.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. The Structural Basis for the Action of the Antibiotics Tetracycline, Pactamycin, and Hygromycin B on the 30S Ribosomal Subunit. Cell 2000, 103, 1143–1154. [Google Scholar] [CrossRef]
- Lintner, N.G.; McClure, K.F.; Petersen, D.; Londregan, A.T.; Piotrowski, D.W.; Wei, L.; Xiao, J.; Bolt, M.; Loria, P.M.; Maguire, B.; et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 2017, 15, e2001882. [Google Scholar] [CrossRef]
- Sun, Y.; Abriola, L.; Niederer, R.O.; Pedersen, S.F.; Alfajaro, M.M.; Silva Monteiro, V.; Wilen, C.B.; Ho, Y.C.; Gilbert, W.V.; Surovtseva, Y.V.; et al. Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 2021, 118, e2023051118. [Google Scholar] [CrossRef]
- Lamb, R.; Ozsvari, B.; Lisanti, C.L.; Tanowitz, H.B.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 2015, 6, 4569–4584. [Google Scholar] [CrossRef]
- Suárez-Rivero, J.M.; Pastor-Maldonado, C.J.; Povea-Cabello, S.; Álvarez-Córdoba, M.; Villalón-García, I.; Talaverón-Rey, M.; Suárez-Carrillo, A.; Munuera-Cabeza, M.; Sánchez-Alcázar, J.A. Mitochondria and Antibiotics: For Good or for Evil? Biomolecules 2021, 11, 1050. [Google Scholar] [CrossRef]
- Myasnikov, A.G.; Natchiar, S.K.; Nebout, M.; Hazemann, I.; Imbert, V.; Khatter, H.; Peyron, J.-F.; Klaholz, B.P. Structure–function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun. 2016, 7, 12856. [Google Scholar] [CrossRef]
- Volarević, S.; Stewart, M.J.; Ledermann, B.; Zilberman, F.; Terracciano, L.; Montini, E.; Grompe, M.; Kozma, S.C.; Thomas, G. Proliferation, But Not Growth, Blocked by Conditional Deletion of 40 S Ribosomal Protein S6. Science 2000, 288, 2045–2047. [Google Scholar] [CrossRef]
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef]
- Lubas, M.; Harder, L.M.; Kumsta, C.; Tiessen, I.; Hansen, M.; Andersen, J.; Lund, A.H.; Frankel, L.B. eIF 5A is required for autophagy by mediating ATG 3 translation. EMBO Rep. 2018, 19, e46072. [Google Scholar] [CrossRef]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018, 6, 282–300.e2. [Google Scholar] [CrossRef]
- Powell, R.; Weisleder, D.; Smith, C.R., Jr.; Rohwedder, W.K. Structures of harringtonine, isoharringtonine, and homoharringtonine. Tetrahedron Lett. 1970, 11, 815–818. [Google Scholar] [CrossRef]
- Tujebajeva, R.M.; Graifer, D.M.; Karpova, G.G.; Ajtkhozhina, N.A. Alkaloid homoharringtonine inhibits polypeptide chain elongation on human ribosomes on the step of peptide bond formation. FEBS Lett. 1989, 257, 254–256. [Google Scholar] [CrossRef]
- Gürel, G.; Blaha, G.; Steitz, T.A.; Moore, P.B. Structures of Triacetyloleandomycin and Mycalamide A Bind to the Large Ribosomal Subunit of Haloarcula marismortui. Antimicrob. Agents Chemother. 2009, 53, 5010–5014. [Google Scholar] [CrossRef] [Green Version]
- Nazha, A.; Kantarjian, H.; Cortes, J.; Quintás-Cardama, A. Omacetaxine mepesuccinate (synribo)—Newly launched in chronic myeloid leukemia. Expert Opin. Pharmacother. 2013, 14, 1977–1986. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Talpaz, M.; Santini, V.; Murgo, A.; Cheson, B.; O’Brien, S.M. Homoharringtonine: History, current research, and future direction. Cancer 2001, 92, 1591–1605. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, Y.; Zhen, T.; Chen, X.; Zhang, M.; Liu, P.; Weng, X.; Chen, B.; Wang, Y. Homoharringtonine synergy with oridonin in treatment of t(8; 21) acute myeloid leukemia. Front. Med. 2019, 13, 388–397. [Google Scholar] [CrossRef]
- Cao, H.; Cheng, Y.; You, L.; Qian, J.; Qian, W. Homoharringtonine and SAHA synergistically enhance apoptosis in human acute myeloid leukemia cells through upregulation of TRAIL and death receptors. Mol. Med. Rep. 2013, 7, 1838–1844. [Google Scholar] [CrossRef]
- Nguyen, T.; Parker, R.; Zhang, Y.; Hawkins, E.; Kmieciak, M.; Craun, W.; Grant, S. Homoharringtonine interacts synergistically with bortezomib in NHL cells through MCL-1 and NOXA-dependent mechanisms. BMC Cancer 2018, 18, 1129. [Google Scholar] [CrossRef]
- Bruno, P.M.; Liu, Y.; Park, G.Y.; Murai, J.; Koch, C.E.; Eisen, T.J.; Pritchard, J.R.; Pommier, Y.; Lippard, S.J.; Hemann, M.T. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 2017, 23, 461–471. [Google Scholar] [CrossRef]
- Sutton, E.C.; DeRose, V.J. Early nucleolar responses differentiate mechanisms of cell death induced by oxaliplatin and cisplatin. J. Biol. Chem. 2021, 296, 100633. [Google Scholar] [CrossRef]
- Piñeiro, D.; González, V.M.; Salinas, M.; Martín, M.E. Analysis of the protein expression changes during taxol-induced apoptosis under translation inhibition conditions. Mol. Cell. Biochem. 2010, 345, 131–144. [Google Scholar] [CrossRef]
- Knight, J.R.P.; Sansom, O.J. Tuning protein synthesis for cancer therapy. Mol. Cell. Oncol. 2021, 8, 1884034. [Google Scholar] [CrossRef]
- Makhale, A.; Nanayakkara, D.; Raninga, P.; Khanna, K.K.; Kalimutho, M. CX-5461 Enhances the Efficacy of APR-246 via Induction of DNA Damage and Replication Stress in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2021, 22, 5782. [Google Scholar] [CrossRef]
- Li, B.; Fang, L.; Wang, B.; Yang, Z.; Zhao, T. Identification of Prognostic RBPs in Osteosarcoma. Technol. Cancer Res. Treat. 2021, 20, 15330338211004918. [Google Scholar] [CrossRef]
- Ogawa, L.M.; Buhagiar, A.F.; Abriola, L.; Leland, B.A.; Surovtseva, Y.V.; Baserga, S.J. Increased numbers of nucleoli in a genome-wide RNAi screen reveal proteins that link the cell cycle to RNA polymerase I transcription. Mol. Biol. Cell 2021, 32, 956–973. [Google Scholar] [CrossRef]
- Rathner, A.; Rathner, P.; Friedrich, A.; Wießner, M.; Kitzler, C.M.; Schernthaner, J.; Karl, T.; Krauß, J.; Lottspeich, F.; Mewes, W.; et al. Drug Development for Target Ribosomal Protein rpL35/uL29 for Repair of LAMB3R635X in Rare Skin Disease Epidermolysis Bullosa. Ski. Pharmacol. Physiol. 2021, 34, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Fan, L.-N.; Han, D.-H.; Yu, Z.; Ma, J.; Liu, Y.-X.; Li, P.-F.; Zhao, D.-H.; Chai, J.; Jiang, L.; et al. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J. Clin. Investig. 2020, 130, 4301–4319. [Google Scholar] [CrossRef] [PubMed]
- Frandemiche, M.L.; De Seranno, S.; Rush, T.; Borel, E.; Elie, A.; Arnal, I.; Lanté, F.; Buisson, A. Activity-Dependent Tau Protein Translocation to Excitatory Synapse Is Disrupted by Exposure to Amyloid-Beta Oligomers. J. Neurosci. 2014, 34, 6084–6097. [Google Scholar] [CrossRef] [PubMed]
- Pallas-Bazarra, N.; Jurado-Arjona, J.; Navarrete, M.; Esteban, J.A.; Hernández, F.; Ávila, J.; Llorens-Martín, M. Novel function of Tau in regulating the effects of external stimuli on adult hippocampal neurogenesis. EMBO J. 2016, 35, 1417–1436. [Google Scholar] [CrossRef] [PubMed]
- Kneynsberg, A.; Combs, B.; Christensen, K.; Morfini, G.; Kanaan, N.M. Axonal Degeneration in Tauopathies: Disease Relevance and Underlying Mechanisms. Front. Neurosci. 2017, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, P.; Mármol, R.M.; Xia, D.; Götz, J.; Meunier, F.A. Frontotemporal dementia mutant Tau promotes aberrant Fyn nanoclustering in hippocampal dendritic spines. eLife 2019, 8, e45040. [Google Scholar] [CrossRef] [PubMed]
- Evans, H.T.; Taylor, D.; Kneynsberg, A.; Bodea, L.-G.; Götz, J. Altered ribosomal function and protein synthesis caused by tau. Acta Neuropathol. Commun. 2021, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Furlan, T.; Kirchmair, A.; Sampson, N.; Puhr, M.; Gruber, M.; Trajanoski, Z.; Santer, F.R.; Parson, W.; Handle, F.; Culig, Z. MYC-Mediated Ribosomal Gene Expression Sensitizes Enzalutamide-resistant Prostate Cancer Cells to EP300/CREBBP Inhibitors. Am. J. Pathol. 2021, 191, 1094–1107. [Google Scholar] [CrossRef]
- Bee, A.; Brewer, D.; Beesley, C.; Dodson, A.; Forootan, S.; Dickinson, T.; Gerard, P.; Lane, B.; Yao, S.; Cooper, C.S.; et al. siRNA Knockdown of Ribosomal Protein Gene RPL19 Abrogates the Aggressive Phenotype of Human Prostate Cancer. PLoS ONE 2011, 6, e22672. [Google Scholar] [CrossRef]
- Arthurs, C.; Murtaza, B.N.; Thomson, C.; Dickens, K.; Henrique, R.; Patel, H.R.H.; Beltran, M.; Millar, M.; Thrasivoulou, C.; Ahmed, A. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS ONE 2017, 12, e0186047. [Google Scholar] [CrossRef]
- Therizols, G.; Laforêts, F.; Marcel, V.; Catez, F.; Bouvet, P.; Diaz, J.-J. Chapter 6—Ribosomal RNA Methylation and Cancer. In Epigenetic Cancer Therapy; Gray, A.G., Ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 115–139. [Google Scholar] [CrossRef]
- Ganot, P.; Bortolin, M.-L.; Kiss, T. Site-Specific Pseudouridine Formation in Preribosomal RNA Is Guided by Small Nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef]
- Marcel, V.; Catez, F.; Diaz, J.J. Ribosome heterogeneity in tumorigenesis: The rRNA point of view. Mol. Cell. Oncol. 2015, 2, e983755. [Google Scholar] [CrossRef]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef]
- Uemura, M.; Zheng, Q.; Koh, C.M.; Nelson, W.G.; Yegnasubramanian, S.; De Marzo, A.M. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene 2012, 31, 1254–1263. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, Y.; Lv, Q.; Zhang, J.; Wang, Q.; Gao, F.; Hou, H.; Zhang, H.; Zhang, W.; Li, L. Overexpression of Ribosomal RNA in the Development of Human Cervical Cancer Is Associated with rDNA Promoter Hypomethylation. PLoS ONE 2016, 11, e0163340. [Google Scholar] [CrossRef]
- Tsoi, H.; Lam, K.C.; Dong, Y.; Zhang, X.; Lee, C.K.; Zhang, J.; Ng, S.C.; Ng, S.S.M.; Zheng, S.; Chen, Y.; et al. Pre-45s rRNA promotes colon cancer and is associated with poor survival of CRC patients. Oncogene 2017, 36, 6109–6118. [Google Scholar] [CrossRef] [Green Version]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA Pseudouridylation Defects Affect Ribosomal Ligand Binding and Translational Fidelity from Yeast to Human Cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.-P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. p53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and rRNA Methylation in Cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Blümke, J.; Wickenhauser, C.; Seliger, B. Relevance of 2′-O-Methylation and Pseudouridylation for the Malignant Melanoma. Cancers 2021, 13, 1167. [Google Scholar] [CrossRef]
- Meyuhas, O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000, 267, 6321–6330. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Kim, J.W.; Lee, B.D.; Kang, H.C.; Xu, J.-C.; Jia, H.; Stankowski, J.; Kim, M.-S.; Zhong, J.; Kumar, M.; et al. Ribosomal Protein s15 Phosphorylation Mediates LRRK2 Neurodegeneration in Parkinson’s Disease. Cell 2014, 157, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Venezia, N.D.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.-J. Emerging Role of Eukaryote Ribosomes in Translational Control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef] [PubMed]
- Watson, G.; Lester, D.; Ren, H.; Forsyth, C.M.; Medina, E.; Perez, D.G.; Darville, L.; Yao, J.; Luca, V.; Koomen, J.; et al. Fucosylated Proteome Profiling Identifies a Fucosylated, Non-Ribosomal, Stress-Responsive Species of Ribosomal Protein S3. Cells 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Kishton, R.J.; Angel, M.; Conn, C.S.; Dalla-Venezia, N.; Marcel, V.; Vincent, A.; Catez, F.; Ferré, S.; Ayadi, L.; et al. Ribosomal Proteins Regulate MHC Class I Peptide Generation for Immunosurveillance. Mol. Cell 2019, 73, 1162–1173.e5. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S. The history of ricin, abrin and related toxins. Toxicon 2004, 44, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Boston, R.S. Ribosome-inactivating proteins: A plant perspective. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 785–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Zhou, Y.-K.; Ji, Z.-L.; Chen, X.-R. The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 2018, 9, 146. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2014; 1104p. [Google Scholar]
- Chen, Y.; Han, L.; Bai, L.; Tang, H.; Zheng, A. Trichosanthin inhibits the proliferation of cervical cancer cells and downregulates STAT-5/C-myc signaling pathway. Pathol. Res. Pract. 2019, 215, 632–638. [Google Scholar] [CrossRef]
- Tuya, N.; Wang, Y.; Tong, L.; Gao, W.; Yu, R.; Xue, L. Trichosanthin enhances the antitumor effect of gemcitabine in non-small cell lung cancer via inhibition of the PI3K/AKT pathway. Exp. Ther. Med. 2017, 14, 5767–5772. [Google Scholar] [CrossRef]
- Li, C.; Zeng, M.; Chi, H.; Shen, J.; Ng, T.-B.; Jin, G.; Lu, D.; Fan, X.; Xiong, B.; Xiao, Z.; et al. Trichosanthin increases Granzyme B penetration into tumor cells by upregulation of CI-MPR on the cell surface. Oncotarget 2017, 8, 26460–26470. [Google Scholar] [CrossRef]
- He, D.; Jin, J.; Zheng, Y.; Bruce, I.C.; Tam, S.; Ma, X. Anti-angiogenesis effect of trichosanthin and the underlying mechanism. Biochem. Biophys. Res. Commun. 2013, 430, 735–740. [Google Scholar] [CrossRef]
- Liu, F.; Wang, B.; Wang, Z.; Yu, S. Trichosanthin down-regulates Notch signaling and inhibits proliferation of the nasopharyngeal carcinoma cell line CNE2 in vitro. Fitoterapia 2012, 83, 838–842. [Google Scholar] [CrossRef]
- Mosinger, M. [Necrosing or clastic effects of ricin on different organs and on experimental sarcomas]. C. R. Seances Soc. Biol. Fil. 1951, 145, 412–415. [Google Scholar]
- Lin, J.-Y.; Chang, Y.-C.; Huang, L.-Y.; Tung, T.-C. The cytotoxic effects of abrin and ricin on Ehrlich ascites tumor cells. Toxicon 1973, 11, 379–380. [Google Scholar] [CrossRef]
- Fodstad, O.; Kvalheim, G.; Godal, A.; Lotsberg, J.; Aamdal, S.; Høst, H.; Pihl, A. Phase I study of the plant protein ricin. Cancer Res. 1984, 44, 862–865. [Google Scholar]
- Licastro, F.; Morini, M.C.; Bolognesi, A.; Stirpe, F. Ricin induces the production of tumour necrosis factor-alpha and interleukin-1 beta by human peripheral-blood mononuclear cells. Biochem. J. 1993, 294, 517–520. [Google Scholar] [CrossRef]
- Gonzalez, T.V.; Farrant, S.A.; Mantis, N.J. Ricin induces IL-8 secretion from human monocyte/macrophages by activating the p38 MAP kinase pathway. Mol. Immunol. 2006, 43, 1920–1923. [Google Scholar] [CrossRef]
- Pervaiz, A.; Adwan, H.; Berger, M.R. Riproximin: A type II ribosome inactivating protein with anti-neoplastic potential induces IL24/MDA-7 and GADD genes in colorectal cancer cell lines. Int. J. Oncol. 2015, 47, 981–990. [Google Scholar] [CrossRef] [Green Version]
- Pervaiz, A.; Zepp, M.; Adwan, H.; Berger, M.R. Riproximin modulates multiple signaling cascades leading to cytostatic and apoptotic effects in human breast cancer cells. J. Cancer Res. Clin. Oncol. 2016, 142, 135–147. [Google Scholar] [CrossRef]
- Cao, D.; Sun, Y.; Wang, L.; He, Q.; Zheng, J.; Deng, F.; Deng, S.; Chang, S.; Yu, X.; Li, M.; et al. Alpha-momorcharin (α-MMC) exerts effective anti-human breast tumor activities but has a narrow therapeutic window in vivo. Fitoterapia 2015, 100, 139–149. [Google Scholar] [CrossRef]
- Deng, N.; Li, M.; Shen, D.; He, Q.; Sun, W.; Liu, M.; Liu, Y.; Zhou, Y.; Zheng, J.; Shen, F. LRP1 receptor-mediated immunosuppression of α-MMC on monocytes. Int. Immunopharmacol. 2019, 70, 80–87. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Li, C.; Ding, M.; Lv, X.; Tao, C.; Yu, H.; Chen, F.; Xu, Y. Curcin C, a novel type I ribosome-inactivating protein from the post-germinating cotyledons of Jatropha curcas. Amino Acids 2017, 49, 1619–1631. [Google Scholar] [CrossRef]
- Mishra, R.; Das, M.K.; Singh, S.; Sharma, R.S.; Sharma, S.; Mishra, V. Articulatin-D induces apoptosis via activation of caspase-8 in acute T-cell leukemia cell line. Mol. Cell. Biochem. 2017, 426, 87–99. [Google Scholar] [CrossRef]

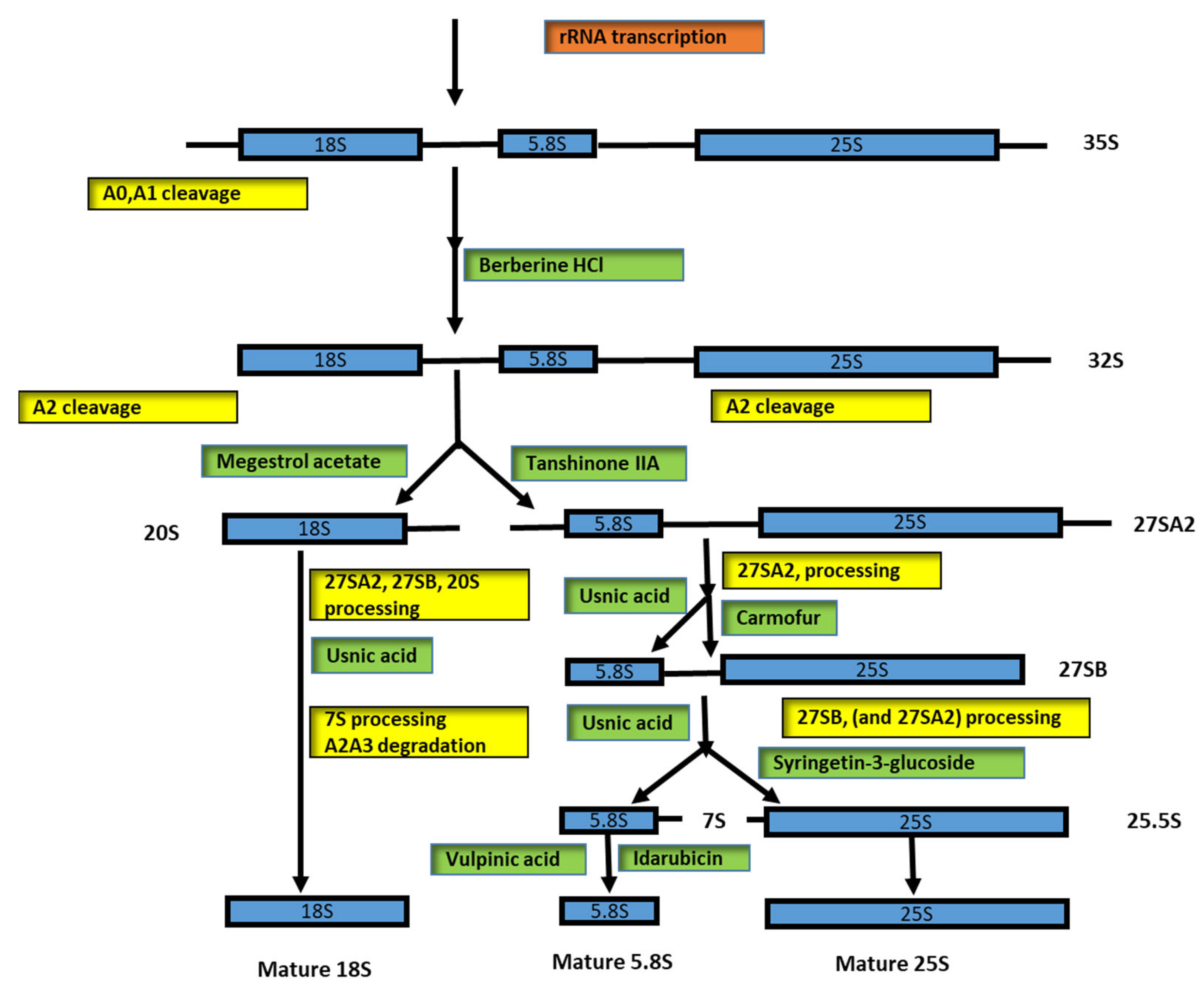
| Type of Substance | Inhibition | Diseases | References |
|---|---|---|---|
| Tanshinone IIA | 27SA2 pre-rRNA; 20SA2-pre-rRNA | Cancer | [61,64,65,66] |
| Diazaborine | Drg1 and block ATP hydrolysis | [62,63] | |
| Megestrol acetate | 20S pre-rRNA; 27S pre-rRNA | [61,67,68,69] | |
| Usnic acid | 20S pre-rRNA | [71,72] | |
| Celastrol | 27S pre-rRNA | [73] | |
| Parecoxib Na | 27S pre-rRNA | [74,75] | |
| Carmofur | 27S pre-rRNA | [76,77,78] | |
| 5-FU (5-fluorouracil) | 7S rRNA | [61] | |
| Syringetine-3-glucoside | 27S, 27SA2, 27SB precursors | [61] | |
| Vulpinic acid | 7S pre-rRNA, small fragments A2-A3, 23S rRNA | [61,83] | |
| Fluphenazine 2HCl | Small fragments A0, A1, A2 | [84] | |
| Idarubicin | 60S pre-ribosome maturation; 7S pre-rRNA accumulation | [85,86] | |
| Doxorubicin/epirubicin | Defects in pre-rRNA maturation | Solid tumors | [61,87,88] |
| Streptonigrin | Transcription and replication of DNA | [61,89] | |
| Cantharidin | 3′-5′ mRNA decay pathway | [90] | |
| Tunicamycin B | Ribosomal protein gene | [91,92,93] | |
| Methotrexate | Transcription of rRNA | [85,95,96] |
| Type of Substance | Inhibition | Diseases | References |
|---|---|---|---|
| CX-3543 | Facilitate Pol I transcription | Neuroendocrine and carcinoid tumors | [112,113] |
| CX-5461 | Inhibit selectively Pol I | Prostate cancer; breast cancer; small lung cancer; ovarian cancer; neuroblastoma | [117,118,119,120,121,122,123] |
| CX-5461/CX-6258 | Pol I | Prostate cancer | [119] |
| BMH-21 | Pol I | Cancer | [124] |
| Type of Substrate | Inhibition | References |
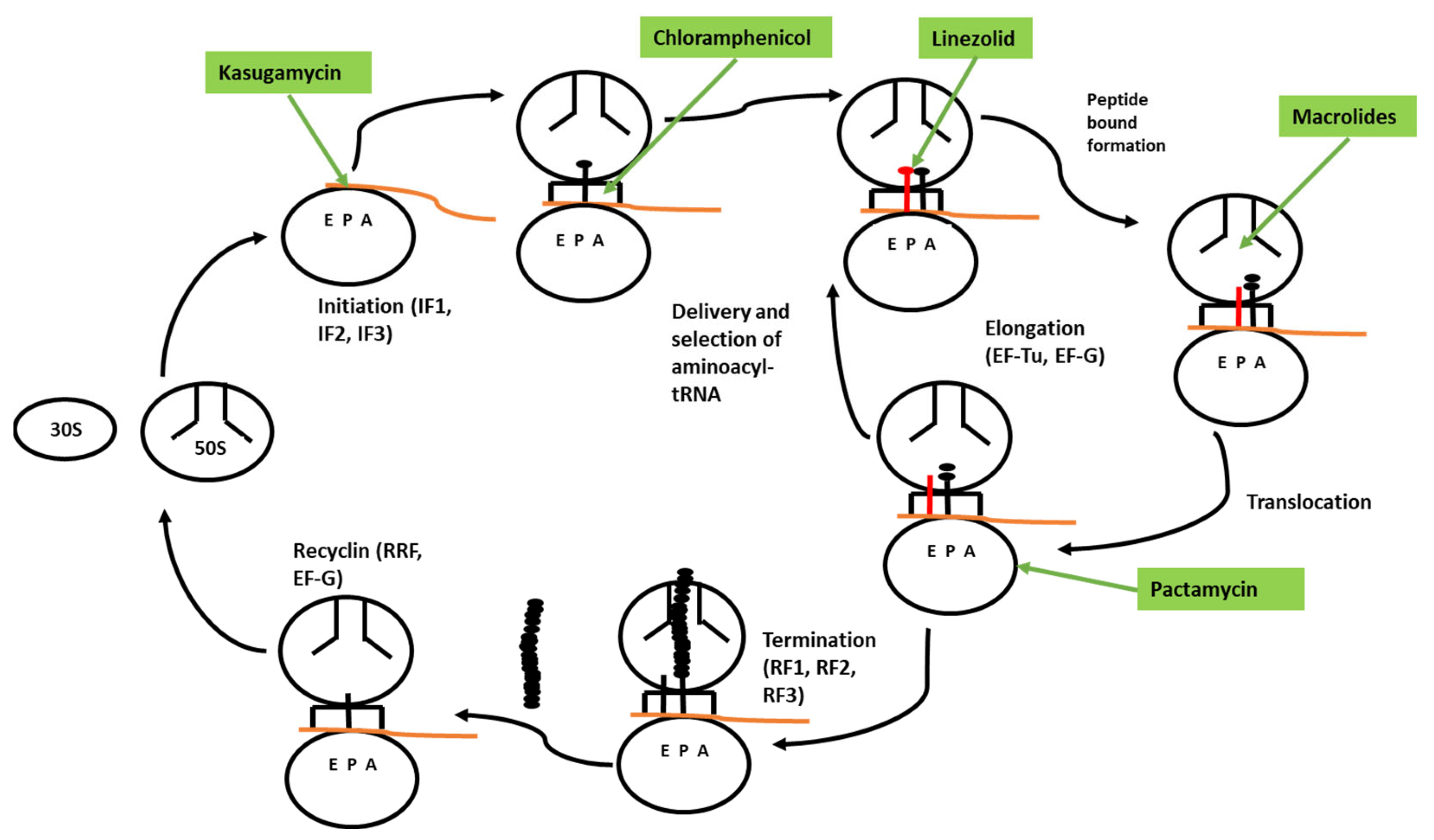
|---|---|---|
| Chloramphenicol (CHL) | Peptide bond formation | [138,139] |
| Linezolid (LZD) | Peptide bond formation | [140,141] |
| Erythromycin (ERY) | Peptide bond formation | [142,143] |
| Azithromycin (AZA) | Peptide bond formation | [142,143] |
| Pikromycin | Arrest ribosome maturation | [145] |
| Kasugamycin (KSG) | Bind 30S subunit | [147,148] |
| Paramycin (PAR) | Protein synthesis | [150] |
| Pactamycin (PAC) | Bind 30S subunit | [150] |
| Small molecule PF06446846 | Not known | [151] |
| Type of Substance | Inhibition | Diseases | References |
|---|---|---|---|
| Harringtonine | 80S | Murine leukemia cells; chronic myeloid leukemia | [164] |
| Bortezomib/ Harringtonine | Diffuse large B cell lymphoma (DLBCL) | [167] | |
| Oxaliplatin | Cell growth via ribosome biogenesis | Tumor/cancer | [168,169] |
| Cisplatine | Ribosome biogenesis | Tumor/cancer | [169] |
| eFT508 (tomivoserbit) | Suppress protein synthesis | Solid tumors | [170,171] |
| 5-Fluorouracil/paclitaxel | Suppress protein synthesis | Breast cancer | [170,171] |
| Cx-5461/APR-246 | Induce apoptosis | Triple-negative breast cancer (TNBC) | [172] |
| Ispinesib, Nocodazole, Paclitaxel, Aurora kinase inhibitor, Hesperidin, MK-5108 | Inhibit DNA replication | Cancers | [174] |
| Atazanavir/artesunate | LAMB3PTC mRNA | Junction epidermolysis bullosa | [175] |
| Tau protein | Decrease protein synthesis | Cell signaling and axonal transport | [181] |
| Enzalutamide | Down regulate BD/HAT | Prostate cancer | [182] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temaj, G.; Chichiarelli, S.; Eufemi, M.; Altieri, F.; Hadziselimovic, R.; Farooqi, A.A.; Yaylim, I.; Saso, L. Ribosome-Directed Therapies in Cancer. Biomedicines 2022, 10, 2088. https://doi.org/10.3390/biomedicines10092088
Temaj G, Chichiarelli S, Eufemi M, Altieri F, Hadziselimovic R, Farooqi AA, Yaylim I, Saso L. Ribosome-Directed Therapies in Cancer. Biomedicines. 2022; 10(9):2088. https://doi.org/10.3390/biomedicines10092088
Chicago/Turabian StyleTemaj, Gazmend, Silvia Chichiarelli, Margherita Eufemi, Fabio Altieri, Rifat Hadziselimovic, Ammad Ahmad Farooqi, Ilhan Yaylim, and Luciano Saso. 2022. "Ribosome-Directed Therapies in Cancer" Biomedicines 10, no. 9: 2088. https://doi.org/10.3390/biomedicines10092088







