Enhancement of Chondrogenic Markers by Exosomes Derived from Cultured Human Synovial Fluid-Derived Cells: A Comparative Analysis of 2D and 3D Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Sample Processing
2.3. Culturing Human Synovial Fluid-Derived Cells (SFDC)
2.4. Cell Proliferation Assay
2.5. Isolation of Exosomes
2.6. Exosome Characterization
2.7. Exosome on Chondrocyte Viability
2.8. Exosomes on Chondrogenic Differentiation
2.9. Real-Time PCR
2.10. Animal Studies
2.11. Statistical Analysis
3. Results
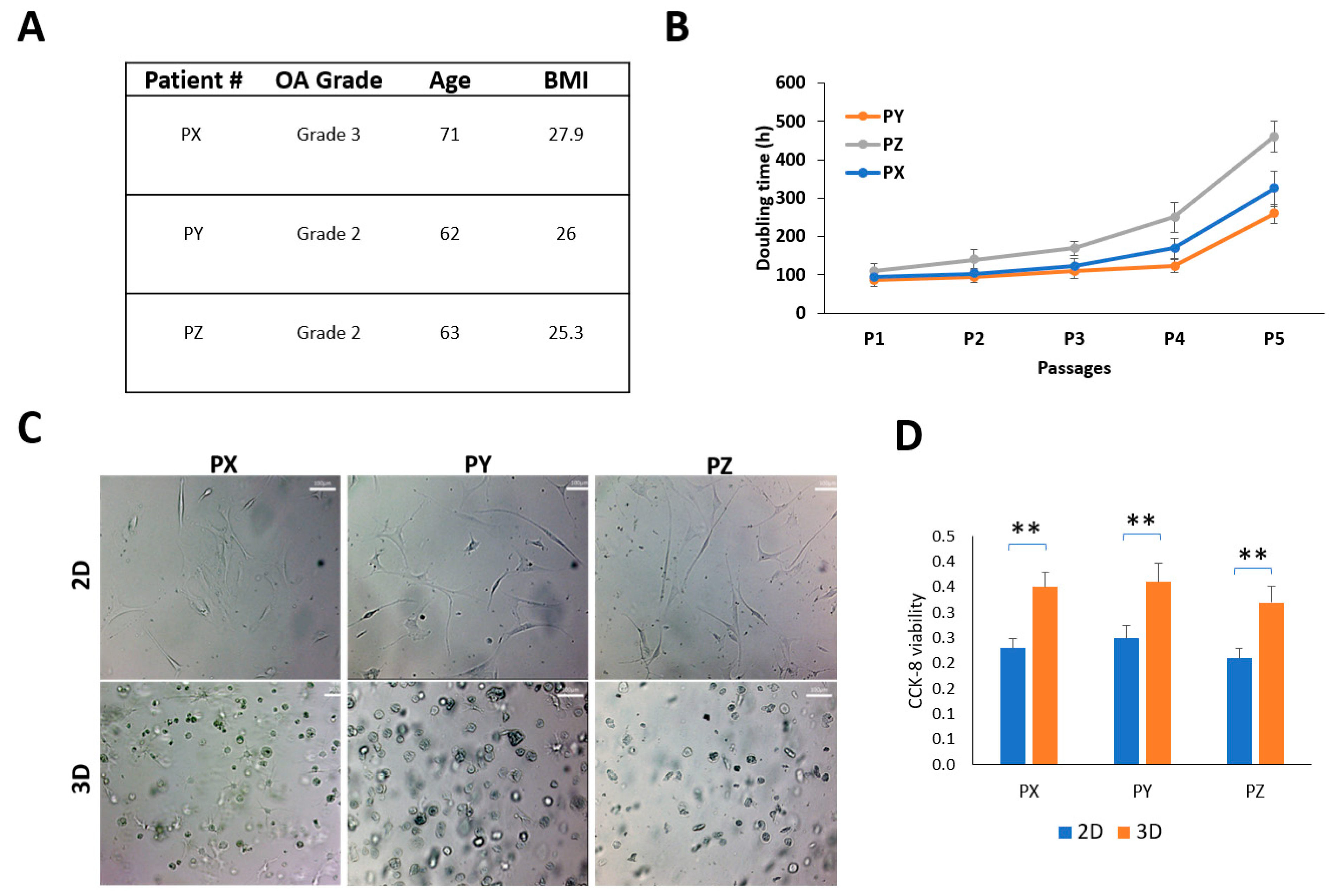
3.1. Patient and Synovial Fluid-Derived Cell Characteristics
3.2. Isolation and Characterization of SFDC Exosomes
3.3. Quantitative and Qualitative Comparison of Exosomes Derived from 2D and 3D Cell Cultures
3.4. The In Vitro Effects of SFDC Exosomes on Viability and Redifferentiation of Dedifferentiated Human Chondrocytes
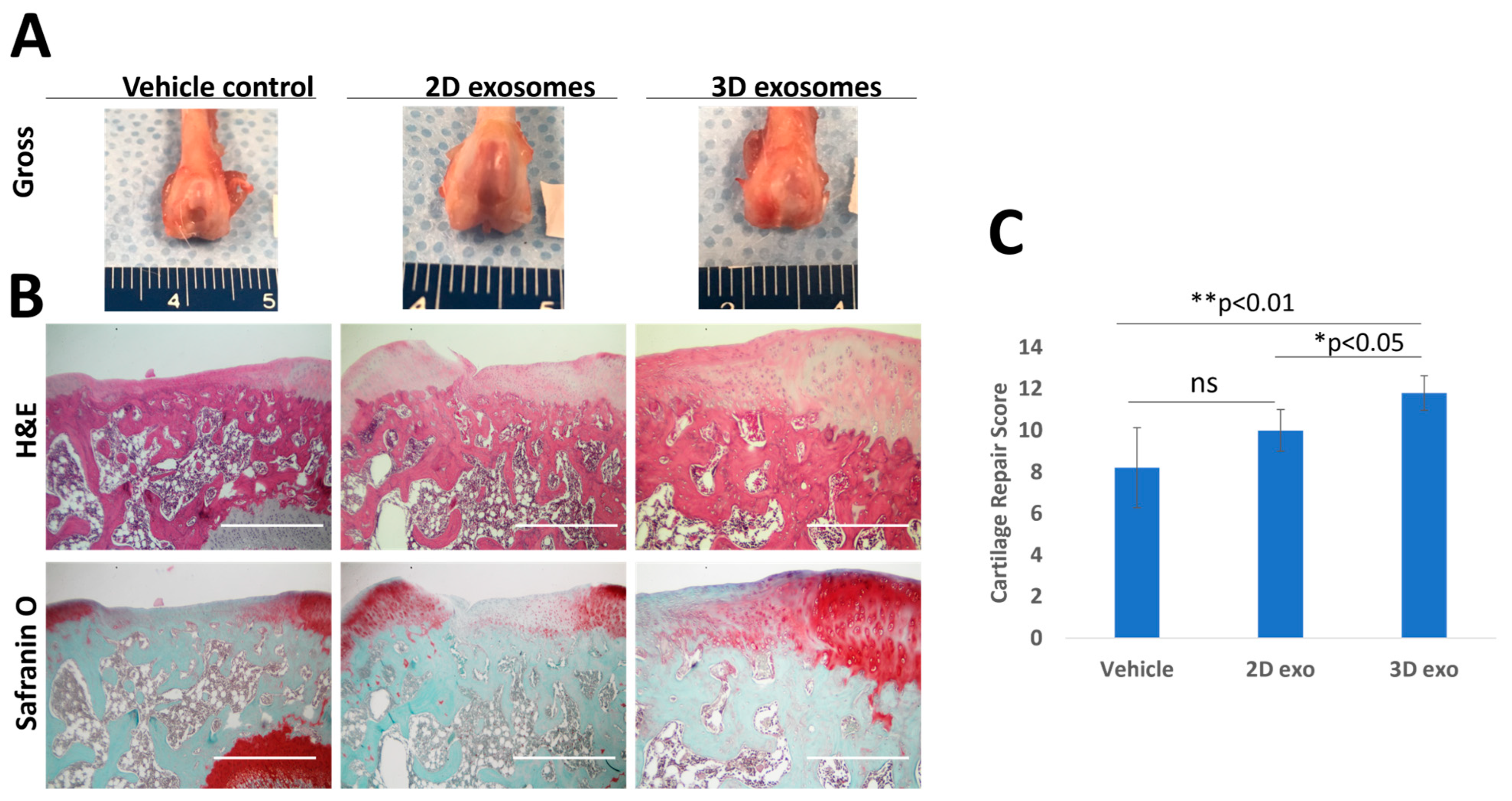
3.5. The In Vivo Effects of SFDC Exosomes on Cartilage Repair
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Varela-Eirin, M.; Loureiro, J.; Fonseca, E.; Corrochano, S.; Caeiro, J.R.; Collado, M.; Mayan, M.D. Cartilage regeneration and ageing: Targeting cellular plasticity in osteoarthritis. Ageing Res. Rev. 2018, 42, 56–71. [Google Scholar] [CrossRef]
- Palazzo, C.; Nguyen, C.; Lefevre-Colau, M.-M.; Rannou, F.; Poiraudeau, S. Risk factors and burden of osteoarthritis. Ann. Phys. Rehabil. Med. 2016, 59, 134–138. [Google Scholar] [CrossRef]
- Vina, E.R.; Kwoh, C.K. Epidemiology of osteoarthritis: Literature update. Curr. Opin. Rheumatol. 2018, 30, 160–167. [Google Scholar] [CrossRef]
- Belluzzi, E.; Stocco, E.; Pozzuoli, A.; Granzotto, M.; Porzionato, A.; Vettor, R.; De Caro, R.; Ruggieri, P.; Ramonda, R.; Rossato, M.; et al. Contribution of infrapatellar fat pad and synovial membrane to knee osteoarthritis pain. BioMed Res. Int. 2019, 2019, 6390182. [Google Scholar] [CrossRef]
- Sargent, A.; Miller, R.H. MSC Therapeutics in Chronic Inflammation. Curr. Stem Cell Rep. 2016, 2, 168–173. [Google Scholar] [CrossRef]
- Fang, W.; Sun, Z.; Chen, X.; Han, B.; Vangsness, C.T. Synovial Fluid Mesenchymal Stem Cells for Knee Arthritis and Cartilage Defects: A Review of the Literature. J. Knee Surg. 2020, 34, 1476–1485. [Google Scholar] [CrossRef]
- Morito, T.; Muneta, T.; Hara, K.; Ju, Y.-J.; Mochizuki, T.; Makino, H.; Umezawa, A.; Sekiya, I. Synovial fluid-derived mesenchymal stem cells increase after intra-articular ligament injury in humans. Rheumatology 2008, 47, 1137–1143. [Google Scholar] [CrossRef]
- Jia, Z.; Liang, Y.; Xu, X.; Li, X.; Liu, Q.; Ou, Y.; Duan, L.; Zhu, W.; Lu, W.; Xiong, J.; et al. Isolation and characterization of human mesenchymal stem cells derived from synovial fluid by magnetic-activated cell sorting (MACS). Cell Biol. Int. 2018, 42, 262–271. [Google Scholar] [CrossRef]
- Isobe, Y.; Koyama, N.; Nakao, K.; Osawa, K.; Ikeno, M.; Yamanaka, S.; Okubo, Y.; Fujimura, K.; Bessho, K. Comparison of human mesenchymal stem cells derived from bone marrow, synovial fluid, adult dental pulp, and exfoliated deciduous tooth pulp. Int. J. Oral Maxillofac. Surg. 2016, 45, 124–131. [Google Scholar] [CrossRef]
- Li, J.; Huang, Y.; Song, J.; Li, X.; Zhang, X.; Zhou, Z.; Chen, D.; Ma, P.X.; Peng, W.; Wang, W.; et al. Cartilage regeneration using arthroscopic flushing fluid-derived mesenchymal stem cells encapsulated in a one-step rapid cross-linked hydrogel. Acta Biomater. 2018, 79, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Liu, Q.; Liang, Y.; Li, X.; Xu, X.; Ouyang, K.; Xiong, J.; Wang, D.; Duan, L. Repair of articular cartilage defects with intra-articular injection of autologous rabbit synovial fluid-derived mesenchymal stem cells. J. Transl. Med. 2018, 16, 123. [Google Scholar] [CrossRef]
- Tan, S.; Fang, W.; Vangsness, C.T., Jr.; Han, B. Influence of Cellular Microenvironment on Human Articular Chondrocyte Cell Signaling. Cartilage 2021, 13, 935S–946S. [Google Scholar] [CrossRef]
- Barreto-Durán, E.; Mejía-Cruz, C.C.; Leal-García, E.; Pérez-Núñez, R.; Rodríguez-Pardo, V.M. Impact of donor characteristics on the quality of bone marrow as a source of mesenchymal stromal cells. Am. J. Stem Cells 2018, 7, 114. [Google Scholar]
- Ankrum, J.A.; Ong, J.F.; Karp, J.M. Mesenchymal stem cells: Immune evasive, not immune privileged. Nat. Biotechnol. 2014, 32, 252–260. [Google Scholar] [CrossRef]
- Oliveira, R.L.; Chagastelles, P.C.; Sesterheim, P.; Pranke, P. In vivo immunogenic response to allogeneic mesenchymal stem cells and the role of preactivated mesenchymal stem cells cotransplanted with allogeneic islets. Stem Cells Int. 2017, 2017, 9824698. [Google Scholar] [CrossRef]
- Joswig, A.-J.; Mitchell, A.; Cummings, K.J.; Levine, G.J.; Gregory, C.A.; Smith, R.; Watts, A.E. Repeated intra-articular injection of allogeneic mesenchymal stem cells causes an adverse response compared to autologous cells in the equine model. Stem Cell Res. Ther. 2017, 8, 42. [Google Scholar] [CrossRef]
- Phan, J.; Kumar, P.; Hao, D.; Gao, K.; Farmer, D.; Wang, A. Engineering mesenchymal stem cells to improve their exosome efficacy and yield for cell-free therapy. J. Extracell. Vesicles 2018, 7, 1522236. [Google Scholar] [CrossRef]
- Latifkar, A.; Hur, Y.H.; Sanchez, J.C.; Cerione, R.A.; Antonyak, M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019, 132, jcs222406. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics characterization of exosome cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.-C.; Yuan, T.; Zhang, Y.-L.; Yin, W.-J.; Guo, S.-C.; Zhang, C.-Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chu, W.C.; Lai, R.C.; Lim, S.K.; Hui, J.H.P.; Toh, W.S. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr. Cartil. 2016, 24, 2135–2140. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, D.; Liu, Z.; Zhou, F.; Dai, J.; Wu, B.; Zhou, J.; Heng, B.C.; Zou, X.H.; Ouyang, H.; et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res. Ther. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 5335. [Google Scholar] [CrossRef]
- Lobb, R.j.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Yan, L.; Wu, X. Exosomes produced from 3D cultures of umbilical cord mesenchymal stem cells in a hollow-fiber bioreactor show improved osteochondral regeneration activity. Cell Biol. Toxicol. 2019, 36, 165–178. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.-C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef]
- Faruqu, F.N.; Liam-Or, R.; Zhou, S.; Nip, R.; Al-Jamal, K.T. Defined serum-free three-dimensional culture of umbilical cord-derived mesenchymal stem cells yields exosomes that promote fibroblast proliferation and migration in vitro. FASEB J. 2021, 35, e21206. [Google Scholar] [CrossRef]
- McWilliam, R.H.; Chang, W.; Liu, Z.; Wang, J.; Han, F.; Black, R.A.; Wu, J.; Luo, X.; Li, B.; Shu, W. Three-dimensional biofabrication of nanosecond laser micromachined nanofibre meshes for tissue engineered scaffolds. Biomater. Transl. 2023, 4, 104–114. [Google Scholar] [CrossRef]
- Sun, Z.; Yang, S.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Xu, J.; Xia, K.; Chang, Y.; et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer 2018, 17, 82. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tian, Y.; Wang, Y.; Xu, L.; Song, G.; Wu, Q.; Wang, W.; Xie, M. Exosomes derived from astrocytes after oxygen-glucose deprivation promote differentiation and migration of oligodendrocyte precursor cells in vitro. Mol. Biol. Rep. 2021, 48, 5473–5484. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; Marturano-Kruik, A.; Ambati, S.R.; Liu, Z.; Godier-Furnemont, A.; Parsa, H.; Lee, B.W.; Moore, M.A.; Vunjak-Novakovic, G. Recapitulating the Size and Cargo of Tumor Exosomes in a Tissue-Engineered Model. Theranostics 2016, 6, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Tan, S.-J.; Yang, Z.; Tayag, C.; Han, B. Tumor Bioengineering Using a Transglutaminase Crosslinked Hydrogel. PLoS ONE 2014, 9, e105616. [Google Scholar] [CrossRef] [PubMed]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Hikita, T.; Miyata, M.; Watanabe, R.; Oneyama, C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep. 2018, 8, 14035. [Google Scholar] [CrossRef]
- Bergholt, N.L.; Lysdahl, H.; Lind, M.; Foldager, C.B. A Standardized Method of Applying Toluidine Blue Metachromatic Staining for Assessment of Chondrogenesis. Cartilage 2019, 10, 370–374. [Google Scholar] [CrossRef]
- O’driscoll, S.W.; Keeley, F.W.; Salter, R.B. The chondrogenic potential of free autogenous periosteal grafts for biological resurfacing of major full-thickness defects in joint surfaces under the influence of continuous passive motion. An experimental investigation in the rabbit. Minerva Anestesiol. 1986, 68, 1017–1035. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Wei, Y.; Bai, L. Recent advances in the understanding of molecular mechanisms of cartilage degeneration, synovitis and subchondral bone changes in osteoarthritis. Connect. Tissue Res. 2016, 57, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-W.; Chen, W.-C.; Liu, H.-W.; Chen, C.-H. Application of Synovial Fluid Mesenchymal Stem Cells: Platelet-rich Plasma Hydrogel for Focal Cartilage Defect. J. Exp. Clin. Med. 2014, 6, 118–124. [Google Scholar] [CrossRef]
- Jess, R.; Ling, T.; Xiong, Y.; Wright, C.J.; Zhao, F. Mechanical environment for in vitro cartilage tissue engineering assisted by in silico models. Biomater. Transl. 2023, 4, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Irawan, V.; Sung, T.-C.; Higuchi, A.; Ikoma, T. Collagen Scaffolds in Cartilage Tissue Engineering and Relevant Approaches for Future Development. Tissue Eng. Regen. Med. 2018, 15, 673–697. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, S.; Chen, W.; Hu, Y.; Geng, Z.; Su, J. Bone/cartilage targeted hydrogel: Strategies and applications. Bioact. Mater. 2023, 23, 156–169. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, X.; Qu, Z.; Hao, J.; Zhang, W. Hypoxia-Preconditioned Extracellular Vesicles from Mesenchymal Stem Cells Improve Cartilage Repair in Osteoarthritis. Membranes 2022, 12, 225. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. In Proteomic Profiling; Springer: Berlin/Heidelberg, Germany, 2015; pp. 179–209. [Google Scholar]
- Bannuru, R.R.; Osani, M.C.; Vaysbrot, E.E.; Arden, N.K.; Bennell, K.; Bierma-Zeinstra, S.M.A.; Kraus, V.B.; Lohmander, L.S.; Abbott, J.H.; Bhandari, M.; et al. OARSI guidelines for the non-surgical management of knee, hip, and polyarticular osteoarthritis. Osteoarthr. Cartil. 2019, 27, 1578–1589. [Google Scholar] [CrossRef]
- Fang, Y.; Wu, N.; Gan, X.; Yan, W.; Morrell, J.C.; Gould, S.J. Higher-Order Oligomerization Targets Plasma Membrane Proteins and HIV Gag to Exosomes. PLoS Biol. 2007, 5, e158. [Google Scholar] [CrossRef]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Zhu, L.; Kalimuthu, S.; Gangadaran, P.; Oh, J.M.; Lee, H.W.; Baek, S.H.; Jeong, S.Y.; Lee, S.-W.; Lee, J.; Ahn, B.-C. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics 2017, 7, 2732–2745. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, J. Synovial fluid-derived exosomal lncRNA PCGEM1 as biomarker for the different stages of osteoarthritis. Int. Orthop. 2018, 42, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Benya, P.D.; Padilla, S.R.; Nimni, M.E. Independent regulation of collagen types by chondrocytes during the loss of differentiated function in culture. Cell 1978, 15, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Tanzil, G.; Mobasheri, A.; de Souza, P.; John, T.; Shakibaei, M. Loss of chondrogenic potential in dedifferentiated chondrocytes correlates with deficient Shc–Erk interaction and apoptosis. Osteoarthr. Cartil. 2004, 12, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Leijten, J.; Wu, L.; Kip, M.; van Blitterswijk, C.; Post, J.; Karperien, M. Gene expression profiling of dedifferentiated human articular chondrocytes in monolayer culture. Osteoarthr. Cartil. 2013, 21, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, R.L.; Kinard, L.A.; Lam, J.; Needham, C.J.; Lu, S.; Kasper, F.K.; Mikos, A.G. Articular chondrocytes and mesenchymal stem cells seeded on biodegradable scaffolds for the repair of cartilage in a rat osteochondral defect model. Biomaterials 2014, 35, 7460–7469. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, J.Y.; Peng, W.M.; Yuan, B.; Bi, Q.; Xu, Y.J. Exosomes from adipose-derived stem cells promote chondrogenesis and suppress inflammation by upregulating miR-145 and miR-221. Mol. Med. Rep. 2020, 21, 1881–1889. [Google Scholar] [CrossRef]
- Mao, G.; Xu, Y.; Long, D.; Sun, H.; Li, H.; Xin, R.; Zhang, Z.; Li, Z.; Yang, Z.; Kang, Y. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res. Ther. 2021, 12, 389. [Google Scholar] [CrossRef]
- Shi, G.; Long, Z.; De la Vega, R.E.; Behfar, A.; Moran, S.L.; Evans, C.; Zhao, C. Purified exosome product enhances chondrocyte survival and regeneration by modulating inflammation and promoting chondrogenesis. Regen. Med. 2023, 18, 55–71. [Google Scholar] [CrossRef]
- Wu, J.; Liu, W.; Bemis, A.; Wang, E.; Qiu, Y.; Morris, E.A.; Flannery, C.R.; Yang, Z. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2007, 56, 3675–3684. [Google Scholar] [CrossRef]
- Tsuchida, A.I.; Beekhuizen, M.; ‘t Hart, M.C.; Radstake, T.; Dhert, W.; Saris, D.; van Osch, G.; Creemers, L.B. Cytokine profiles in the joint depend on pathology, but are different between synovial fluid, cartilage tissue and cultured chondrocytes. Arthritis Res. Ther. 2014, 16, 441. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, B.; Fang, W.; Yang, Z.; Wang, Y.; Zhao, S.; Hoang, B.X.; Vangsness, C.T., Jr. Enhancement of Chondrogenic Markers by Exosomes Derived from Cultured Human Synovial Fluid-Derived Cells: A Comparative Analysis of 2D and 3D Conditions. Biomedicines 2023, 11, 3145. https://doi.org/10.3390/biomedicines11123145
Han B, Fang W, Yang Z, Wang Y, Zhao S, Hoang BX, Vangsness CT Jr. Enhancement of Chondrogenic Markers by Exosomes Derived from Cultured Human Synovial Fluid-Derived Cells: A Comparative Analysis of 2D and 3D Conditions. Biomedicines. 2023; 11(12):3145. https://doi.org/10.3390/biomedicines11123145
Chicago/Turabian StyleHan, Bo, William Fang, Zhi Yang, Yuntao Wang, Shuqing Zhao, Ba Xuan Hoang, and C. Thomas Vangsness, Jr. 2023. "Enhancement of Chondrogenic Markers by Exosomes Derived from Cultured Human Synovial Fluid-Derived Cells: A Comparative Analysis of 2D and 3D Conditions" Biomedicines 11, no. 12: 3145. https://doi.org/10.3390/biomedicines11123145




