Circular RNA CircFOXO3 Functions as a Competitive Endogenous RNA for Acid-Sensing Ion Channel Subunit 1 Mediating Oxeiptosis in Nucleus Pulposus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. CircRNA Microarray and Bioinformatics Analysis
2.3. Clinical Samples
2.4. Isolation and Culture of Human NPCs
2.5. Adenovirus and Adeno-Associated Virus (AAV) Construction
2.6. Surgical Procedures and In Vivo Transduction
2.7. RNA Extraction and Quantitative PCR (qPCR)
2.8. Western Blotting
2.9. Cell Viability Assay
2.10. Annexin V/Propidium Iodide (PI) Staining Assay
2.11. ROS Detection Assay
2.12. Quantification of Mitochondrial Membrane Potential (MMP; ΔΨm)
2.13. Immunofluorescence Assay
2.14. Transmission Electron Microscopy (TEM)
2.15. RNA Fluorescence In Situ Hybridization (RNA FISH)
2.16. RNA Immunoprecipitation (RIP) Assay
2.17. RNA Pull-Down Assay
2.18. Dual-Luciferase Reporter Gene Assay
2.19. Intracellular Calcium Ion (Ca2+) Measurement
2.20. Magnetic Resonance Imaging (MRI) Analyses
2.21. In Vivo Bioluminescence Imaging
2.22. Histological Evaluations and Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Staining
2.23. Immunohistochemistry (IHC)
2.24. Statistical Analysis
3. Results
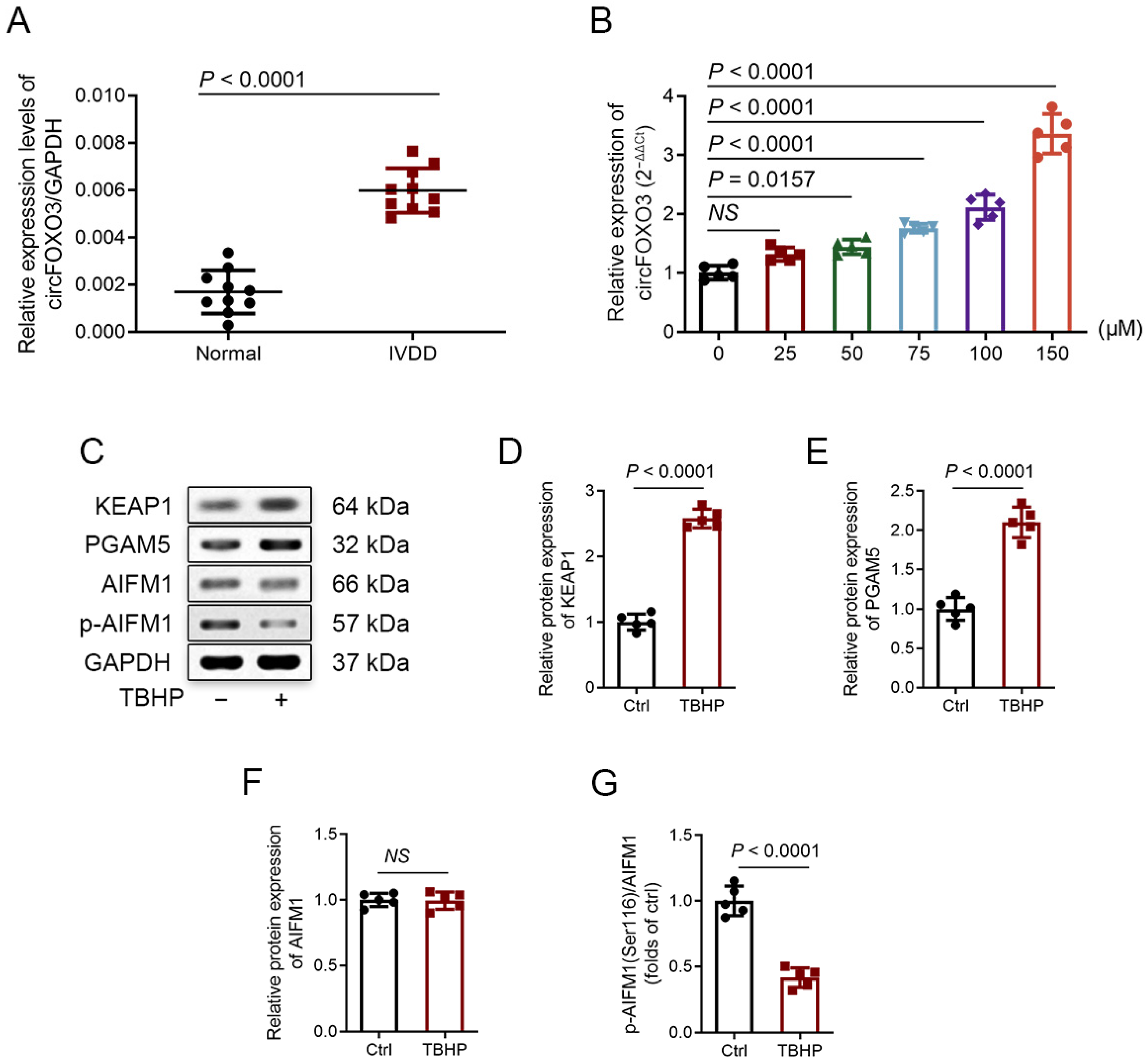
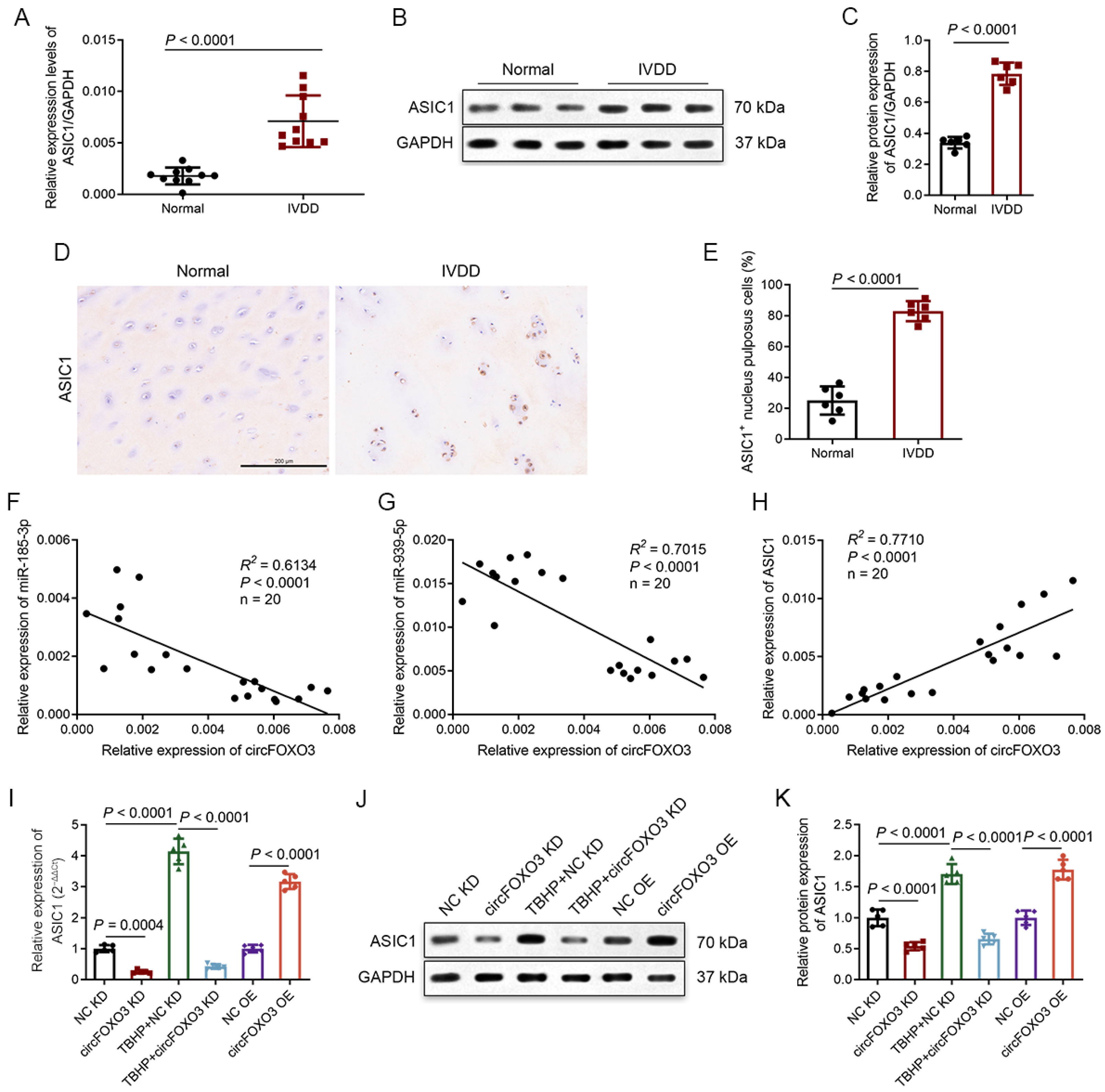
3.1. Degenerative NP Tissues Express a Higher Level of circFOXO3
3.2. TBHP Treatment Activates Oxeiptosis-Related Pathway in NPCs In Vitro
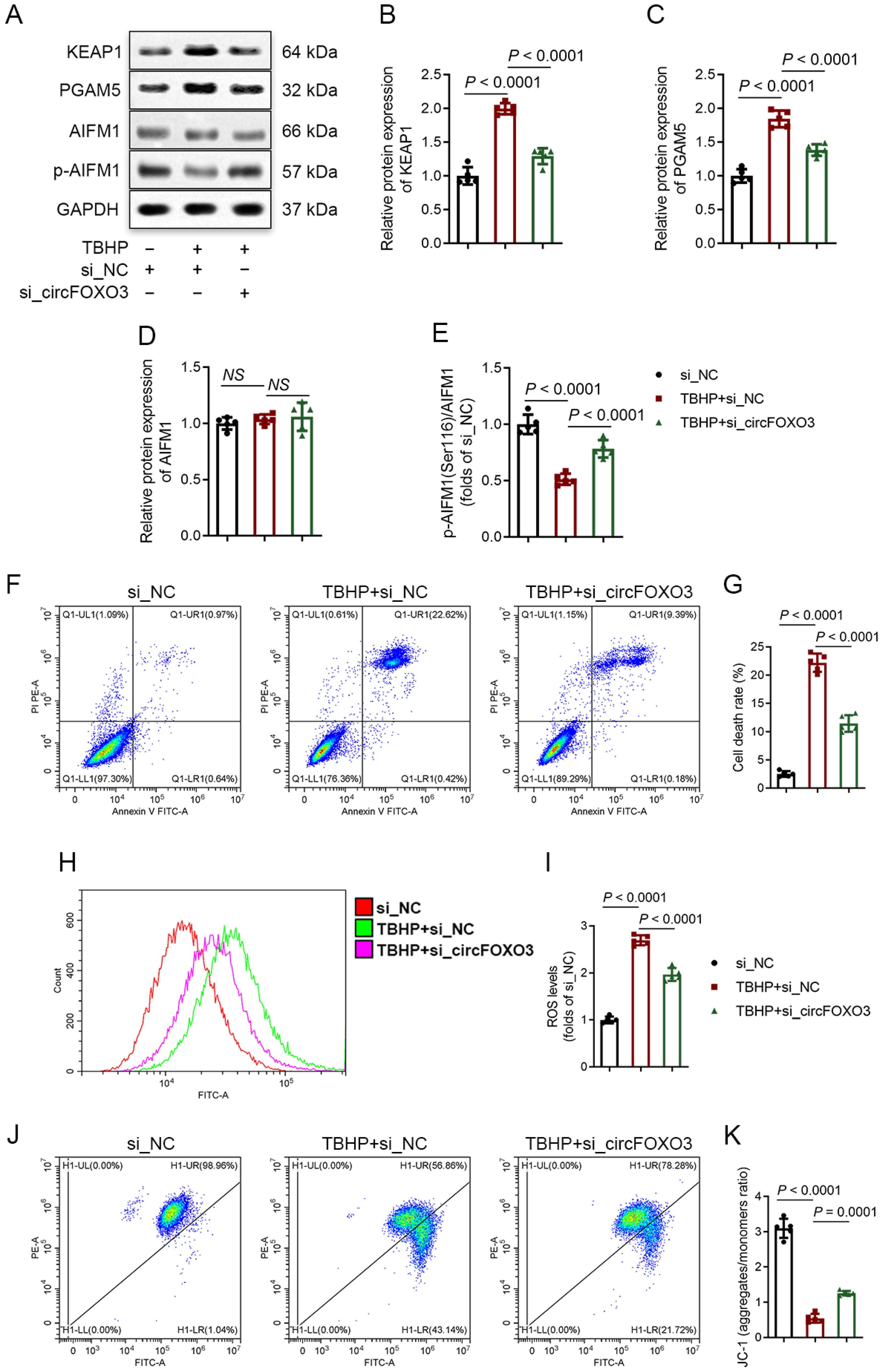
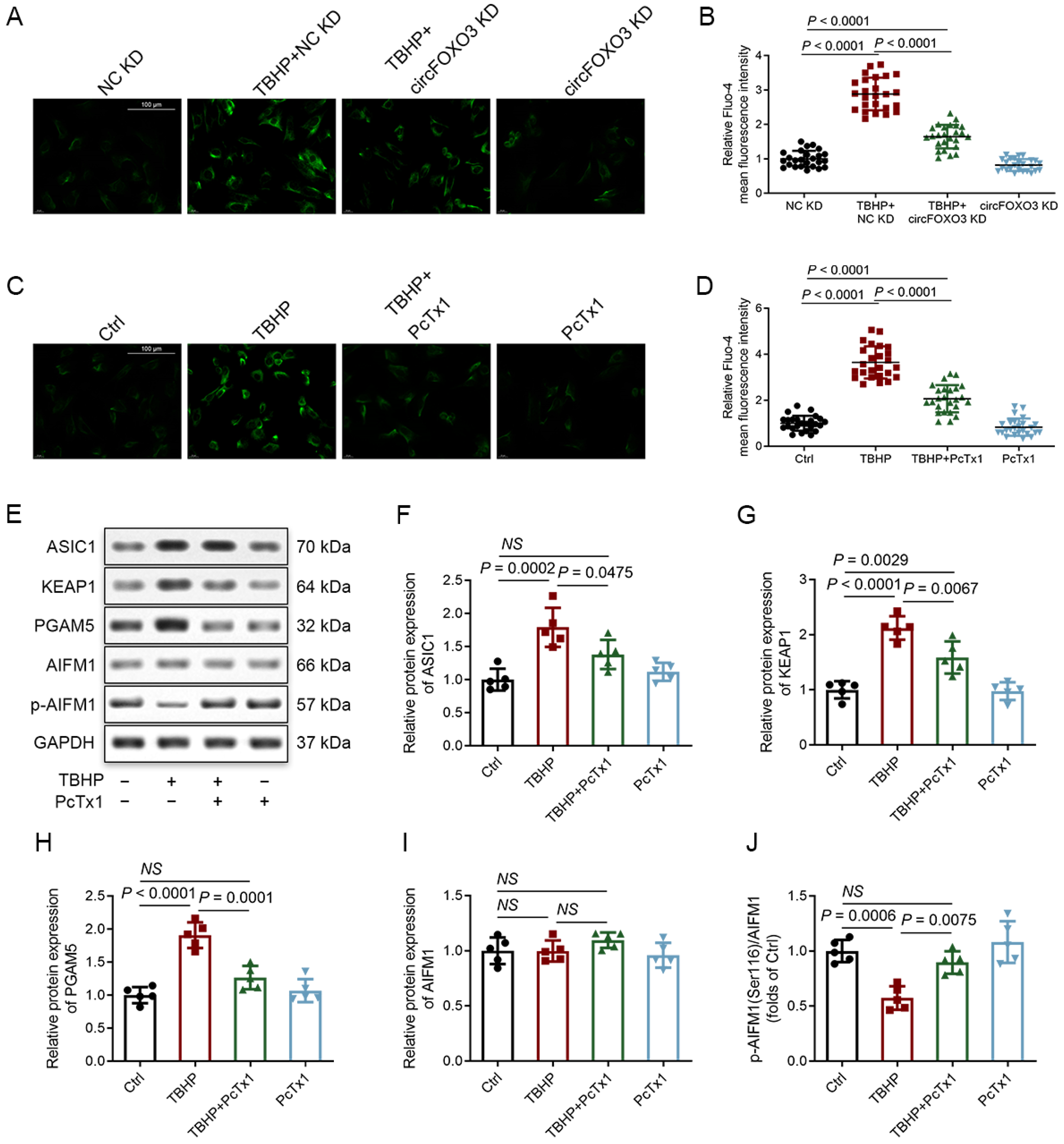
3.3. Knocking down circFOXO3 Suppresses Oxidative Stress-Induced Oxeiptosis in NPCs
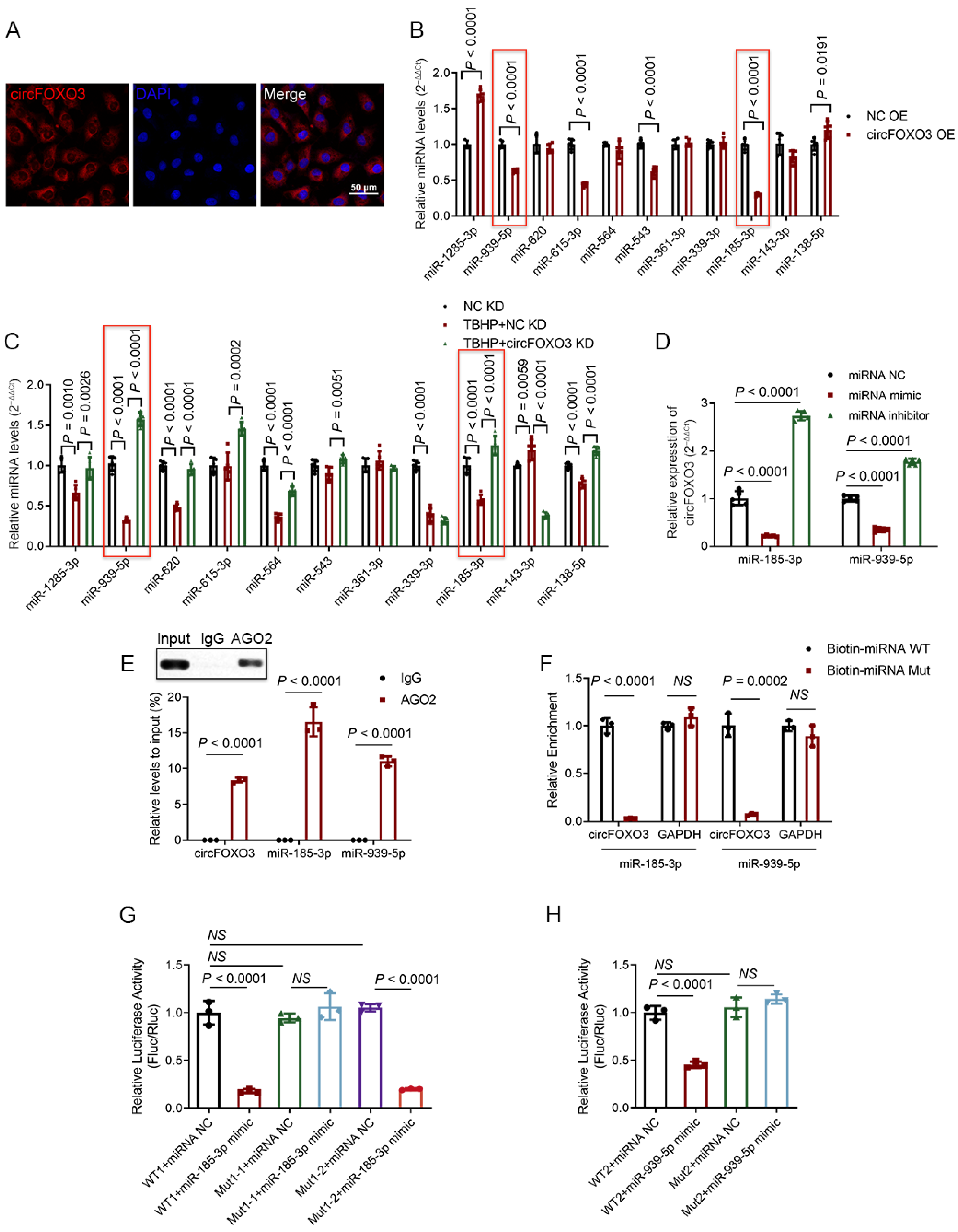
3.4. CircFOXO3 Functions as a Molecular Sponge to Target and Downregulate miR-185-3p and miR-939-5p in NPCs
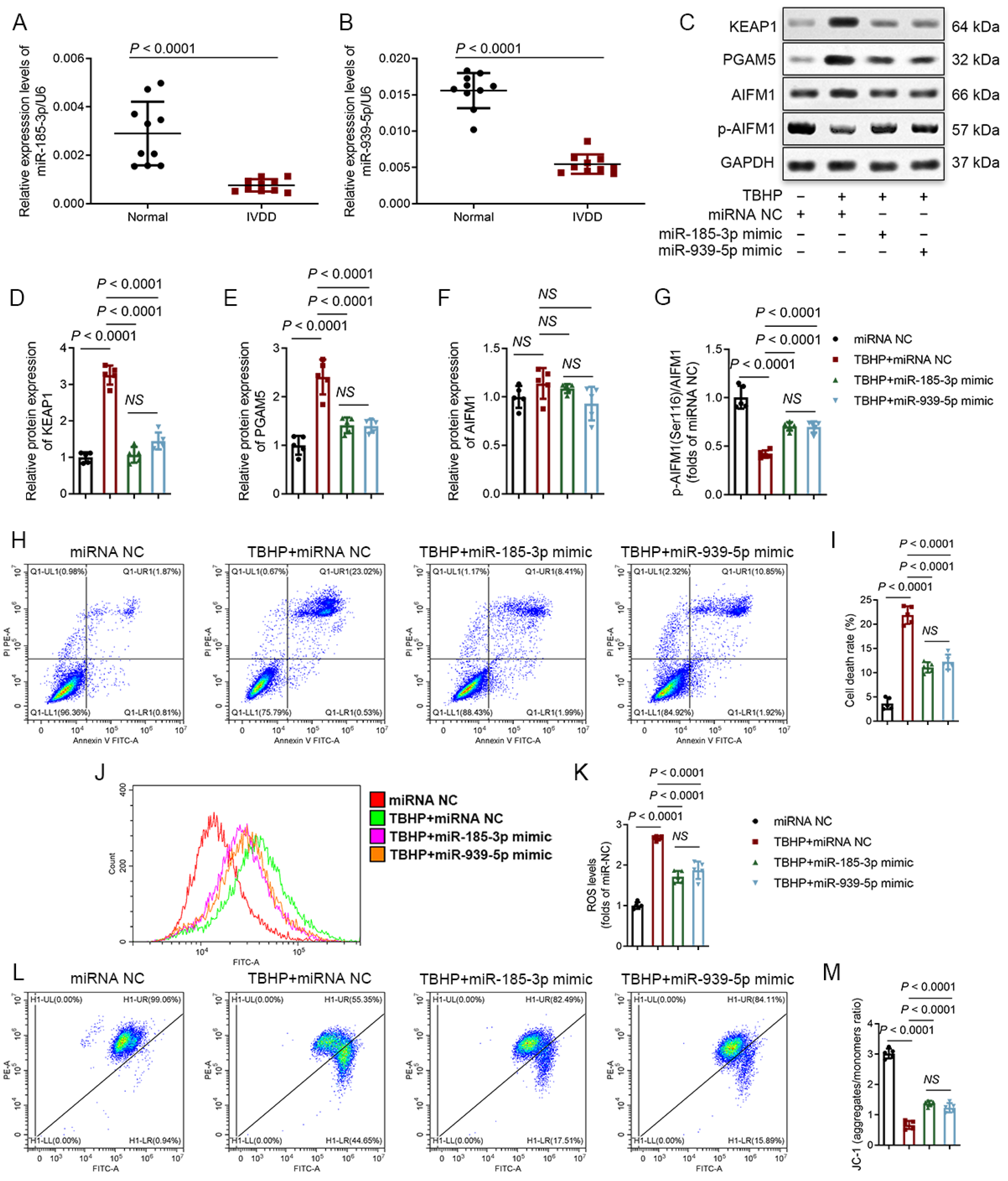
3.5. MiR-185-3p and miR-939-5p Are Downregulated in Degenerative NP Tissues and Participate in the Regulation of Oxeiptosis
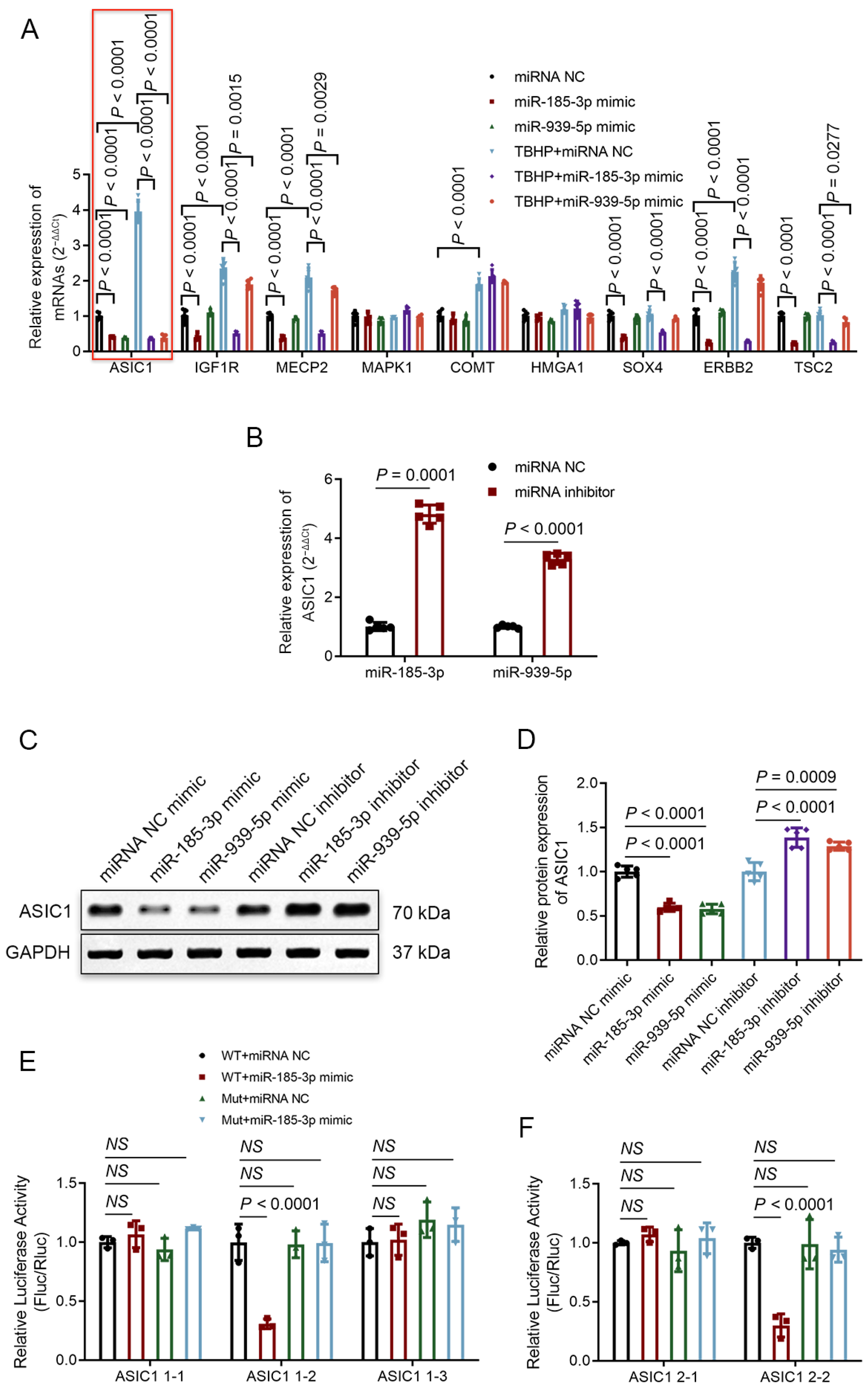
3.6. ASIC1 Is a Common Target of miR-185-3p and miR-939-5p
3.7. Degenerative NP Tissues Express a Higher Level of ASIC1, and circFOXO3 Intervenes with Oxeiptosis in NPCs by Regulating ASIC1 through Sponge miR-185-3p and miR-939-5p
3.8. CircFOXO3 Affects the Susceptibility of NPCs to TBHP-Induced Oxeiptosis by Regulating the Expression of ASIC1
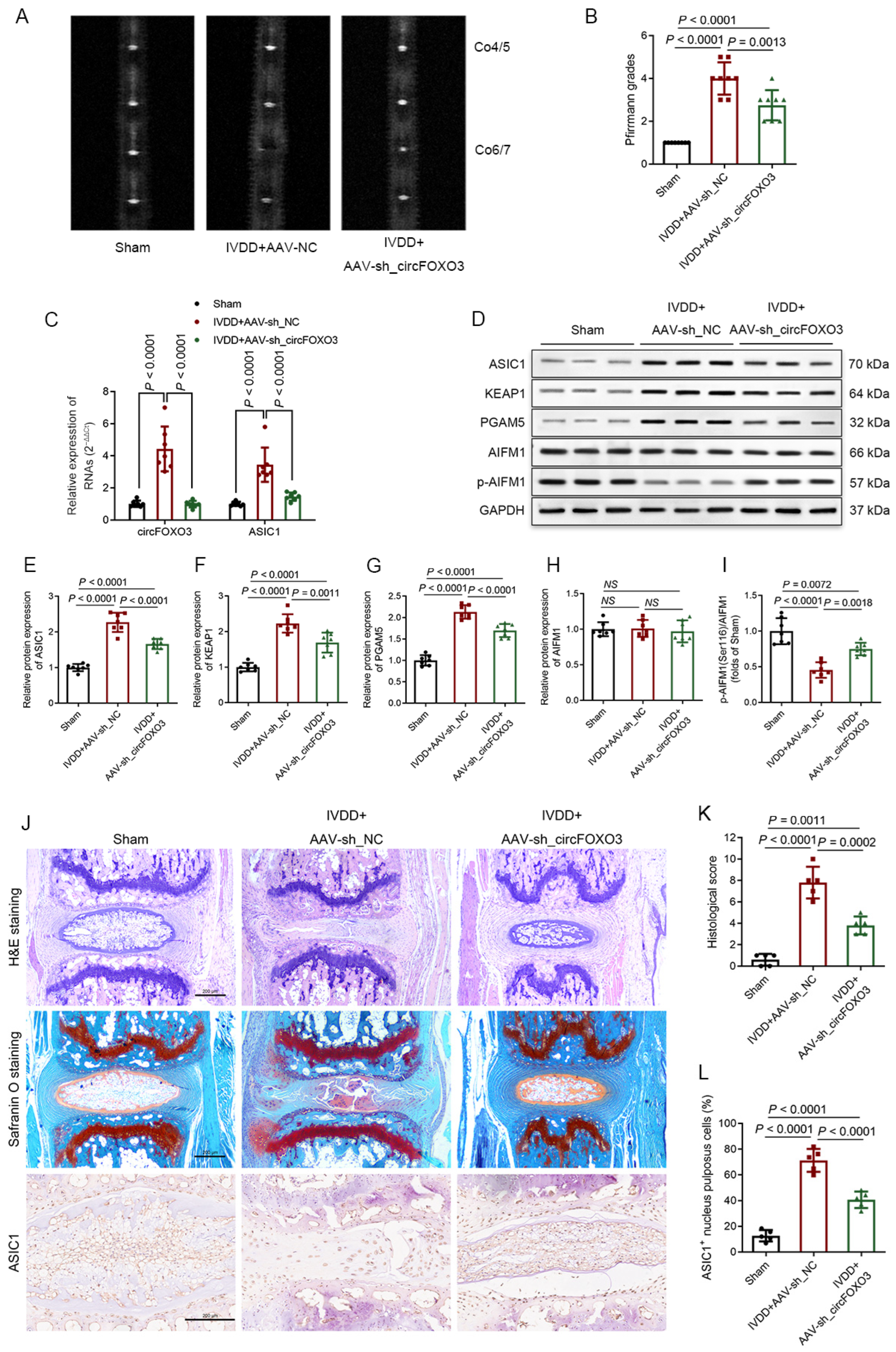
3.9. Inhibition of circFOXO3 Alleviates IVDD In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katz, J.N.; Zimmerman, Z.E.; Mass, H.; Makhni, M.C. Diagnosis and Management of Lumbar Spinal Stenosis: A Review. JAMA 2022, 327, 1688–1699. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, X.; Gao, W.; Hu, H.; Wang, X.; Hao, D. lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol. Med. Rep. 2019, 20, 3160–3174. [Google Scholar] [CrossRef]
- Alvarez-Garcia, O.; Matsuzaki, T.; Olmer, M.; Miyata, K.; Mokuda, S.; Sakai, D.; Masuda, K.; Asahara, H.; Lotz, M.K. FOXO are required for intervertebral disk homeostasis during aging and their deficiency promotes disk degeneration. Aging Cell 2018, 17, e12800. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qiu, C.; Wang, W.; Peng, J.; Cheng, X.; Shangguan, Y.; Xu, M.; Li, J.; Qu, R.; Chen, X.; et al. Cortistatin protects against intervertebral disc degeneration through targeting mitochondrial ROS-dependent NLRP3 inflammasome activation. Theranostics 2020, 10, 7015–7033. [Google Scholar] [CrossRef]
- Kang, L.; Liu, S.; Li, J.; Tian, Y.; Xue, Y.; Liu, X. The mitochondria-targeted anti-oxidant MitoQ protects against intervertebral disc degeneration by ameliorating mitochondrial dysfunction and redox imbalance. Cell Prolif. 2020, 53, e12779. [Google Scholar] [CrossRef]
- Tschoeke, S.K.; Hellmuth, M.; Hostmann, A.; Robinson, Y.; Ertel, W.; Oberholzer, A.; Heyde, C.-E. Apoptosis of human intervertebral discs after trauma compares to degenerated discs involving both receptor-mediated and mitochondrial-dependent pathways. J. Orthop. Res. 2008, 26, 999–1006. [Google Scholar] [CrossRef]
- Holze, C.; Michaudel, C.; Mackowiak, C.; Haas, D.A.; Benda, C.; Hubel, P.; Pennemann, F.L.; Schnepf, D.; Wettmarshausen, J.; Braun, M.; et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 2018, 19, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Aramouni, K.; Assaf, R.; Shaito, A.; Fardoun, M.; Al-Asmakh, M.; Sahebkar, A.; Eid, A.H. Biochemical and cellular basis of oxidative stress: Implications for disease onset. J. Cell. Physiol. 2023, 238, 1951–1963. [Google Scholar] [CrossRef] [PubMed]
- Scaturro, P.; Pichlmair, A. Oxeiptosis—A cell death pathway to mitigate damage caused by radicals. Cell Death Differ. 2018, 25, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Lin, N.; Dong, D.; Ma, J.; Su, J.; Sun, L. PGAM5: A crucial role in mitochondrial dynamics and programmed cell death. Eur. J. Cell Biol. 2021, 100, 151144. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, R.-f.; Li, J.; Yu, K.-d.; Bi, K.-x. Alloimperatorin activates apoptosis, ferroptosis, and oxeiptosis to inhibit the growth and invasion of breast cancer cells in vitro. Biochem. Cell Biol. 2022, 100, 213–222. [Google Scholar] [CrossRef]
- Tsui, K.-H.; Li, C.-J. Mitoquinone shifts energy metabolism to reduce ROS-induced oxeiptosis in female granulosa cells and mouse oocytes. Aging 2023, 15, 246–260. [Google Scholar] [CrossRef]
- Wang, C.-X.; Chen, L.-H.; Zhuang, H.-B.; Shi, Z.-S.; Chen, Z.C.; Pan, J.-P.; Hong, Z.-S. Auriculasin enhances ROS generation to regulate colorectal cancer cell apoptosis, ferroptosis, oxeiptosis, invasion and colony formation. Biochem. Biophys. Res. Commun. 2022, 587, 99–106. [Google Scholar] [CrossRef]
- Pallichankandy, S.; Thayyullathil, F.; Cheratta, A.R.; Subburayan, K.; Alakkal, A.; Sultana, M.; Drou, N.; Arshad, M.; Tariq, S.; Galadari, S. Targeting oxeiptosis-mediated tumor suppression: A novel approach to treat colorectal cancers by sanguinarine. Cell Death Discov. 2023, 9, 94. [Google Scholar] [CrossRef]
- Kang, P.; Chen, J.; Zhang, W.; Guo, N.; Yi, X.; Cui, T.; Chen, J.; Yang, Y.; Wang, Y.; Du, P.; et al. Oxeiptosis: A novel pathway of melanocytes death in response to oxidative stress in vitiligo. Cell Death Discov. 2022, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Ma, X.; Sun, C.; Han, J.; Zhou, C.; Wong, S.H.; Chan, M.T.V.; Wu, W.K.K. Circular RNAs in Intervertebral Disc Degeneration: An Updated Review. Front. Mol. Biosci. 2022, 8, 781424. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wu, T.; Ji, G.; Wu, H.; Lai, Y.; Wei, B.; Huang, W. Circular RNA and intervertebral disc degeneration: Unravelling mechanisms and implications. Front. Mol. Biosci. 2023, 10, 1302017. [Google Scholar] [CrossRef]
- Jiang, C.; Chen, Z.; Wang, X.; Zhang, Y.; Guo, X.; Xu, Z.; Yang, H.; Hao, D. The potential mechanisms and application prospects of non-coding RNAs in intervertebral disc degeneration. Front. Endocrinol. 2022, 13, 1081185. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tian, L.; Li, J.; Li, Y.; Du, L.; Huo, Z.; Xu, B. The Roles of circRNAs in Intervertebral Disc Degeneration: Inflammation, Extracellular Matrix Metabolism, and Apoptosis. Anal. Cell. Pathol. 2022, 2022, 9550499. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-L.; Song, G.; Guo, J.-B.; Su, X.; Chen, Y.-M.; Yang, Z.; Chen, P.-J.; Wang, X.-Q. Interactions Among lncRNA/circRNA, miRNA, and mRNA in Musculoskeletal Degenerative Diseases. Front. Cell Dev. Biol. 2021, 9, 753931. [Google Scholar] [CrossRef]
- Lin, S.P.; Hu, J.; Wei, J.X.; Ye, S.; Bu, J.; Xu, W.; Chen, S.; Wu, Y.; Wu, G.; Zhu, L.; et al. Silencing of circFoxO3 Protects HT22 Cells from Glutamate-Induced Oxidative Injury via Regulating the Mitochondrial Apoptosis Pathway. Cell. Mol. Neurobiol. 2020, 40, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhu, C.; Wang, B.; Zheng, H.; McAlister, V.; Lacefield, J.C.; Quan, D.; Mele, T.; Greasley, A.; Liu, K.; et al. Circular RNA Foxo3 in cardiac ischemia-reperfusion injury in heart transplantation: A new regulator and target. Am. J. Transplant. 2021, 21, 2992–3004. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, C.; Wen, X.; Liu, W.; Huang, X.; Li, Y.; Zang, J.; Weng, Z.; Lu, D.; Tsang, C.K.; et al. Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol. Ther. 2022, 30, 1275–1287. [Google Scholar] [CrossRef]
- Zhou, L.; Wu, B.; Yang, J.; Wang, B.; Pan, J.; Xu, D.; Du, C. Knockdown of circFOXO3 ameliorates cigarette smoke-induced lung injury in mice. Respir. Res. 2021, 22, 294. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, L.; Fang, J.; Zhang, N.; Li, D.; Sheng, X.; Zhou, J.; Wang, S.; Wang, J. Circular RNA circFoxo3 Promotes Granulosa Cell Apoptosis under Oxidative Stress through Regulation of FOXO3 Protein. DNA Cell Biol. 2022, 41, 1026–1037. [Google Scholar] [CrossRef]
- Kim, C.H.; Oliver, C.; Dar, H.; Drissi, H.; Presciutti, S.M. AAV6 as an effective gene delivery vector for prolonged transgene expression in intervertebral disc cells in vivo. Genes Dis. 2022, 9, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Chan, W.C.W.; Leung, V.Y.L.; Cheah, K.S.E.; Cheung, K.M.C.; Sakai, D.; McCann, M.R.; Bedore, J.; Séguin, C.A.; Chan, D. Histological and reference system for the analysis of mouse intervertebral disc. J. Orthop. Res. 2018, 36, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jing, X.; Guo, J.; Yao, X.; Guo, F. Mitophagy in degenerative joint diseases. Autophagy 2021, 17, 2082–2092. [Google Scholar] [CrossRef]
- Fan, C.; Chu, G.; Yu, Z.; Ji, Z.; Kong, F.; Yao, L.; Wang, J.; Geng, D.; Wu, X.; Mao, H. The role of ferroptosis in intervertebral disc degeneration. Front. Cell Dev. Biol. 2023, 11, 1219840. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, H.; Yang, R.; Hou, Y.; Li, Y.; Zhu, J.; Fu, C. Biomaterials delivery strategies to repair degenerated intervertebral discs by regulating the inflammatory microenvironment. Front. Immunol. 2023, 14, 1051606. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, K.; Miao, Z.; Wang, Q.; Miao, Y.; Guo, Z.; Qiu, Y.; Chen, B.; Ren, L.; Wei, Z.; et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro-Oncology 2019, 21, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Xie, X.; Lin, L. circFOXO3 protects cardiomyocytes against radiation-induced cardiotoxicity. Mol. Med. Rep. 2021, 23, 177. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zong, M.; Lu, Y.; Guo, Y.; Lv, H.; Xie, L.; Fu, Z.; Cheng, Y.; Si, Y.; Ye, B.; et al. Differentially expressed lnc-NOS2P3-miR-939-5p axis in chronic heart failure inhibits myocardial and endothelial cells apoptosis via iNOS/TNFα pathway. J. Cell. Mol. Med. 2020, 24, 11381–11396. [Google Scholar] [CrossRef]
- Chen, Z.-W.; Hu, J.-F.; Wang, Z.-W.; Liao, C.-Y.; Kang, F.-P.; Lin, C.-F.; Huang, Y.; Huang, L.; Tian, Y.-F.; Chen, S. Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 153. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ma, J.; Fan, H.; Liu, M.; Zhu, Y.; Li, Y.; Tang, H. TNF-α-induced lncRNA LOC105374902 promotes the malignant behavior of cervical cancer cells by acting as a sponge of miR-1285-3p. Biochem. Biophys. Res. Commun. 2019, 513, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Hu, B.; Ding, B.; Zhao, Q.; Zhang, Y.; Xiao, L. N6-Methyladenosine-induced miR-143-3p promotes intervertebral disc degeneration by regulating SOX5. Bone 2022, 163, 116503. [Google Scholar] [CrossRef]
- Hendgen-Cotta, U.B.; Messiha, D.; Esfeld, S.; Deenen, R.; Rassaf, T.; Totzeck, M. Inorganic nitrite modulates miRNA signatures in acute myocardial in vivo ischemia/reperfusion. Free Radic. Res. 2017, 51, 91–102. [Google Scholar] [CrossRef]
- Ru, N.; Zhang, F.; Liang, J.; Du, Y.; Wu, W.; Wang, F.; Liu, X. MiR-564 is down-regulated in osteosarcoma and inhibits the proliferation of osteosarcoma cells via targeting Akt. Gene 2018, 645, 163–169. [Google Scholar] [CrossRef]
- Wang, B.; Wang, D.; Yan, T.; Yuan, H. MiR-138-5p promotes TNF-α-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp. Cell Res. 2016, 345, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yuan, J.; Li, J.; Du, Z.; Yin, M.; Cheng, X.; Liu, X.; Jia, J. Hsa_circ_0008870 suppresses bone formation of growth plate through inhibition of miR-185-3p/MAPK1 axis in idiopathic short stature. Front. Bioeng. Biotechnol. 2022, 10, 1022830. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Zhang, Y.; Sun, D. miR-217 and miR-543 downregulation mitigates inflammatory response and myocardial injury in children with viral myocarditis by regulating the SIRT1/AMPK/NF-κB signaling pathway. Int. J. Mol. Med. 2020, 45, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Song, H.; Hu, H.; Zhou, C.; Chen, Q.; Hong, L.; Huang, M.; Zhu, H. MiR-361-3p alleviates cerebral ischemia–reperfusion injury by targeting NACC1 through the PINK1/Parkin pathway. J. Mol. Histol. 2022, 53, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, F.; Wan, Y.; Liang, H.; Li, S.; Peng, B.; Shao, L.; Xu, Y.; Jiang, D. Cancer-Secreted Exosomal MiR-620 Inhibits ESCC Aerobic Glycolysis via FOXM1/HER2 Pathway and Promotes Metastasis. Front. Oncol. 2022, 12, 756109. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, R.; Li, H.; Ji, Q.; Cheng, X.; Thorne, R.F.; Hondermarck, H.; Liu, X.; Shen, C. ASIC1 and ASIC3 mediate cellular senescence of human nucleus pulposus mesenchymal stem cells during intervertebral disc degeneration. Aging 2021, 13, 10703–10723. [Google Scholar] [CrossRef]
- Zhao, K.; An, R.; Xiang, Q.; Li, G.; Wang, K.; Song, Y.; Liao, Z.; Li, S.; Hua, W.; Feng, X.; et al. Acid-sensing ion channels regulate nucleus pulposus cell inflammation and pyroptosis via the NLRP3 inflammasome in intervertebral disc degeneration. Cell Prolif. 2021, 54, e12941. [Google Scholar] [CrossRef]
- Cherif, H.; Mannarino, M.; Pacis, A.S.; Ragoussis, J.; Rabau, O.; Ouellet, J.A.; Haglund, L. Single-Cell RNA-Seq Analysis of Cells from Degenerating and Non-Degenerating Intervertebral Discs from the Same Individual Reveals New Biomarkers for Intervertebral Disc Degeneration. Int. J. Mol. Sci. 2022, 23, 3993. [Google Scholar] [CrossRef]
- Guo, W.; Mu, K.; Zhang, B.; Sun, C.; Zhao, L.; Li, H.-R.; Dong, Z.-Y.; Cui, Q. The circular RNA circ-GRB10 participates in the molecular circuitry inhibiting human intervertebral disc degeneration. Cell Death Dis. 2020, 11, 612. [Google Scholar] [CrossRef]
- Kawarai, Y.; Jang, S.H.; Lee, S.; Millecamps, M.; Kang, H.; Gregoire, S.; Suzuki-Narita, M.; Ohtori, S.; Stone, L.S. Exercise attenuates low back pain and alters epigenetic regulation in intervertebral discs in a mouse model. Spine J. 2021, 21, 1938–1949. [Google Scholar] [CrossRef]
- Li, S.; Liao, Z.; Yin, H.; Liu, O.; Hua, W.; Wu, X.; Zhang, Y.; Gao, Y.; Yang, C. G3BP1 coordinates lysophagy activity to protect against compression-induced cell ferroptosis during intervertebral disc degeneration. Cell Prolif. 2023, 56, e13368. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhao, H.; Li, Z.; Xue, H.; Lu, J.; Ma, W. A common polymorphism of COMT was associated with symptomatic lumbar disc herniation based on a large sample with Chinese Han ancestry. Sci. Rep. 2018, 8, 13000. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-C.; Zheng, B.; Sun, R.-X.; Meng, Y.-K.; Wang, S.-M.; Yang, H.-S.; Chen, Y.; Shi, J.-G.; Guo, Y.-F. MiR-499a-5p suppresses apoptosis of human nucleus pulposus cells and degradation of their extracellular matrix by targeting SOX4. Biomed. Pharmacother. 2019, 113, 108652. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; He, S.-J.; Liu, W.; Yang, M.-J.; Hou, Z.-Y.; Meng, Q.; Qian, Z.-L. EZH2 upregulates the expression of MAPK1 to promote intervertebral disc degeneration via suppression of miR-129-5p. J. Gene Med. 2022, 24, e3395. [Google Scholar] [CrossRef]
- Zhuang, Y.; Song, S.; Xiao, D.; Liu, X.; Han, X.; Du, S.; Li, Y.; He, Y.; Zhang, S. Exosomes Secreted by Nucleus Pulposus Stem Cells Derived from Degenerative Intervertebral Disc Exacerbate Annulus Fibrosus Cell Degradation via Let-7b-5p. Front. Mol. Biosci. 2022, 8, 766115. [Google Scholar] [CrossRef]
- Storozhuk, M.; Cherninskyi, A.; Maximyuk, O.; Isaev, D.; Krishtal, O. Acid-Sensing Ion Channels: Focus on Physiological and Some Pathological Roles in the Brain. Curr. Neuropharmacol. 2021, 19, 1570–1589. [Google Scholar] [PubMed]
- Wang, Y.; Zhou, H.; Sun, Y.; Huang, Y. Acid-sensing ion channel 1: Potential therapeutic target for tumor. Biomed. Pharmacother. 2022, 155, 113835. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, F. Acid-Sensing Ion Channel-1a in Articular Chondrocytes and Synovial Fibroblasts: A Novel Therapeutic Target for Rheumatoid Arthritis. Front. Immunol. 2021, 11, 580936. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, X.; Wang, L.; Zhang, J.; Pan, X.; Li, Y.; Cao, R.; Li, B.; Lin, H.; Wang, Y.; et al. Recent Advances in Acid-sensitive Ion Channels in Central Nervous System Diseases. Curr. Pharm. Des. 2022, 28, 1406–1411. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, Y.-Q.; Li, C.; He, M.; Rusyniak, W.G.; Annamdevula, N.; Ochoa, J.; Leavesley, S.J.; Xu, J.; Rich, T.C.; et al. Human ASIC1a mediates stronger acid-induced responses as compared with mouse ASICIa. FASEB J. 2018, 32, 3832–3843. [Google Scholar] [CrossRef]
- Li, B.; Zhou, J.; Luo, Y.; Tao, K.; Zhang, L.; Zhao, Y.; Lin, Y.; Zeng, X.; Yu, H. Suppressing circ_0008494 inhibits HSCs activation by regulating the miR-185-3p/Col1a1 axis. Front. Pharmacol. 2022, 13, 1050093. [Google Scholar] [CrossRef]
- Liang, S.; Ning, R.; Zhang, J.; Liu, J.; Zhang, J.; Shen, H.; Chen, R.; Duan, J.; Sun, Z. MiR-939-5p suppresses PM2.5-induced endothelial injury via targeting HIF-1α in HAECs. Nanotoxicology 2021, 15, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 2018, 37, 289. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, G.; Gao, H.; Li, Y.; Zhuang, L.; Meng, Z.; Liu, L. miR-939-5p Contributes to the Migration and Invasion of Pancreatic Cancer by Targeting ARHGAP4. Onco Targets Ther. 2020, 13, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Zai, Z.; Xu, Y.; Qian, X.; Li, Z.; Ou, Z.; Zhang, T.; Wang, L.; Ling, Y.; Peng, X.; Zhang, Y.; et al. Estrogen antagonizes ASIC1a-induced chondrocyte mitochondrial stress in rheumatoid arthritis. J. Transl. Med. 2022, 20, 561. [Google Scholar] [CrossRef]
- Wu, B.-m.; Bargaineer, J.; Zhang, L.; Yang, T.; Xiong, Z.; Leng, T. Upregulation of acid sensing ion channel 1a (ASIC1a) by hydrogen peroxide through the JNK pathway. Acta Pharmacol. Sin. 2021, 42, 1248–1255. [Google Scholar] [CrossRef]
- Cai, F.; Hong, X.; Tang, X.; Liu, N.-C.; Wang, F.; Zhu, L.; Xie, X.-H.; Xie, Z.-Y.; Wu, X.-T. ASIC1a activation induces calcium-dependent apoptosis of BMSCs under conditions that mimic the acidic microenvironment of the degenerated intervertebral disc. Biosci. Rep. 2019, 39, BSR20192708. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, F.R.; Xu, R.S.; Hu, W.; Jiang, D.L.; Ji, C.; Chen, F.H.; Yuan, F.L. Acid-sensing ion channel 1a-mediated calcium influx regulates apoptosis of endplate chondrocytes in intervertebral discs. Expert Opin. Ther. Targets 2014, 18, 1–14. [Google Scholar] [CrossRef]
- Murphy, E.; Liu, J.C. Mitochondrial calcium and reactive oxygen species in cardiovascular disease. Cardiovasc. Res. 2022, 119, 1105–1116. [Google Scholar] [CrossRef]
- Wang, S.; Li, B.; Solomon, V.; Fonteh, A.; Rapoport, S.I.; Bennett, D.A.; Arvanitakis, Z.; Chui, H.C.; Sullivan, P.M.; Yassine, H.N. Calcium-dependent cytosolic phospholipase A2 activation is implicated in neuroinflammation and oxidative stress associated with ApoE4. Mol. Neurodegener. 2022, 17, 42. [Google Scholar] [CrossRef]
- Yan, J.; Wang, Y.; Zhang, J.; Liu, X.; Yu, L.; He, Z. Rapidly Blocking the Calcium Overload/ROS Production Feedback Loop to Alleviate Acute Kidney Injury via Microenvironment-Responsive BAPTA-AM/BAC Co-Delivery Nanosystem. Small 2023, 19, 2206936. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Song, Y.; Chen, G.; Zhang, B.; Bai, Y.; Sun, C.; Fan, D.; Chen, Z. Circular RNA CircFOXO3 Functions as a Competitive Endogenous RNA for Acid-Sensing Ion Channel Subunit 1 Mediating Oxeiptosis in Nucleus Pulposus. Biomedicines 2024, 12, 678. https://doi.org/10.3390/biomedicines12030678
Chen X, Song Y, Chen G, Zhang B, Bai Y, Sun C, Fan D, Chen Z. Circular RNA CircFOXO3 Functions as a Competitive Endogenous RNA for Acid-Sensing Ion Channel Subunit 1 Mediating Oxeiptosis in Nucleus Pulposus. Biomedicines. 2024; 12(3):678. https://doi.org/10.3390/biomedicines12030678
Chicago/Turabian StyleChen, Xi, Ying Song, Guanghui Chen, Baoliang Zhang, Yang Bai, Chuiguo Sun, Dongwei Fan, and Zhongqiang Chen. 2024. "Circular RNA CircFOXO3 Functions as a Competitive Endogenous RNA for Acid-Sensing Ion Channel Subunit 1 Mediating Oxeiptosis in Nucleus Pulposus" Biomedicines 12, no. 3: 678. https://doi.org/10.3390/biomedicines12030678




