Weighted Gene Co-Expression Network Analysis (WGCNA) Discovered Novel Long Non-Coding RNAs for Polycystic Ovary Syndrome
Abstract
:1. Introduction
2. Materials and Methods
2.1. PCOS Data Acquisition
2.2. Processing of the Dataset and Identification of (Differential Expression Genes) DEGs
2.3. WGCNA and Module Identification
2.4. Enrichment Analysis of the Modules and Identification of the Interaction Networks and Hub Genes
3. Results
3.1. Identification of Differentially Expressed lncRNAs
3.2. Analysis of Weighted Gene Co-Expression Networks and Classification of the Desired Module
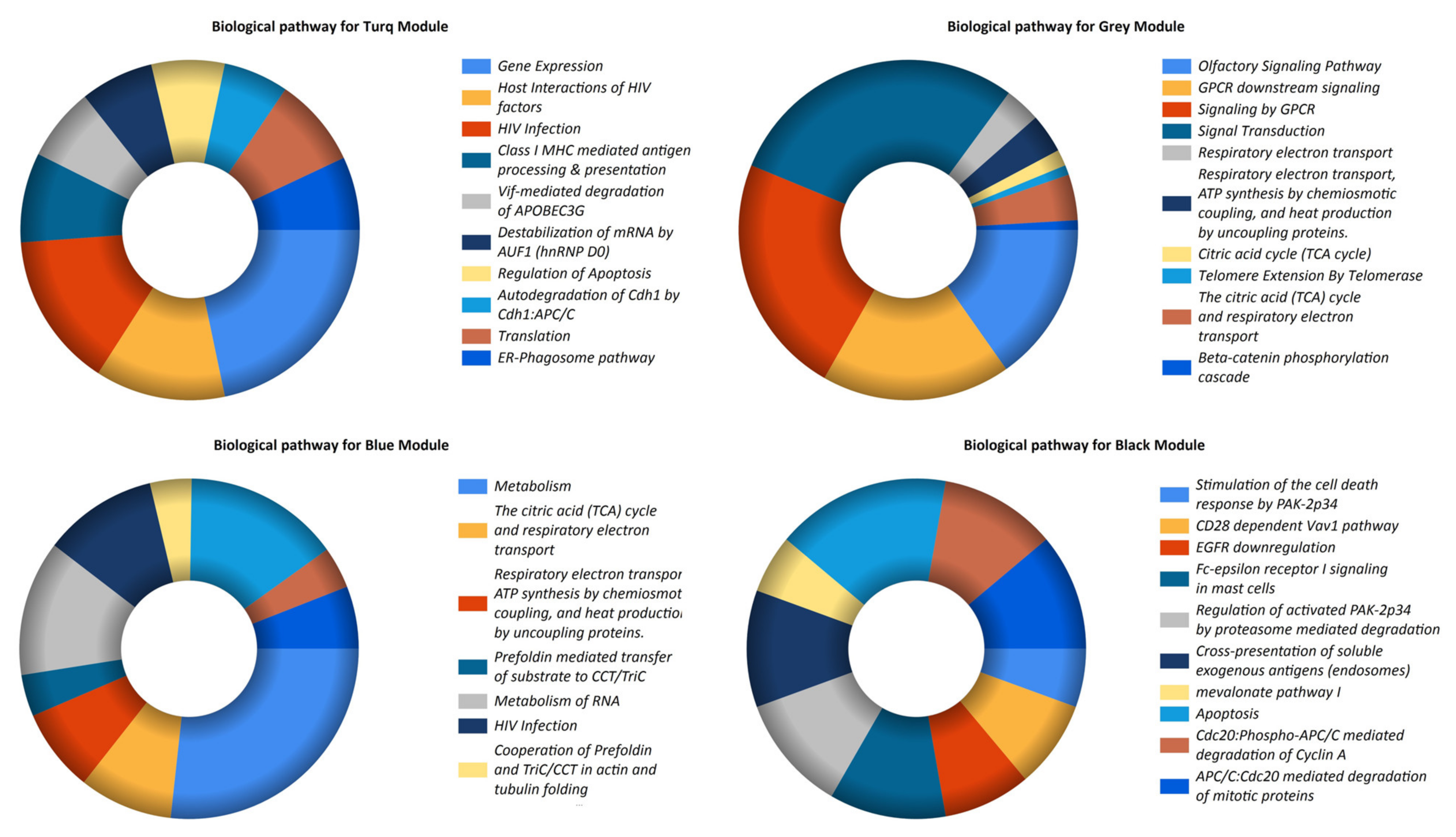
3.3. Pathway Enrichment and Establishment of the PPI Network and Hub Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ye, W.; Xie, T.; Song, Y.; Zhou, L. The role of androgen and its related signals in PCOS. J. Cell. Mol. Med. 2021, 25, 1825–1837. [Google Scholar] [CrossRef]
- Chang, S.; Dunaif, A. Diagnosis of Polycystic Ovary Syndrome: Which Criteria to Use and When? Endocrinol. Metab. Clin. 2021, 50, 11–23. [Google Scholar] [CrossRef]
- Luo, Y.; Cui, C.; Han, X.; Wang, Q.; Zhang, C. The role of miRNAs in polycystic ovary syndrome with insulin resistance. J. Assist. Reprod. Genet. 2021, 38, 289. [Google Scholar] [CrossRef]
- Gnanadass, S.A.; Prabhu, Y.D.; Gopalakrishnan, A.V. Association of metabolic and inflammatory markers with polycystic ovarian syndrome (PCOS): An update. Arch. Gynecol. Obstet. 2021, 303, 631–643. [Google Scholar] [CrossRef]
- Heidarzadehpilehrood, R.; Pirhoushiaran, M.; Abdollahzadeh, R.; Binti Osman, M.; Sakinah, M.; Nordin, N.; Abdul Hamid, H. A Review on CYP11A1, CYP17A1, and CYP19A1 Polymorphism Studies: Candidate Susceptibility Genes for Polycystic Ovary Syndrome (PCOS) and Infertility. Genes 2022, 13, 302. [Google Scholar] [CrossRef]
- Sirmans, S.M.; Pate, K.A. Epidemiology, diagnosis, and management of polycystic ovary syndrome. Clin. Epidemiol. 2014, 6, 1–13. [Google Scholar] [CrossRef]
- Dapas, M.; Lin, F.T.J.; Nadkarni, G.N.; Sisk, R.; Legro, R.S.; Urbanek, M.; Hayes, M.G.; Dunaif, A. Distinct subtypes of polycystic ovary syndrome with novel genetic associations: An unsupervised, phenotypic clustering analysis. PLoS Med. 2020, 17, e1003132. [Google Scholar] [CrossRef]
- Oguz, S.H.; Yildiz, O. An Update on Contraception in Polycystic Ovary Syndrome. Endocrinol. Metab. 2021, 36, 296–311. [Google Scholar] [CrossRef]
- Burks, H.R.; Wild, R.A. Diagnostic criteria and epidemiology of PCOS. In Polycystic Ovary Syndrome: Current and Emerging Concepts; Springer: New York, NY, USA, 2014; pp. 3–10. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Johnson, R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell 2019, 73, 869–883. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, L.; Zhong, T.; Mueller, M.; Men, Y.; Zhang, N.; Xie, J.; Giang, K.; Chung, H.; Sun, X.; et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 2015, 6, 10221. [Google Scholar] [CrossRef]
- Han, P.; Chang, C.P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef]
- Liang, W.C.; Ren, J.L.; Wong, C.W.; Chan, S.O.; Waye, M.M.; Fu, W.M.; Zhang, J.F. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene 2018, 37, 1445–1456. [Google Scholar] [CrossRef]
- Han, Q.; Xu, L.; Lin, W.; Yao, X.; Jiang, M.; Zhou, R.; Sun, X.; Zhao, L. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene 2018, 38, 3019–3032. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic Discovery of Xist RNA Binding Proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Luo, J.; Luan, S.; He, C.; Li, Z. Long non-coding RNAs involved in cancer metabolic reprogramming. Cell. Mol. Life Sci. 2019, 76, 495–504. [Google Scholar] [CrossRef]
- Rahimpour, A.; Heidarzadehpilehrood, R.; Abdollahi, S.; Ranjbari, H.; Shams, Z.; Ghasemi, S.A.; Najmaei, S.; Pirhoushiaran, M. A comprehensive bioinformatic analysis revealed novel MicroRNA biomarkers of Parkinson’s disease. Cell Biol. Int. 2022, 46, 1841–1851. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11, 1485. [Google Scholar] [CrossRef]
- Li, L.; Zhu, J.; Ye, F.; Duan, Z.; Zhou, J.; Huang, Z.; Wang, L. Upregulation of the lncRNA SRLR in polycystic ovary syndrome regulates cell apoptosis and IL-6 expression. Cell Biochem. Funct. 2020, 38, 880–885. [Google Scholar] [CrossRef]
- Qin, L.; Huang, C.C.; Yan, X.M.; Wang, Y.; Li, Z.Y.; Wei, X.C. Long non-coding RNA h19 is associated with polycystic ovary syndrome in Chinese women: A preliminary study. Endocr. J. 2019, 66, 587–595. [Google Scholar] [CrossRef]
- Liu, M.; Zhu, H.; Li, Y.; Zhuang, J.; Cao, T.; Wang, Y. Expression of serum lncRNA-Xist in patients with polycystic ovary syndrome and its relationship with pregnancy outcome. Taiwan. J. Obstet. Gynecol. 2020, 59, 372–376. [Google Scholar] [CrossRef]
- Wang, J.; Xia, S.; Arand, B.; Zhu, H.; Machiraju, R.; Huang, K.; Ji, H.; Qian, J. Single-Cell Co-expression Analysis Reveals Distinct Functional Modules, Co-regulation Mechanisms and Clinical Outcomes. PLoS Comput. Biol. 2016, 12, e4892. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Sean, D.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Smyth, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Sørensen, A.E.; Wissing, M.L.; Salö, S.; Lis, A.; Englund, M.; Dalgaard, L.T. MicroRNAs related to polycystic ovary syndrome (PCOS). Genes 2014, 5, 684–708. [Google Scholar] [CrossRef]
- McCartney, C.R.; Marshall, J.C. CLINICAL PRACTICE. Polycystic Ovary Syndrome. N. Engl. J. Med. 2016, 375, 54–64. [Google Scholar] [CrossRef] [Green Version]
- Mutter, G.L.; Zaino, R.J.; Baak, J.P.A.; Bentley, R.C.; Robboy, S.J. Benign endometrial hyperplasia sequence and endometrial intraepithelial neoplasia. Int. J. Gynecol. Pathol. 2007, 26, 103–114. [Google Scholar] [CrossRef]
- Gompel, A. Progesterone and endometrial cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 69, 95–107. [Google Scholar] [CrossRef]
- Khan, G.H.; Galazis, N.; Docheva, N.; Layfield, R.; Atiomo, W. Overlap of proteomics biomarkers between women with pre-eclampsia and PCOS: A systematic review and biomarker database integration. Hum. Reprod. 2015, 30, 133–148. [Google Scholar] [CrossRef]
- De Leo, V.; Musacchio, M.C.; Cappelli, V.; Massaro, M.G.; Morgante, G.; Petraglia, F. Genetic, hormonal and metabolic aspects of PCOS: An update. Reprod. Biol. Endocrinol. 2016, 14, 38. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Lim, L.J.; Wong, S.Y.S.; Huang, F.; Lim, S.; Chong, S.S.; Ooi, L.L.; Kon, O.L.; Lee, C.G. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019, 79, 5131–5140. [Google Scholar] [CrossRef]
- Zhang, X.; Lv, X.; Li, X.; Wang, Y.; Lin, H.Y.; Zhang, J.; Peng, C. Dysregulated circulating SOCS3 and haptoglobin expression associated with stable coronary artery disease and acute coronary syndrome: An integrated study based on bioinformatics analysis and case-control validation. Anatol. J. Cardiol. 2020, 24, 160. [Google Scholar] [CrossRef]
- Manna, I.; Quattrone, A.; De Benedittis, S.; Iaccino, E.; Quattrone, A. Roles of Non-Coding RNAs as Novel Diagnostic Biomarkers in Parkinson’s Disease. J. Park. Dis. 2021, 11, 1475. [Google Scholar] [CrossRef]
- Chi, L.M.; Wang, L.P.; Jiao, D. Identification of Differentially Expressed Genes and Long Noncoding RNAs Associated with Parkinson’s Disease. Park. Dis. 2019, 2019, 6078251. [Google Scholar] [CrossRef]
- Moreno-García, L.; López-Royo, T.; Calvo, A.C.; Toivonen, J.M.; de la Torre, M.; Moreno-Martínez, L.; Molina, N.; Aparicio, P.; Zaragoza, P.; Manzano, R.; et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9582. [Google Scholar] [CrossRef]
- Gambineri, A.; Pelusi, C.; Vicennati, V.; Pagotto, U.; Pasquali, R. Obesity and the polycystic ovary syndrome. Int. J. Obes. 2002, 26, 883–896. [Google Scholar] [CrossRef]
- Cai, H.; Yu, Y.; Ni, X.; Li, C.; Hu, Y.; Wang, J.; Chen, F.; Xi, S.; Chen, Z. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020, 11, 1032. [Google Scholar] [CrossRef]
- Fang, X.; Pan, X.; Mai, H.; Yuan, X.; Liu, S.; Wen, F. LINC00998 functions as a novel tumor suppressor in acute myeloid leukemia via regulating the ZFP36 ring finger protein/mammalian target of rapamycin complex 2 axis. Bioengineered 2021, 12, 10363–10372. [Google Scholar] [CrossRef]
- Ye, N.; Rao, S.; Du, T.; Hu, H.; Liu, Z.; Shen, Y.; Xu, Q. Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene 2017, 601, 21–26. [Google Scholar] [CrossRef]
- Jurgec, S.; Jezernik, G.; Gorenjak, M.; Büdefeld, T.; Potočnik, U. Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies. Cancers 2022, 14, 4681. [Google Scholar] [CrossRef]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.J.; Chen, S.; Wang, Q.; Pan, H. Application of Weighted Gene Co-expression Network Analysis for Data from Paired Design. Sci. Rep. 2018, 8, 622. [Google Scholar] [CrossRef]
- Signor, S.A.; Nuzhdin, S.V. The Evolution of Gene Expression in cis and trans. Trends Genet. 2018, 34, 532–544. [Google Scholar] [CrossRef]
- Johnsen, S.; Dolan, S.E.; Fitch, K.V.; Killilea, K.M.; Shifren, J.L.; Grinspoon, S.K. Absence of Polycystic Ovary Syndrome Features in Human Immunodeficiency Virus-Infected Women Despite Significant Hyperinsulinemia and Truncal Adiposity. J. Clin. Endocrinol. Metab. 2005, 90, 5596–5604. [Google Scholar] [CrossRef]
- Diamanti-Kandarakis, E.; Dunaif, A. Insulin resistance and the polycystic ovary syndrome revisited: An update on mechanisms and implications. Endocr. Rev. 2012, 33, 981–1030. [Google Scholar] [CrossRef]
- Nilsson, E.; Benrick, A.; Kokosar, M.; Krook, A.; Lindgren, E.; Källman, T.; Martis, M.M.; Højlund, K.; Ling, C.; Stener-Victorin, E. Transcriptional and Epigenetic Changes Influencing Skeletal Muscle Metabolism in Women With Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2018, 103, 4465–4477. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Wang, Y.; Xu, S.; Qi, G.; Wu, X. Bioinformatics identification of microRNAs involved in polycystic ovary syndrome based on microarray data. Mol. Med. Rep. 2019, 20, 281. [Google Scholar] [CrossRef]
- Lines, M.A.; Huang, L.; Schwartzentruber, J.; Douglas, S.L.; Lynch, D.C.; Beaulieu, C.; Guion-Almeida, M.L.; Zechi-Ceide, R.M.; Gener, B.; Gillessen-Kaesbach, G.; et al. Haploinsufficiency of a Spliceosomal GTPase Encoded by EFTUD2 Causes Mandibulofacial Dysostosis with Microcephaly. Am. J. Hum. Genet. 2012, 90, 369. [Google Scholar] [CrossRef]
- Guo, R.; Zheng, L.; Park, J.W.; Lv, R.; Chen, H.; Jiao, F.; Xu, W.; Mu, S.; Wen, H.; Qiu, J.; et al. BS69/ZMYND11 Reads and Connects Histone H3.3 Lysine 36 Trimethylation-Decorated Chromatin to Regulated Pre-mRNA Processing. Mol. Cell 2014, 56, 298–310. [Google Scholar] [CrossRef]
- Zhu, C.; Xiao, F.; Hong, J.; Wang, K.; Liu, X.; Cai, D.; Fusco, D.N.; Zhao, L.; Jeong, S.W.; Brisac, C.; et al. EFTUD2 Is a Novel Innate Immune Regulator Restricting Hepatitis C Virus Infection through the RIG-I/MDA5 Pathway. J. Virol. 2015, 89, 6608–6618. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Wang, Z.; Luo, L.; Chen, Y.; Han, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Feng, J.; et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. 2019, 12, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Lang, Q.; Yidong, X.; Xueguang, Z.; Sixian, W.; Wenming, X.; Tao, Z. ETA-mediated anti-TNF-α therapy ameliorates the phenotype of PCOS model induced by letrozole. PLoS ONE 2019, 14, e0217495. [Google Scholar] [CrossRef]
- Zhu, K.; Li, S.; Liu, J.; Hong, Y.; Chen, Z.J.; Du, Y. Role of RAB5A in FSHR-mediated signal transduction in human granulosa cells. Reproduction 2018, 155, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Huang, Z.Y.; Yu, K.; Li, J.; Fu, X.W.; Deng, S.L. Estrogen Biosynthesis and Signal Transduction in Ovarian Disease. Front. Endocrinol. 2022, 13, 827032. [Google Scholar] [CrossRef]
- Kuang, H.; Duan, Y.; Li, D.; Xu, Y.; Ai, W.; Li, W.; Wang, Y.; Liu, S.; Li, M.; Liu, X.; et al. The role of serum inflammatory cytokines and berberine in the insulin signaling pathway among women with polycystic ovary syndrome. PLoS ONE 2020, 15, e0235404. [Google Scholar] [CrossRef]
- Makker, A.; Goel, M.M.; Das, V.; Agarwal, A. PI3K-Akt-mTOR and MAPK signaling pathways in polycystic ovarian syndrome, uterine leiomyomas and endometriosis: An update. Gynecol. Endocrinol. 2012, 28, 175–181. [Google Scholar] [CrossRef]
- Zheng, Q.; Liu, M.; Fu, J. ALG2 inhibits the epithelial-to-mesenchymal transition and stemness of ovarian granulosa cells through the Wnt/β-catenin signaling pathway in polycystic ovary syndrome. Reprod. Biol. 2022, 22, 100706. [Google Scholar] [CrossRef]
- Chen, Q.; Zheng, B.; Du, S.; Lin, Y. Explore the potential molecular mechanism of polycystic ovarian syndrome by protein–protein interaction network analysis. Taiwan. J. Obstet. Gynecol. 2021, 60, 807–815. [Google Scholar] [CrossRef]
- Azziz, R.; Carmina, E.; Chen, Z.; Dunaif, A.; Laven, J.S.; Legro, R.S.; Lizneva, D.; Natterson-Horowtiz, B.; Teede, H.J.; Yildiz, B.O. Polycystic ovary syndrome. Nat. Rev. Dis. Prim. 2016, 2, 16057. [Google Scholar] [CrossRef]
- Lee, S.; Kang, D.W.; Hudgins-Spivey, S.; Krust, A.; Lee, E.Y.; Koo, Y.; Cheon, Y.; Gye, M.C. Theca-Specific Estrogen Receptor-α Knockout Mice Lose Fertility Prematurely. Endocrinology 2009, 150, 3855. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Huang, Y.L.; Liu, J.Q.; Huang, Q. Transcription factor-microRNA synergistic regulatory network revealing the mechanism of polycystic ovary syndrome. Mol. Med. Rep. 2016, 13, 3920–3928. [Google Scholar] [CrossRef] [PubMed]
- Cortón, M.; Botella-Carretero, J.I.; López, J.A.; Camafeita, E.; San Millán, J.L.; Escobar-Morreale, H.F.; Peral, B. Proteomic analysis of human omental adipose tissue in the polycystic ovary syndrome using two-dimensional difference gel electrophoresis and mass spectrometry. Hum. Reprod. 2008, 23, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Yoon, T.; Choi, E.C.; Lee, K. Interaction of cofilin with triose-phosphate isomerase contributes glycolytic fuel for Na,K-ATPase via Rho-mediated signaling pathway. J. Biol. Chem. 2002, 277, 48931–48937. [Google Scholar] [CrossRef]
- Nasri, F.; Zare, M.; Doroudchi, M.; Gharesi-Fard, B. Proteome Analysis of CD4+ T Cells Reveals Differentially Expressed Proteins in Infertile Polycystic Ovary Syndrome Patients. Endocr. Metab. Immune Disord. Drug Targets 2020, 21, 1998–2004. [Google Scholar] [CrossRef]
- Hou, T.; Zhang, R.; Jian, C.; Ding, W.; Wang, Y.; Ling, S.; Ma, Q.; Hu, X.; Cheng, H.; Wang, X. NDUFAB1 confers cardio-protection by enhancing mitochondrial bioenergetics through coordination of respiratory complex and supercomplex assembly. Cell Res. 2019, 29, 754–766. [Google Scholar] [CrossRef]
- Feng, D.; Witkowski, A.; Smith, S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J. Biol. Chem. 2009, 284, 11436–11445. [Google Scholar] [CrossRef] [Green Version]
- Brody, S.; Oh, C.; Hoja, U.; Schweizer, E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997, 408, 217–220. [Google Scholar] [CrossRef]
- Tari, A.M. GRB2: A pivotal protein in signal transduction. Semin. Oncol. 2001, 28 (Suppl. 16), 142–147. [Google Scholar] [CrossRef]
- Corbould, A.; Zhao, H.; Mirzoeva, S.; Aird, F.; Dunaif, A. Enhanced mitogenic signaling in skeletal muscle of women with polycystic ovary syndrome. Diabetes 2006, 55, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.; Tronser, M.; Sers, C.; Ahadova, A.; Endris, V.; Mamlouk, S.; Horst, D.; Möbs, M.; Bischoff, P.; Kloor, M.; et al. The majority of β-catenin mutations in colorectal cancer is homozygous. BMC Cancer 2020, 20, 1038. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Pang, Y.L.; Wang, X.; Li, Y.H. Computational characterization and identification of human polycystic ovary syndrome genes. Sci. Rep. 2018, 8, 12949. [Google Scholar] [CrossRef] [PubMed]
- Manti, M.; Stener-Victorin, E.; Benrick, A. Skeletal Muscle Immunometabolism in Women With Polycystic Ovary Syndrome: A Meta-Analysis. Front. Physiol. 2020, 11, 1331. [Google Scholar] [CrossRef]


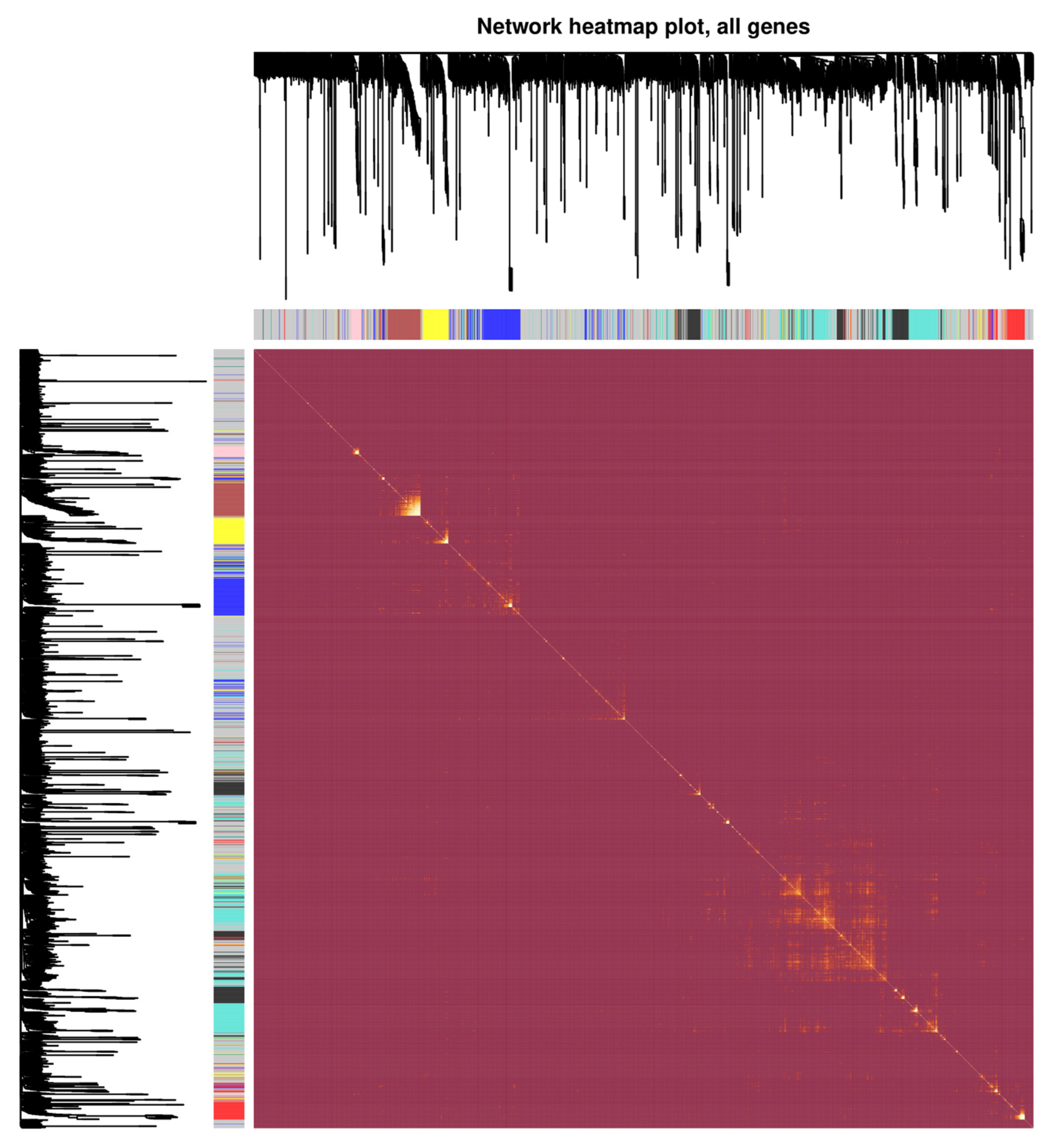
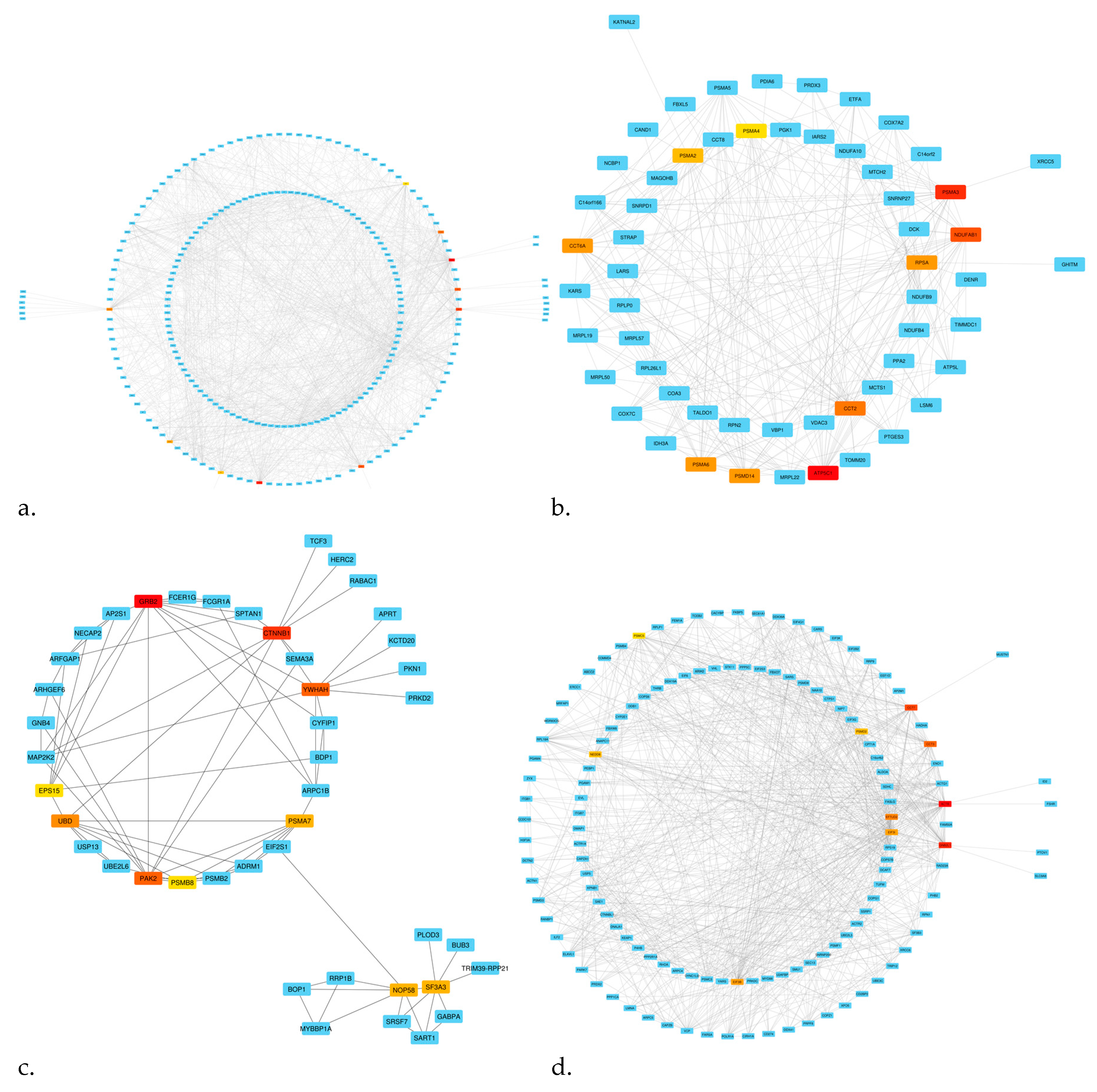
| LncRNA | LogFC | p-Value | Expression | Co-Expression Module | PPI Networks | Hub Genes |
|---|---|---|---|---|---|---|
| LOC102725104 | 1.825633999 | 0.005721501 | Upregulated | Turquoise | 134 node and 842 edge | ACTB, GNB2L1, CCT7, CCT3, EFTUD2, EIF3B, EIF3I, NEDD8, PSMD2, PSMC5 |
| LINC00328 | 1.722041383 | 0.014404303 | ||||
| LOC102725070 | 1.629786576 | 0.01119095 | ||||
| LOC101059935 | 1.621249989 | 0.004591495 | ||||
| LOC100127951 | 1.577888412 | 0.000306974 | Upregulated | Gray | 238 node and 1142 edge | HSP90AA1, GART, ESR1, RPS3, JUN, POLR2C, ALDH18A1, RUVBL1, TPI1, NHP2 |
| LOC648987 | −1.222919534 | 0.005645454 | Downregulated | |||
| LOC93622 | −1.24184329 | 0.006655765 | Downregulated | |||
| LOC644936 | −1.288710709 | 0.001962644 | Downregulated | Black | 44 node and 85 edge | GRB2, CTNNB1, YWHAH, PAK2, UBD, PSMA7, NOP58, SF3A3, PSMB8, EPS15 |
| LINC00998 | −1.383262604 | 0.012944269 | Downregulated | Blue | 58 node and 296 edge | ATP5C1, PSMA3, NDUFAB1, CCT2, PSMA6, CCT6A, PSMD14, RPSA, PSMA2, PSMA4 |
| LncRNAs | Co-Expressed Modules | Pathways |
|---|---|---|
| LOC102725104 LINC00328 LOC102725070 LOC101059935 | Turquoise | Gene expression |
| Host interactions of HIV factors | ||
| HIV infection | ||
| Class I MHC mediated antigen processing and presentation | ||
| Vif-mediated degradation of APOBEC3G | ||
| Destabilization of mRNA by AUF1 (hnRNP D0) | ||
| Regulation of apoptosis | ||
| Autodegradation of Cdh1 by Cdh1: APC/C | ||
| Translation | ||
| ER–phagosome pathway | ||
| LOC100127951 LOC648987 LOC93622 | Gray | Olfactory signaling pathway |
| GPCR downstream signaling | ||
| Signaling by GPCR | ||
| Signal transduction | ||
| Respiratory electron transport | ||
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | ||
| Citric acid cycle (TCA cycle) | ||
| Telomere extension by telomerase | ||
| The citric acid (TCA) cycle and respiratory electron transport | ||
| Beta-catenin phosphorylation cascade | ||
| LINC00998 | Blue | Metabolism |
| The citric acid (TCA) cycle and respiratory electron transport | ||
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | ||
| Prefoldin mediated transfer of substrate to CCT/TriC | ||
| Metabolism of RNA | ||
| HIV infection | ||
| Cooperation of prefoldin and TriC/CCT in actin and tubulin folding | ||
| Gene expression | ||
| Chaperonin-mediated protein folding | ||
| Autodegradation of Cdh1 by Cdh1: APC/C | ||
| LOC644936 | Black | Stimulation of the cell death response by PAK-2p34 |
| CD28-dependent Vav1 pathway | ||
| EGFR downregulation | ||
| Fc-epsilon receptor I signaling in mast cells | ||
| Regulation of activated PAK-2p34 by proteasome mediated degradation | ||
| Cross-presentation of soluble exogenous antigens (endosomes) | ||
| Mevalonate pathway I | ||
| Apoptosis | ||
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | ||
| APC/C:Cdc20 mediated degradation of mitotic proteins |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heidarzadehpilehrood, R.; Pirhoushiaran, M.; Binti Osman, M.; Abdul Hamid, H.; Ling, K.-H. Weighted Gene Co-Expression Network Analysis (WGCNA) Discovered Novel Long Non-Coding RNAs for Polycystic Ovary Syndrome. Biomedicines 2023, 11, 518. https://doi.org/10.3390/biomedicines11020518
Heidarzadehpilehrood R, Pirhoushiaran M, Binti Osman M, Abdul Hamid H, Ling K-H. Weighted Gene Co-Expression Network Analysis (WGCNA) Discovered Novel Long Non-Coding RNAs for Polycystic Ovary Syndrome. Biomedicines. 2023; 11(2):518. https://doi.org/10.3390/biomedicines11020518
Chicago/Turabian StyleHeidarzadehpilehrood, Roozbeh, Maryam Pirhoushiaran, Malina Binti Osman, Habibah Abdul Hamid, and King-Hwa Ling. 2023. "Weighted Gene Co-Expression Network Analysis (WGCNA) Discovered Novel Long Non-Coding RNAs for Polycystic Ovary Syndrome" Biomedicines 11, no. 2: 518. https://doi.org/10.3390/biomedicines11020518






