Cellular Responses Induced by NCT-503 Treatment on Triple-Negative Breast Cancer Cell Lines: A Proteomics Approach
Abstract
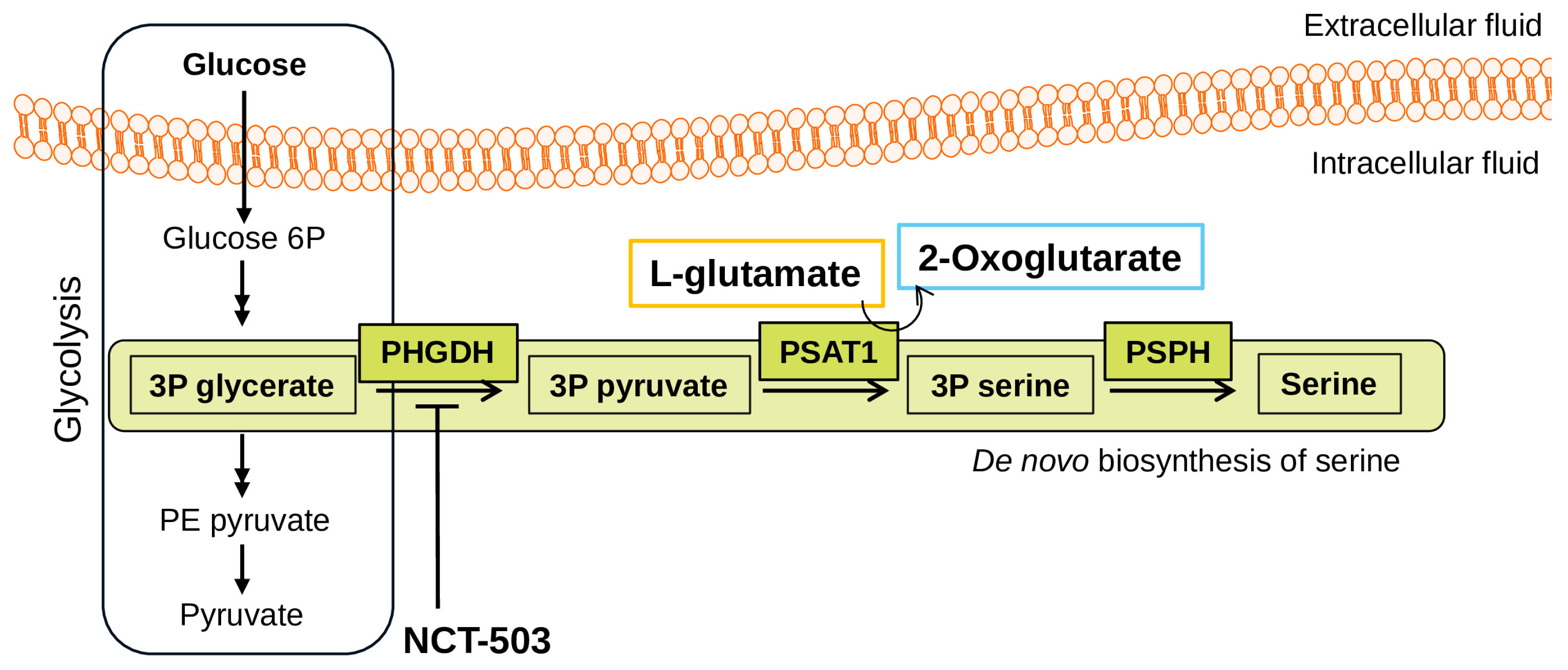
1. Introduction
2. Materials and Methods
2.1. Cell Culturing and Assays
2.1.1. Cell Viability Testing by MTT Assay
2.1.2. Colony Formation Assay
2.1.3. Wound Healing Assay
2.1.4. Morphological Analysis by Fluorescence Microscopy
2.1.5. Cell Culture Procedures and Protein Extraction for Mass Spectrometry Analysis
2.2. Sample Preparation for Mass Spectrometry
2.2.1. Protein Extraction
2.2.2. Protein Digestion and Peptide Purification
2.3. Label Free Nano-LC-IMS-MS and Data Analysis
2.3.1. Data Acquisition and Primary Data Processing
2.3.2. Data Analysis
3. Results
3.1. Proteome Heterogenity of Untreated TNBC Cell Lines
3.2. TNBC Cells Response to NCT-503 Treatment
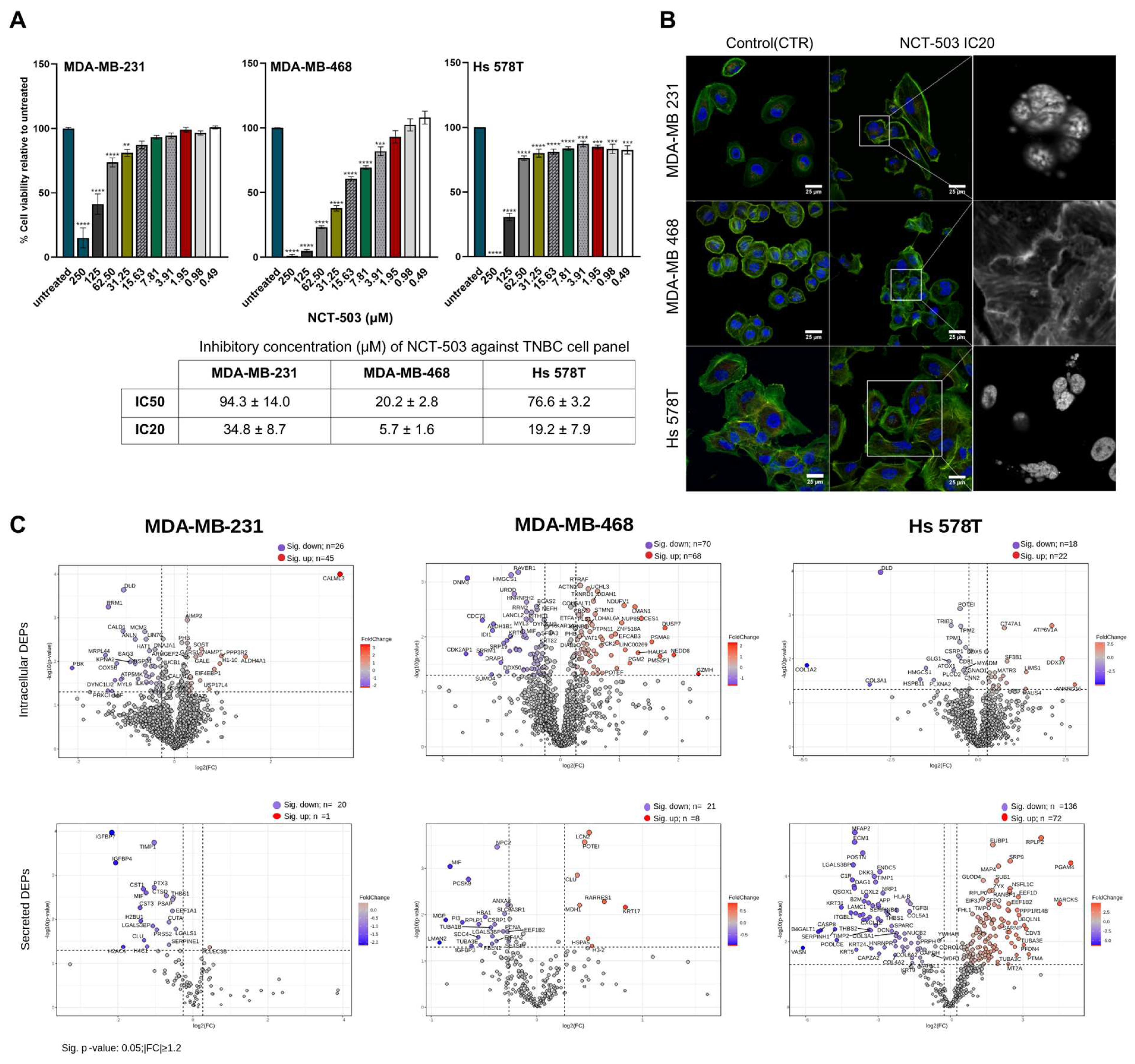
3.2.1. Results of In Vitro Assays
3.2.2. Differentially Expressed Proteins Induced by NCT-503 Treatment
3.2.3. Enrichment Analysis
3.2.4. Reactome GSEA Analysis
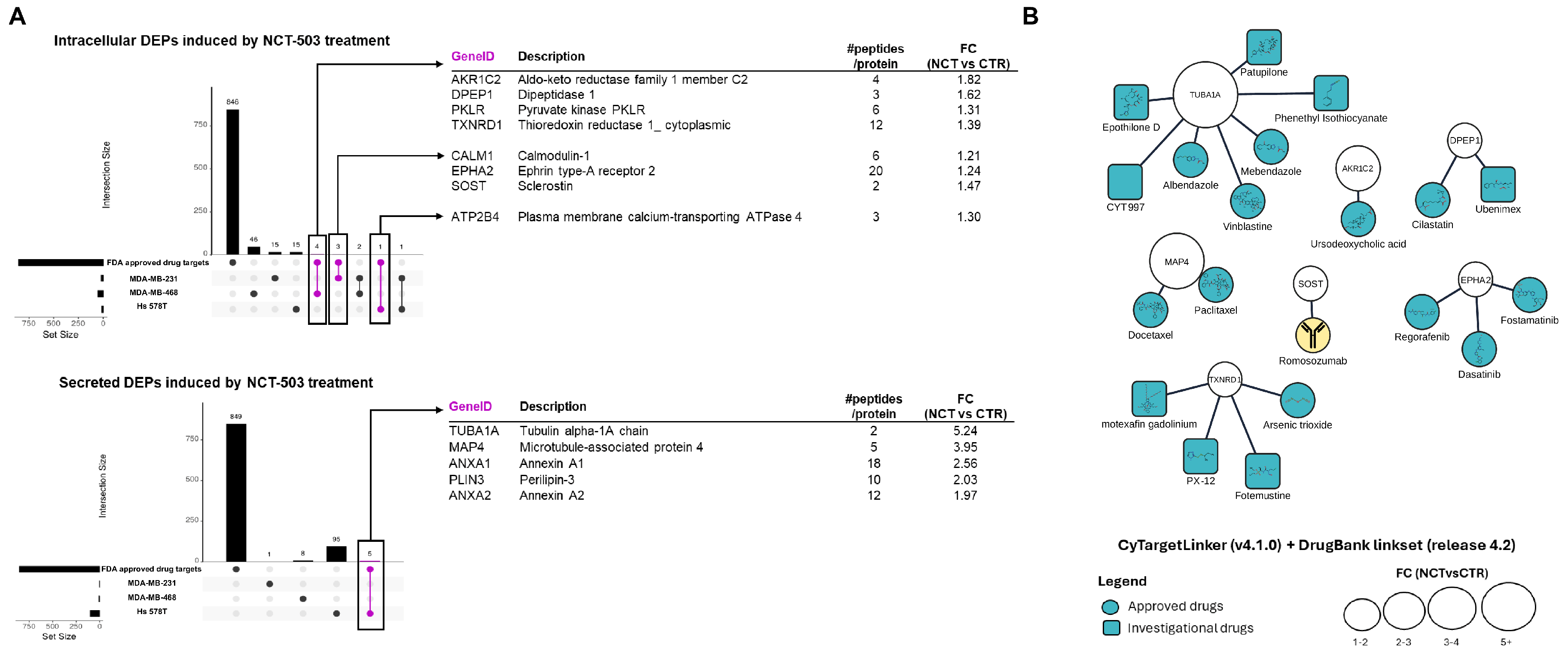
3.3. Targetable Proteins Induced by NCT-503 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today (Version 1.1). Available online: https://gco.iarc.who.int/today (accessed on 4 March 2024).
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast Cancer Heterogeneity and Its Implication in Personalized Precision Therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef]
- Kirkby, M.; Popatia, A.M.; Lavoie, J.R.; Wang, L. The Potential of Hormonal Therapies for Treatment of Triple-Negative Breast Cancer. Cancers 2023, 15, 4702. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Z.H.; Ma, L.X.; Wu, S.Y.; Wu, J.; Yu, K.D.; Sui, X.Y.; Xu, Y.; Liu, X.Y.; Chen, L.; et al. Optimising First-Line Subtyping-Based Therapy in Triple-Negative Breast Cancer (FUTURE-SUPER): A Multi-Cohort, Randomised, Phase 2 Trial. Lancet Oncol. 2024, 25, 184–197. [Google Scholar] [CrossRef]
- Pralea, I.-E.; Moldovan, R.-C.; Țigu, A.-B.; Ionescu, C.; Iuga, C. Mass Spectrometry-Based Omics for the Characterization of Triple-Negative Breast Cancer Bio-Signature. J. Pers. Med. 2020, 10, 277. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Beatty, A.; Fink, L.S.; Singh, T.; Strigun, A.; Peter, E.; Ferrer, C.M.; Nicolas, E.; Cai, K.Q.; Moran, T.P.; Reginato, M.J.; et al. Metabolite Profiling Reveals the Glutathione Biosynthetic Pathway as a Therapeutic Target in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2018, 17, 264–275. [Google Scholar] [CrossRef]
- Gong, Y.; Ji, P.; Yang, Y.S.; Xie, S.; Yu, T.J.; Xiao, Y.; Jin, M.L.; Ma, D.; Guo, L.W.; Pei, Y.C.; et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021, 33, 51–64.e9. [Google Scholar] [CrossRef]
- Wang, S.; Yuan, X.; Song, G.; Yang, L.; Jin, J.; Liu, W. Serine Metabolic Reprogramming in Tumorigenesis, Tumor Immunity, and Clinical Treatment. Adv. Nutr. 2023, 14, 1050–1066. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, M.; Wang, M.; Yu, X.; Guo, J.; Sun, T.; Li, X.; Yao, L.; Dong, H.; Xu, Y. Metabolic Reprogramming in Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Conger, K.O.; Chidley, C.; Ozgurses, M.E.; Zhao, H.; Kim, Y.; Semina, S.E.; Burns, P.; Rawat, V.; Sheldon, R.; Ben-Sahra, I.; et al. ASCT2 Is the Primary Serine Transporter in Cancer Cells. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ravez, S.; Spillier, Q.; Marteau, R.; Feron, O.; Frédérick, R. Challenges and Opportunities in the Development of Serine Synthetic Pathway Inhibitors for Cancer Therapy. J. Med. Chem. 2017, 60, 1227–1237. [Google Scholar] [CrossRef]
- Dong, J.K.; Lei, H.M.; Liang, Q.; Tang, Y.B.; Zhou, Y.; Wang, Y.; Zhang, S.; Li, W.B.; Tong, Y.; Zhuang, G.; et al. Overcoming Erlotinib Resistance in EGFR Mutation-Positive Lung Adenocarcinomas through Repression of Phosphoglycerate Dehydrogenase. Theranostics 2018, 8, 1808–1823. [Google Scholar] [CrossRef]
- Xia, Y.; Ye, B.; Ding, J.; Yu, Y.; Alptekin, A.; Thangaraju, M.; Prasad, P.D.; Ding, Z.C.; Park, E.J.; Choi, J.H.; et al. Metabolic Reprogramming by MYCN Confers Dependence on the Serine-Glycine-One-Carbon Biosynthetic Pathway. Cancer Res. 2019, 79, 3837–3850. [Google Scholar] [CrossRef]
- Wei, L.; Lee, D.; Law, C.T.; Zhang, M.S.; Shen, J.; Chin, D.W.C.; Zhang, A.; Tsang, F.H.C.; Wong, C.L.S.; Ng, I.O.L.; et al. Genome-Wide CRISPR/Cas9 Library Screening Identified PHGDH as a Critical Driver for Sorafenib Resistance in HCC. Nat. Commun. 2019, 10, 4681. [Google Scholar] [CrossRef]
- Issaq, S.H.; Mendoza, A.; Kidner, R.; Rosales, T.I.; Duveau, D.Y.; Heske, C.M.; Rohde, J.M.; Boxer, M.B.; Thomas, C.J.; DeBerardinis, R.J.; et al. EWS-FLI1-Regulated Serine Synthesis and Exogenous Serine Are Necessary for Ewing Sarcoma Cellular Proliferation and Tumor Growth. Mol. Cancer Ther. 2020, 19, 1520–1529. [Google Scholar] [CrossRef]
- Wu, X.; Xia, J.; Zhang, J.; Zhu, Y.; Wu, Y.; Guo, J.; Chen, S.; Lei, Q.; Meng, B.; Kuang, C.; et al. Phosphoglycerate Dehydrogenase Promotes Proliferation and Bortezomib Resistance through Increasing Reduced Glutathione Synthesis in Multiple Myeloma. Br. J. Haematol. 2020, 190, 52–66. [Google Scholar] [CrossRef]
- Yoshino, H.; Enokida, H.; Osako, Y.; Nohata, N.; Yonemori, M.; Sugita, S.; Kuroshima, K.; Tsuruda, M.; Tatarano, S.; Nakagawa, M. Characterization of PHGDH Expression in Bladder Cancer: Potential Targeting Therapy with Gemcitabine/Cisplatin and the Contribution of Promoter DNA Hypomethylation. Mol. Oncol. 2020, 14, 2190–2202. [Google Scholar] [CrossRef]
- Jeon, M.J.; You, M.H.; Han, J.M.; Sim, S.; Yoo, H.J.; Lee, W.K.; Kim, T.Y.; Song, D.E.; Shong, Y.K.; Kim, W.G.; et al. High Phosphoglycerate Dehydrogenase Expression Induces Stemness and Aggressiveness in Thyroid Cancer. Thyroid 2020, 30, 1625–1638. [Google Scholar] [CrossRef]
- Mullarky, E.; Lucki, N.C.; Zavareh, R.B.; Anglin, J.L.; Gomes, A.P.; Nicolay, B.N.; Wong, J.C.Y.; Christen, S.; Takahashi, H.; Singh, P.K.; et al. Identification of a Small Molecule Inhibitor of 3-Phosphoglycerate Dehydrogenase to Target Serine Biosynthesis in Cancers. Proc. Natl. Acad. Sci. USA 2016, 113, 1778–1783. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Ham Chan, S.; Rohde, J.M.; Lewis, C.A.; JYM Swier, L.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH Inhibitor Reveals Coordination of Serine Synthesis and 1-Carbon Unit Fate HHS Public Access Author Manuscript. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef]
- Białopiotrowicz, E.; Noyszewska-Kania, M.; Kachamakova-Trojanowska, N.; Łoboda, A.; Cybulska, M.; Grochowska, A.; Kopczyński, M.; Mikula, M.; Prochorec-Sobieszek, M.; Firczuk, M.; et al. Serine Biosynthesis Pathway Supports MYC–MiR-494–EZH2 Feed-Forward Circuit Necessary to Maintain Metabolic and Epigenetic Reprogramming of Burkitt Lymphoma Cells. Cancers 2020, 12, 580. [Google Scholar] [CrossRef]
- Rafehi, H.; Orlowski, C.; Georgiadis, G.T.; Ververis, K.; El-Osta, A.; Karagiannis, T.C. Clonogenic Assay: Adherent Cells. J. Vis. Exp. 2011, 49, e2573. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In Vitro Scratch Assay: A Convenient and Inexpensive Method for Analysis of Cell Migration in Vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Ţigu, A.B.; Toma, V.A.; Mot, A.C.; Jurj, A.; Moldovan, C.S.; Fischer-Fodor, E.; Berindan-Neagoe, I.; Pârvu, M. The Synergistic Antitumor Effect of 5-Fluorouracil Combined with Allicin against Lung and Colorectal Carcinoma Cells. Molecules 2020, 25, 1947. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift Time-Specific Collision Energies Enable Deep-Coverage Data-Independent Acquisition Proteomics. Nat. Methods 2014, 11, 167–170. [Google Scholar] [CrossRef]
- Watson, J.; Smith, M.; Francavilla, C.; Schwartz, J.M. SubcellulaRVis: A Web-Based Tool to Simplify and Visualise Subcellular Compartment Enrichment. Nucleic Acids Res. 2022, 50, W718–W725. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Griss, J.; Viteri, G.; Sidiropoulos, K.; Nguyen, V.; Fabregat, A.; Hermjakob, H. ReactomeGSA—Efficient Multi-Omics Comparative Pathway Analysis. Mol. Cell Proteom. 2020, 19, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph Receptors and Ephrins in Cancer Progression. Nat. Rev. Cancer 2024, 24, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Wang, Z.; Yang, C. Rho GTPases: Big Players in Breast Cancer Initiation, Metastasis and Therapeutic Responses. Cells 2020, 9, 2167. [Google Scholar] [CrossRef] [PubMed]
- Wojtukiewicz, M.Z.; Sierko, E.; Hempel, D.; Tucker, S.C.; Honn, K.V. Platelets and Cancer Angiogenesis Nexus. Cancer Metastasis Rev. 2017, 36, 249–262. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Z.; Tian, Y.; Li, Z.; Liu, Z.; Zhu, S. The Critical Role of Platelet in Cancer Progression and Metastasis. Eur. J. Med. Res. 2023, 28, 385. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and Clinical Characterization of the Basal-like Subtype of Invasive Breast Carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef]
- Mehanna, J.; Haddad, F.G.H.; Eid, R.; Lambertini, M.; Kourie, H.R. Triple-Negative Breast Cancer: Current Perspective on the Evolving Therapeutic Landscape. Int. J. Women’s Health 2019, 11, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-Negative Breast Cancer Molecular Subtyping and Treatment Progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Liao, M.; Zhang, J.; Wang, G.; Wang, L.; Liu, J.; Ouyang, L.; Liu, B. Small-Molecule Drug Discovery in Triple Negative Breast Cancer: Current Situation and Future Directions. J. Med. Chem. 2021, 64, 2382–2418. [Google Scholar] [CrossRef]
- Coussy, F.; Lavigne, M.; de Koning, L.; Botty, R.E.; Nemati, F.; Naguez, A.; Bataillon, G.; Ouine, B.; Dahmani, A.; Montaudon, E.; et al. Response to MTOR and PI3K Inhibitors in Enzalutamide-Resistant Luminal Androgen Receptor Triple-Negative Breast Cancer Patient-Derived Xenografts. Theranostics 2020, 10, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.K.; Abramson, V.; Tan, T.; Dent, R. The Evolution of Triple-Negative Breast Cancer: From Biology to Novel Therapeutics. Am. Soc. Clin. Oncol. Educ. B 2016, 36, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef] [PubMed]
- Morris, P.G.; Rota, S.; Cadoo, K.; Zamora, S.; Patil, S.; D’Andrea, G.; Gilewski, T.; Bromberg, J.; Dang, C.; Dickler, M.; et al. Phase II Study of Paclitaxel and Dasatinib in Metastatic Breast Cancer. Clin. Breast Cancer 2018, 18, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Dering, J.; Ginther, C.; Wilson, C.A.; Glaspy, P.; Tchekmedyian, N.; Slamon, D.J. Dasatinib, an Orally Active Small Molecule Inhibitor of Both the Src and Abl Kinases, Selectively Inhibits Growth of Basal-Type/“Triple-Negative” Breast Cancer Cell Lines Growing in Vitro. Breast Cancer Res. Treat. 2007, 105, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.L.; Baurain, J.F.; Sparano, J.; Strauss, L.; Campone, M.; Fumoleau, P.; Rugo, H.; Awada, A.; Sy, O.; Llombart-Cussac, A. A Phase 2 Trial of Dasatinib in Patients with Advanced HER2-Positive and/or Hormone Receptor-Positive Breast Cancer. Clin. Cancer Res. 2011, 17, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Skoczynska, A.; Skoczynska, M. Breast Cancer and Arsenic Anticancer Effects: Systematic Review of the Experimental Data from in Vitro Studies. Genet. Res. 2022, 2022, 8030931. [Google Scholar] [CrossRef] [PubMed]
- Salahuddin, M.M.; Omran, G.A.; Helmy, M.W.; Houssen, M.E. Effect of Regorafenib on P2X7 Receptor Expression and Different Oncogenic Signaling Pathways in a Human Breast Cancer Cell Line: A Potential of New Insight of the Antitumor Effects of Regorafenib. Curr. Issues Mol. Biol. 2021, 43, 2199–2209. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fan, C.; Tang, J. Regorafenib Suppresses Migration of and Induces Cell Cycle Arrest and Apoptosis in Mcf-7 Cells. Braz. J. Pharm. Sci. 2021, 57, e18122. [Google Scholar] [CrossRef]
- Joe, N.S.; Godet, I.; Milki, N.; Ain, N.U.I.; Oza, H.H.; Riggins, G.J.; Gilkes, D.M. Mebendazole Prevents Distant Organ Metastases in Part by Decreasing ITGβ4 Expression and Cancer Stemness. Breast Cancer Res. 2022, 24, 98. [Google Scholar] [CrossRef]
- Priotti, J.; Baglioni, M.V.; García, A.; Rico, M.J.; Leonardi, D.; Lamas, M.C.; Menacho Márquez, M. Repositioning of Anti-Parasitic Drugs in Cyclodextrin Inclusion Complexes for Treatment of Triple-Negative Breast Cancer. AAPS PharmSciTech 2018, 19, 3734–3741. [Google Scholar] [CrossRef] [PubMed]
- Im, E.K.; Choi, Y.H.; Paik, K.J.; Suh, H.; Jin, Y.; Kim, K.W.; Yoo, Y.H.; Kim, N.D. Novel Bile Acid Derivatives Induce Apoptosis via a P53-Independent Pathway in Human Breast Carcinoma Cells. Cancer Lett. 2001, 163, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Hesse, E.; Schröder, S.; Brandt, D.; Pamperin, J.; Saito, H.; Taipaleenmäki, H. Sclerostin Inhibition Alleviates Breast Cancer-Induced Bone Metastases and Muscle Weakness. JCI Insight 2019, 4, e125543. [Google Scholar] [CrossRef] [PubMed]
- Issa, F.; Ioppolo, J.A.; Rendina, L.M. Boron and Gadolinium Neutron Capture Therapy; Elsevier Ltd.: Amsterdam, The Netherlands, 2013; Volume 3, ISBN 9780080965291. [Google Scholar]
- Avendaño, C.; Menéndez, J.C. Anticancer Strategies Involving Radical Species. In Medicinal Chemistry of Anticancer Drugs, 3rd ed.; Avendaño, C., Menéndez, J.C., Eds.; Elsevier: Boston, MA, USA, 2023; pp. 165–235. ISBN 978-0-12-818549-0. [Google Scholar]
- Yang, F.; Xiao, Y.; Ding, J.H.; Jin, X.; Ma, D.; Li, D.Q.; Shi, J.X.; Huang, W.; Wang, Y.P.; Jiang, Y.Z.; et al. Ferroptosis Heterogeneity in Triple-Negative Breast Cancer Reveals an Innovative Immunotherapy Combination Strategy. Cell Metab. 2023, 35, 84–100.e8. [Google Scholar] [CrossRef]
- Koschorke, A.; Faraci, S.; Giani, D.; Chiodoni, C.; Iorio, E.; Canese, R.; Colombo, M.P.; Lamolinara, A.; Iezzi, M.; Ladomery, M.; et al. Phenethyl Isothiocyanate Hampers Growth and Progression of HER2-Positive Breast and Ovarian Carcinoma by Targeting Their Stem Cell Compartment. Cell Oncol. 2019, 42, 815–828. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pralea, I.-E.; Moldovan, R.-C.; Țigu, A.-B.; Moldovan, C.-S.; Fischer-Fodor, E.; Iuga, C.-A. Cellular Responses Induced by NCT-503 Treatment on Triple-Negative Breast Cancer Cell Lines: A Proteomics Approach. Biomedicines 2024, 12, 1087. https://doi.org/10.3390/biomedicines12051087
Pralea I-E, Moldovan R-C, Țigu A-B, Moldovan C-S, Fischer-Fodor E, Iuga C-A. Cellular Responses Induced by NCT-503 Treatment on Triple-Negative Breast Cancer Cell Lines: A Proteomics Approach. Biomedicines. 2024; 12(5):1087. https://doi.org/10.3390/biomedicines12051087
Chicago/Turabian StylePralea, Ioana-Ecaterina, Radu-Cristian Moldovan, Adrian-Bogdan Țigu, Cristian-Silviu Moldovan, Eva Fischer-Fodor, and Cristina-Adela Iuga. 2024. "Cellular Responses Induced by NCT-503 Treatment on Triple-Negative Breast Cancer Cell Lines: A Proteomics Approach" Biomedicines 12, no. 5: 1087. https://doi.org/10.3390/biomedicines12051087
APA StylePralea, I.-E., Moldovan, R.-C., Țigu, A.-B., Moldovan, C.-S., Fischer-Fodor, E., & Iuga, C.-A. (2024). Cellular Responses Induced by NCT-503 Treatment on Triple-Negative Breast Cancer Cell Lines: A Proteomics Approach. Biomedicines, 12(5), 1087. https://doi.org/10.3390/biomedicines12051087








