Comparative Therapeutic Effects of Plant-Extract Synthesized and Traditionally Synthesized Gold Nanoparticles on Alcohol-Induced Inflammatory Activity in SH-SY5Y Cells In Vitro
Abstract
:1. Introduction
- (i)
- Pharmacotherapy is a one-size-fits-all approach of addiction treatment that has not been largely successful, possibly because alcoholics constitute varying subtypes with differing biological and psychosocial contributions to the disease [10].
- (ii)
- Alcoholism is a multifaceted disorder associated with neurochemical heterogeneity and behavioral complexities, but current and prospective anti-alcoholism drugs selectively target specific receptors or transporters [8,9]. Therefore, patients may have to take many pills daily for comprehensive protection/treatment of alcoholism. This may be partially responsible for the patients’ non-adherence to pharmacotherapy regimens.
- (iii)
- The current therapeutic drugs exhibit poor bioavailability, serious side effects and probability of development of addiction (for benzodiazepines). Therefore, there is an urgent need to develop potent, effective and safer medications to treat alcohol-related disorders.
- Herbal Medications: Although many Asian countries have used medicinal plants such as Tabernanthe iboga, Panax ginseng, Salvia miltiorrhiza, Hypericum perforatum, Pelargonium graveolens, Lippia citriodora, Punica granatum, Morinda citrifolia L., Mirabilis jalapa, Aloysia triphylla, etc., to treat addiction for centuries, only recently has the West begun to understand their pharmacological possibilities and clinical applications in alcoholism treatment [9,10,11]. Despite large volume of research data in support of medicinal plants’ anti-alcoholism effects, their clinical applications are hindered by poor solubility and bioavailability of the active ingredients, inability to cross the blood–brain barrier, and high variability [12]. Early work by Benlhabib et al. [13,14] showed that Pueraria lobata (kudzu root) contains three major isoflavones, puerarin (PU), genistein (GE), and daidzein (DE), exhibiting the following pattern: PU >> GE > DE. Either an aqueous extract of kudzu root or purified PU alone reduced (1) alcohol consumption (50% suppression) without affecting water intake, and (2) severity of the alcohol withdrawal symptoms in of alcohol-preferring rats when administered orally. In the studies where the isoflavones were given over the course of several days, maximal suppression of alcohol intake occurred in 2 to 3 days [14].
- Traditionally Synthesized Nanoparticles: Development of nanoparticle (NP)-based pharmacotherapy is a promising development in diagnosing and designing personalized treatment of addiction and other diseases [15]. Studies have used colloidal gold and silver NPs, functionalized with multiple pharmaceuticals and other active ligands, such as a blood–brain barrier permeant peptide, in treatment of alcoholism [16,17]. NPs, because of their unique properties, may circumvent the disadvantages of current pharmacotherapy discussed above and/or listed below [18,19]. Some of the advantages of NPs are (i) improved bioavailability and therapeutic efficacy; (ii) multiple drugs loaded in a single nanocarriers, resulting in improved compliance because patients will not have to take multiple pills; (iii) on-demand drug release—nanocarriers may be designed to release drugs as needed via external (ultrasound) or internal (pH or selected enzymes) cues. However, the traditionally synthesized gold and silver NPs have some disadvantages: they require stabilization to prevent rapid aggregation, difficult to functionalize with certain ligands, and undergo defunctionalization, releasing toxic NPs. Since the NPs contain the FDA approved drugs listed above, they thus exhibit the same limitations listed above for pharmaceutical preparation.
- Plant Extract Synthesized Nanoparticles: Earlier studies have described “green” synthesis of gold and silver NPs using plant extracts that are environment friendly, cost effective, easily scaled up for large scale syntheses of nanoparticles, and do not require stabilizers such as polyethylene glycols [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34]. Most importantly, the “green” nanoparticles may retain the therapeutic potency of the plant and the unique properties of NPs. The key problems associated with the “green” NPs are lack of (i) methodology to identify the surface ligands; (ii) dose-response studies, and (iii) established therapeutic doses.
2. Materials and Methods
2.1. Nanoparticle Synthesis and Characterization
2.1.1. The Traditional Synthesis
2.1.2. Plant-Extract Based Synthesis
2.2. Nanoparticles Were Characterized by Measuring the Following Indices
2.2.1. UV–vis Spectrum
2.2.2. FT-IR Spectrum
2.2.3. Surface Topology by Transmission Electron Microscope (TEM)
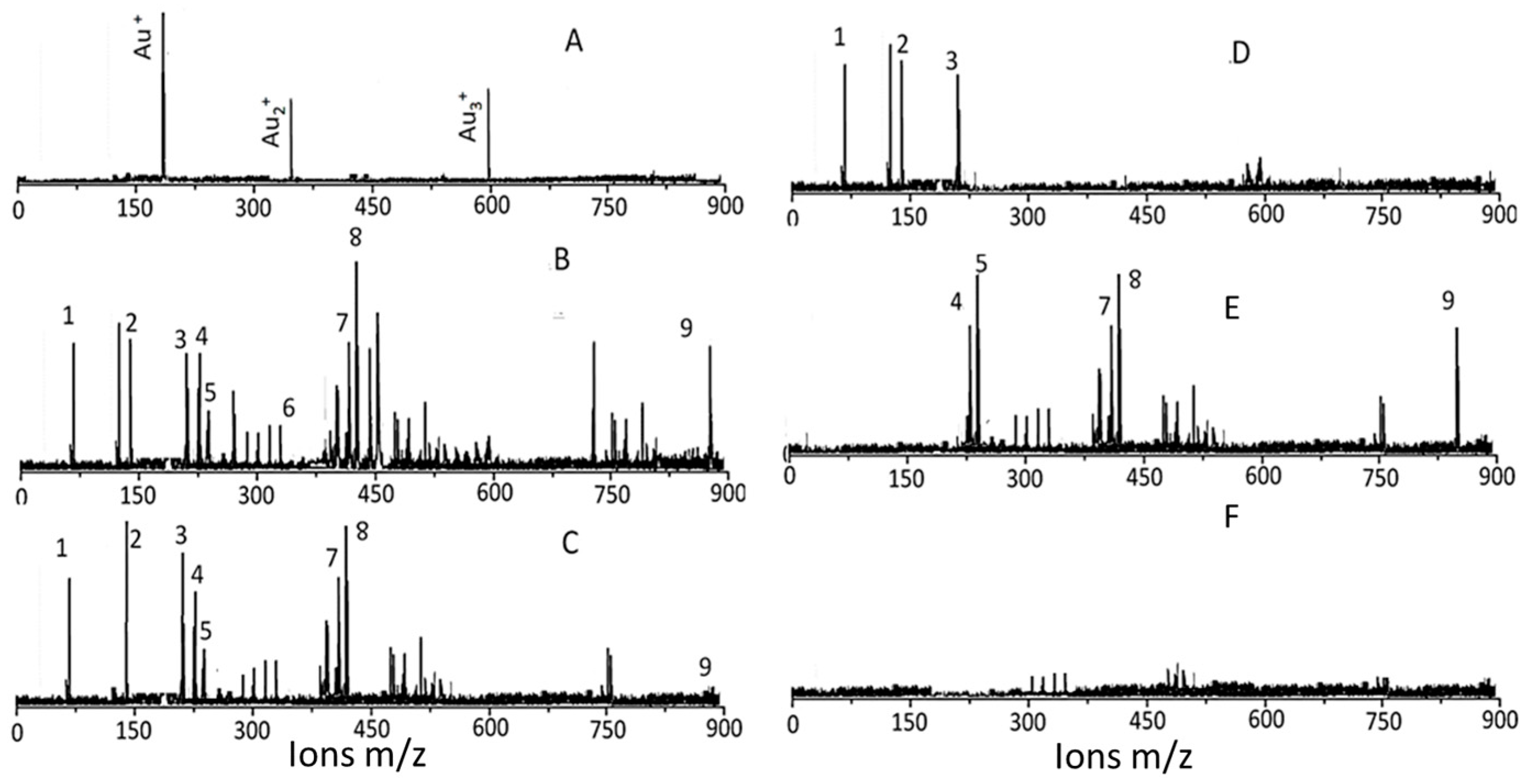
2.2.4. Identification of AuNPs and the Surface Functional Groups
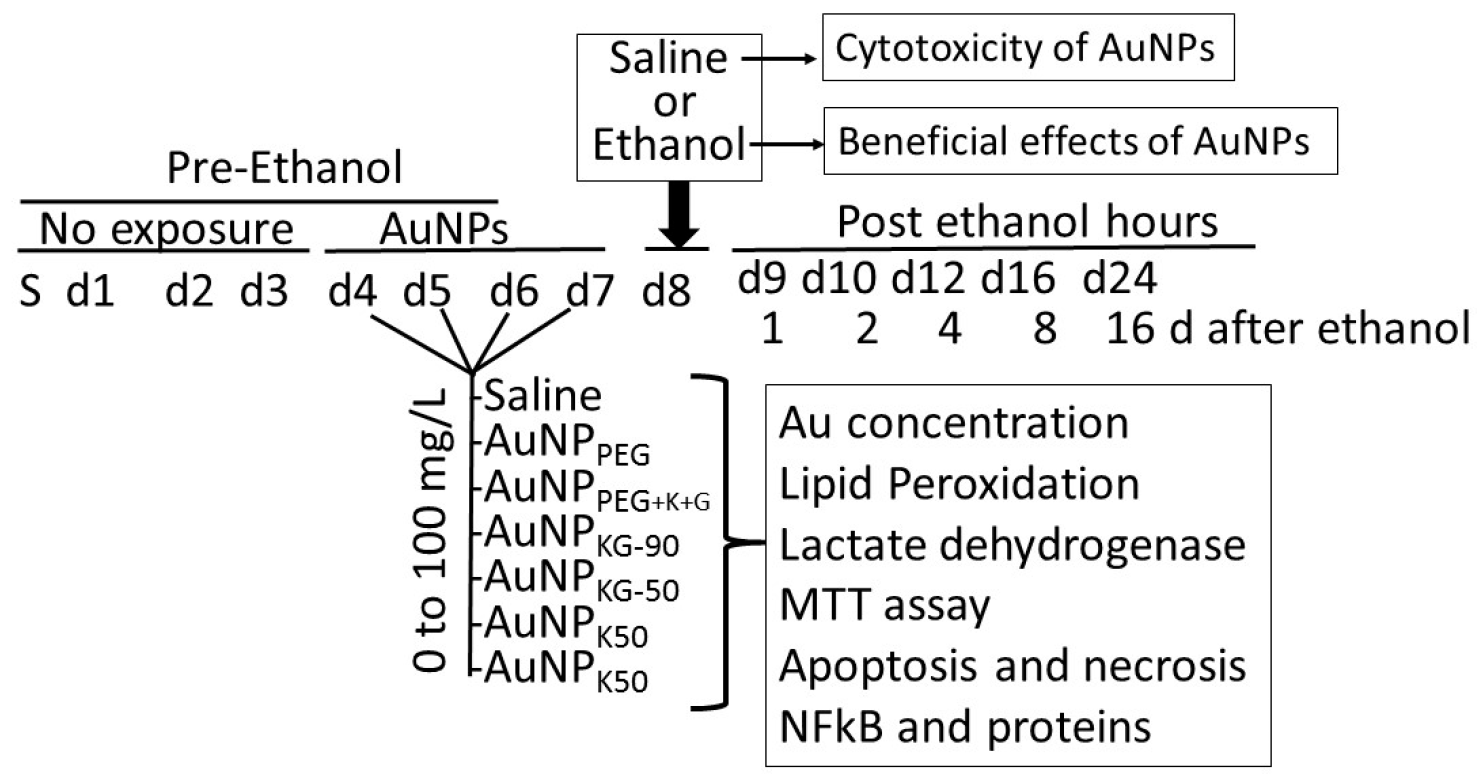
2.3. Beneficial Efficacy and Adverse Effects of AuNPs
- (i)
- Determination of internalized AuNPs by measuring intracellular Au concentrations (iAuCs): the atomic absorption spectrometric method described by Pedersen and Graabaek [36] and Benlhabib, et al. [13,14] was used for analysis of Au in tissue samples. In brief, the cells were harvested, treated with trypsin, etched with potassium iodide (KI) and iodine (I2) at 1:6 ratio (0.34 mM I2) to remove the surface AuNPs [37], washed with plasma buffered saline, and mixed with 5 mL of aqua regia, and digested for 24 h. The digest was diluted 1:1 with a solution of 2 ppm yttrium in dilute nitric acid, serving as an internal standard. The instrument was calibrated using a solution of 1 ppm Au and 1 ppm yttrium in 50% aqua regia. The samples were introduced in a segment flow mode of a flameless atomic absorption spectrophotometer (Beckman model 485, Beckman, Indianapolis, IN, USA). Au concentration was determined using a calibration curve. The brain Au concentrations were normalized to total tissue weight.
- (ii)
- Lactate dehydrogenase release assay [38]: 50 μL of clear cell media was mixed with the reconstituted 2× LDH assay buffer (223 mg 2-p-iodophenyl-3-p-nitrophenyl-5-phenl tetrazolium chloride, 57 mg N-methylphenazonium methyl sulfate, 575 mg nicotinamide adenine nucleotide (NAD), and 3.2 g lactic acid in 480 mL 200 mM Tris buffer solution, pH 8.0). The mixture was gently shaken for 30 seconds and incubated in dark for 10 to 30 min at room temperature. Reaction was stopped with 50 μL of Stop Solution (1 M acetic acid), mixed, and absorbance was measured between 490–520 nm.
- (iii)
- Lipid peroxidase: the cells (3.6 × 106) were suspended in 2 mL of a solution containing 15% trichloroacetic acid, 0.25 N HCl and 0.5% thiobarbituric acid, and the samples were heated for 25 min in a boiling water bath. The samples were cooled and centrifuged for 10 min at 4000 rpm. The absorbance of the supernatant fraction was determined at a wavelength of 535 nm. An extinction coefficient of 1.56 × 105 M−1 cm−1 was used to calculate the concentration of malondialdehyde. Values were expressed as pmol of MDA per mg protein.
- (iv)
- Cell viability: the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium reduction assay kit from Millipore (Millipore Sigma, St Louis, MO, USA) was used to assess cell viability.
- (v)
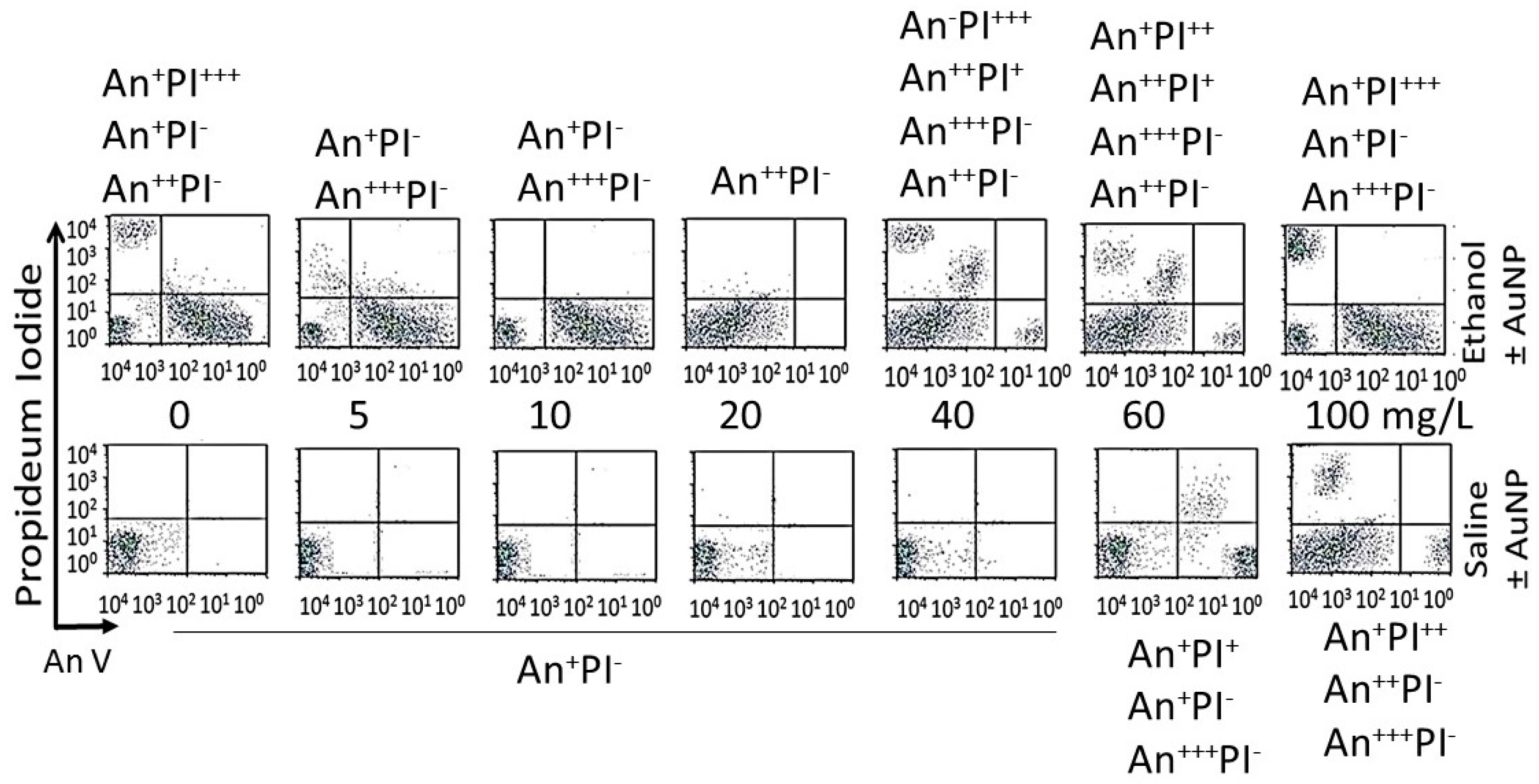
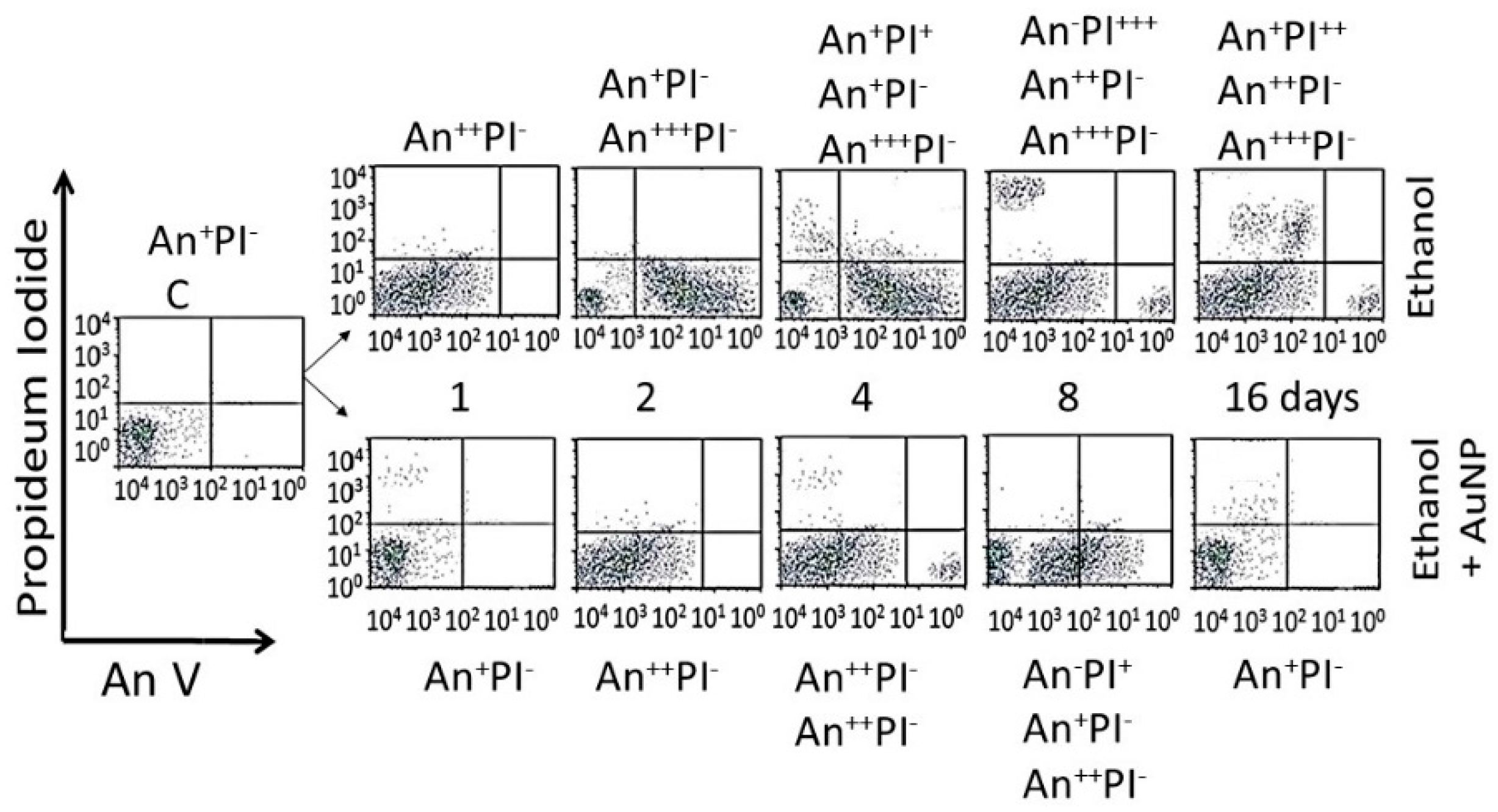
- Analysis of cells undergoing apoptosis and necrosis: cells were exposed to the AuNPs followed by ethanol as shown in Figure 1. At different time intervals after cessation ethanol, the cells were harvested and double stained with annexin V-FITC (An) and propidium iodide (PI), as described previously [15,38], and were subjected to fluorescence activated cell sorting (FACS) analysis using on a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA). All the experiments were performed in triplicate. Cells were classified as An−PI− or An+ PI− (normal cells), An−PI+++ (necrotic cells), An++PI− (early apoptosis stage), An+++PI+ (late apoptosis stage).
- (vi)
- NFκB activation by electrophoretic mobility shift assay (EMSA) [39]: cell cultures from in vitro studies or finely chopped tissue samples from in vivo studies were suspended in HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) hypotonic buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA (ethylenediaminetetraacetic acid), 0.1 mM EGTA (ethylene glycol tetraacetic acid_, 1 mM Dm, 0.5 mM phenylmethylsulfonyl fluoride, and 10 pg/mL leupeptin, antipain, aprotinin, and pepstatin) for 15 min on ice. Samples were vortexed for 10 s with 0.6% Nonidet P-40, and centrifuged at 12,000× g for 60 s. The pellets containing nuclei were resuspended in nucleus buffer (20 mM HEPES, pH 7.9, 25% glycerol, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 10 pg/mL leupeptin, antipain, aprotinin, and pepstatin) and briefly sonicated on ice. For EMSA, nuclear extracts (10 pg of protein) were incubated in 25 µL of total reaction volume containing 20 mM HEPES, pH 7.9, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5% glycerol, 200 pg/mL bovine serum albumin, and 2.5 pg of poly(d1-dC) for 15 min at 4 °C. The carboxytetramethyl-rhodamine (TMR)-labeled oligonucleotide (0.5 ng) was then added to the reaction mixture, and incubated for 20 min at room temperature.
2.4. Characterization of AuNP–Protein Interaction
2.5. Statistical Analysis
3. Results
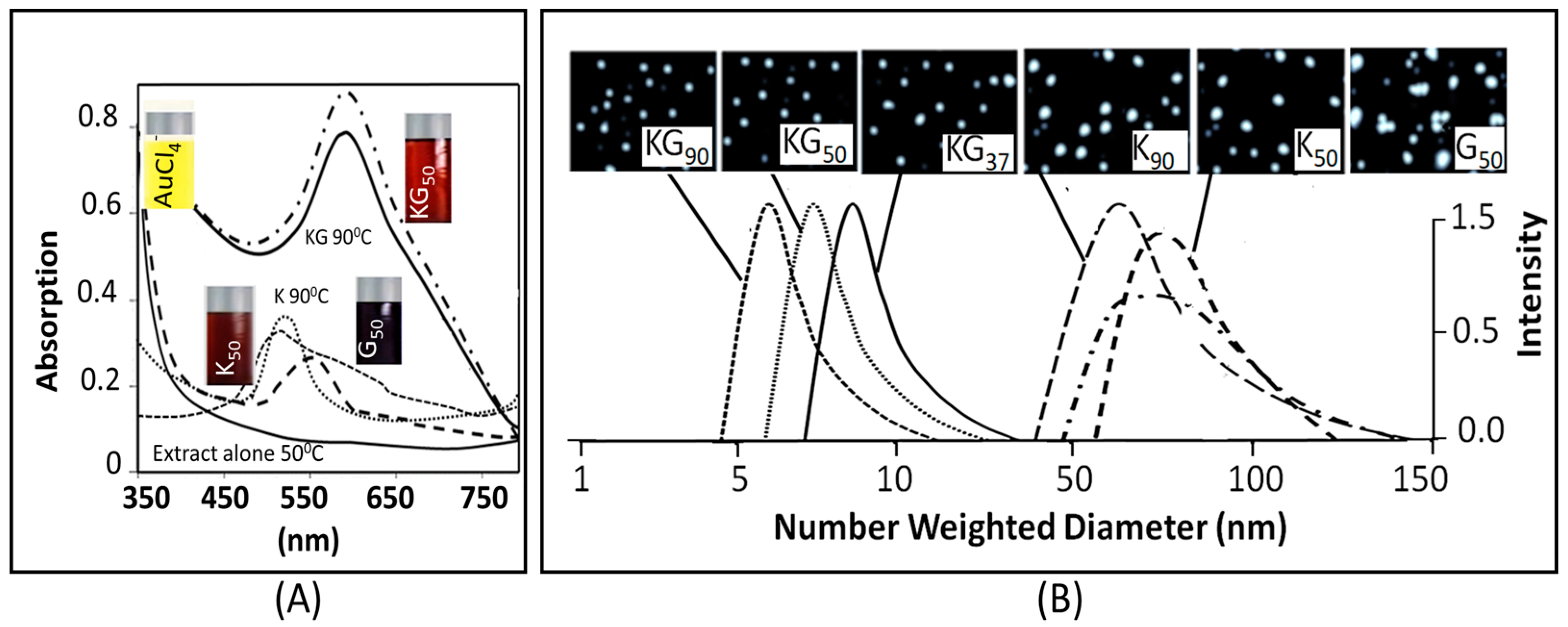
3.1. Nanoparticle Synthesis and Characterization
3.1.1. Physicochemical Characterization
3.1.2. The Surface Characterization
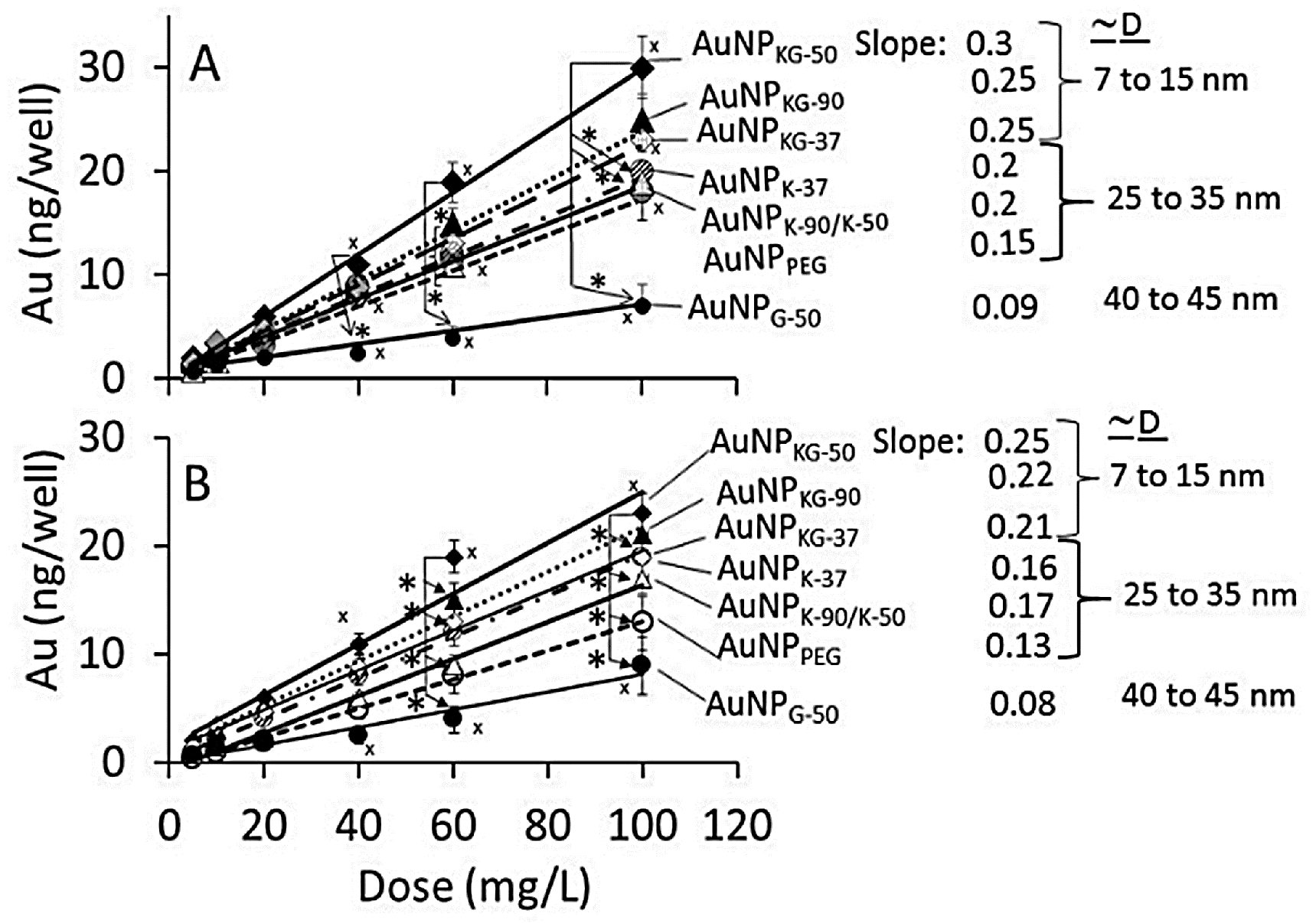
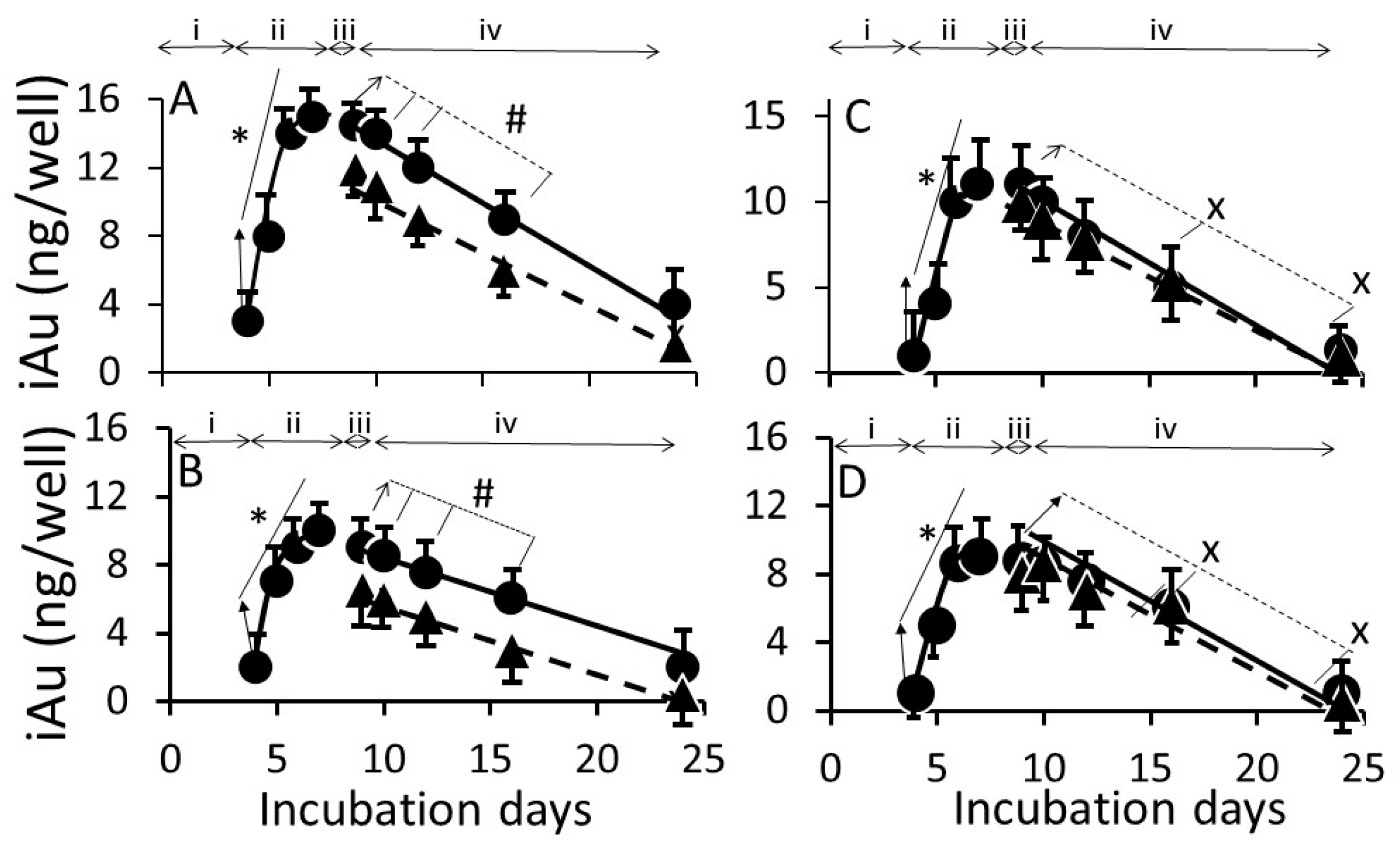
3.2. AuNP Internalization in Cells
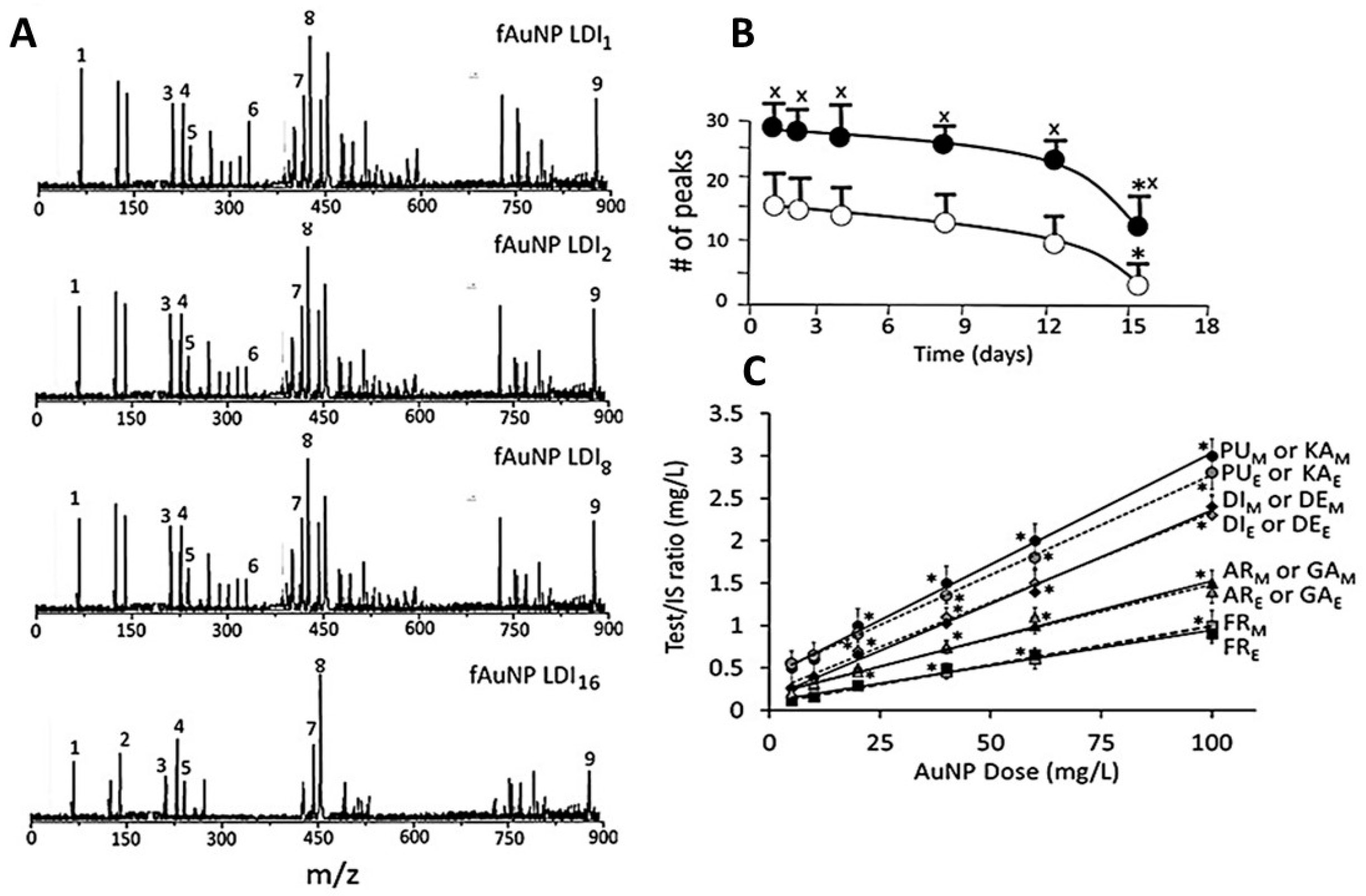
3.3. Characterization of Surface Ligands in AuNPs Collected from Exposed Cells
3.4. Beneficial and Adverse Effects of AuNPs
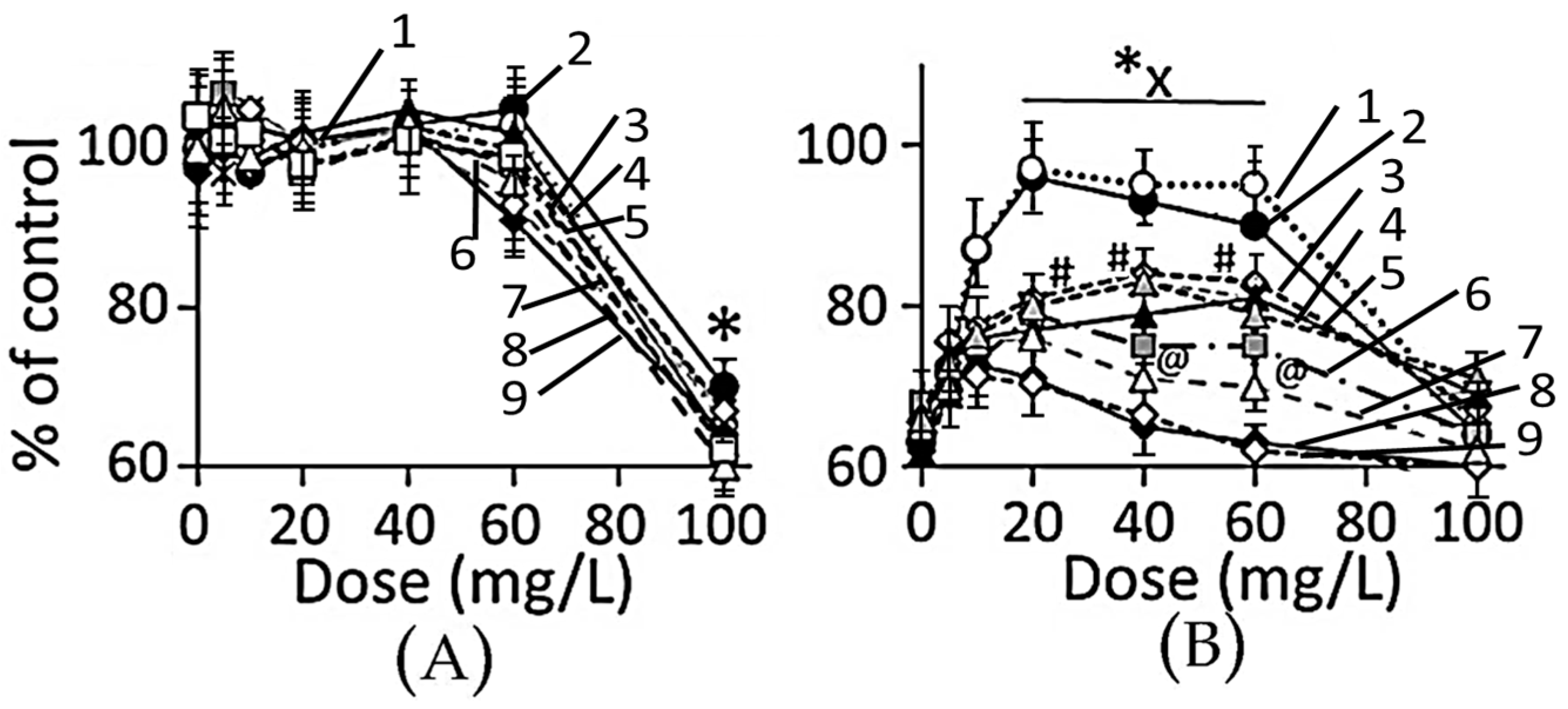
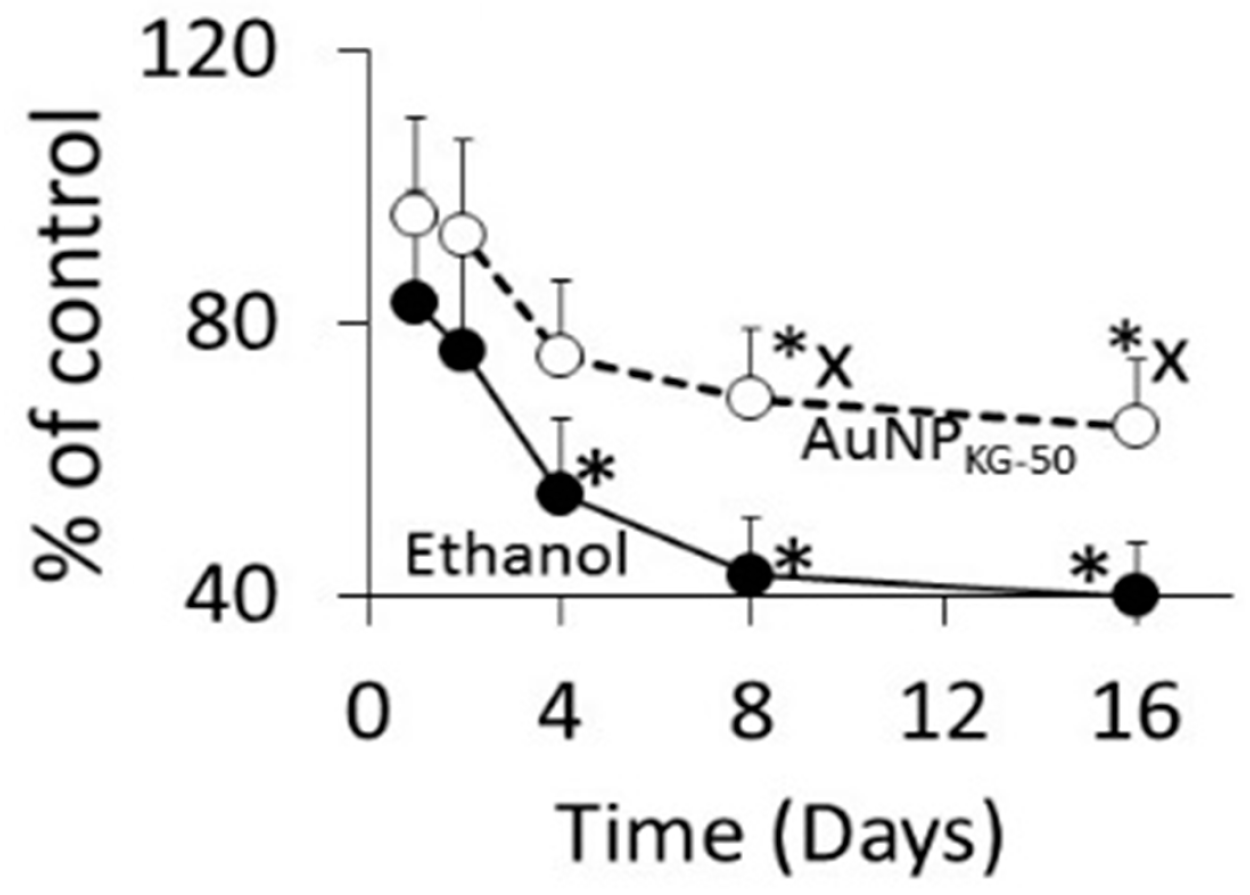
3.4.1. Cell Viability
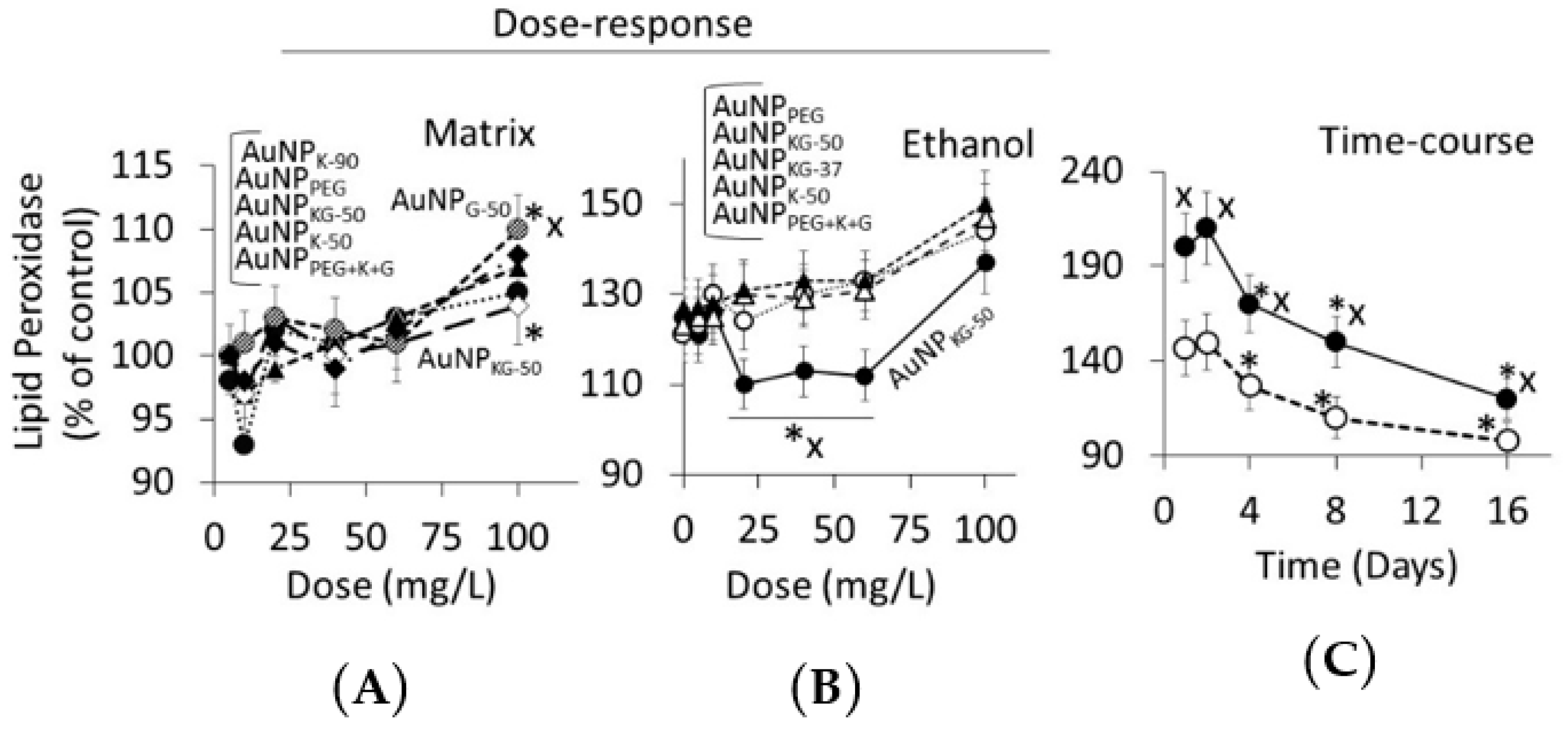
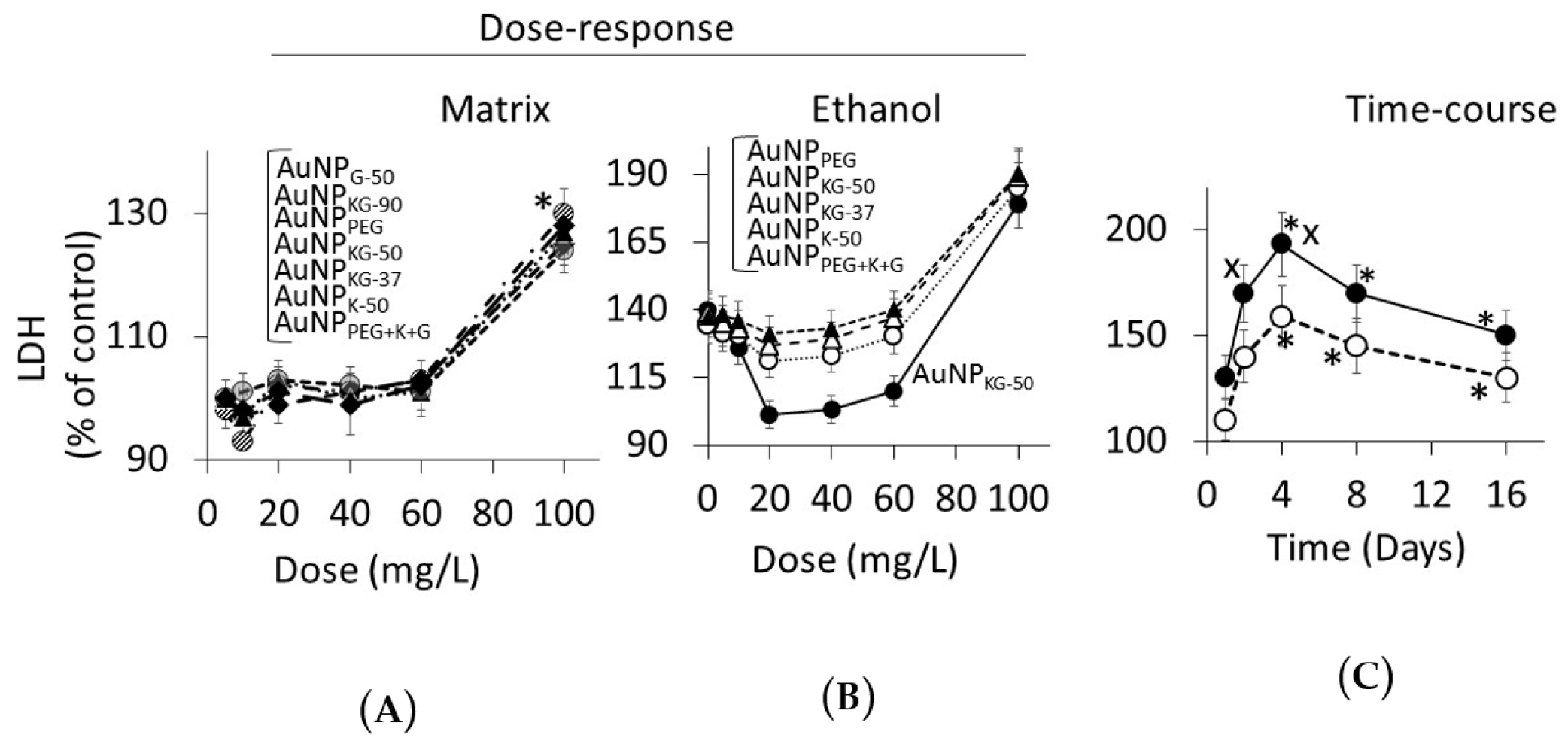
3.4.2. Lipid Peroxidase and Lactate Dehydrogenase Activities
3.4.3. Enumeration of Apoptotic and Necrotic Cells
3.4.4. NFκB Activation
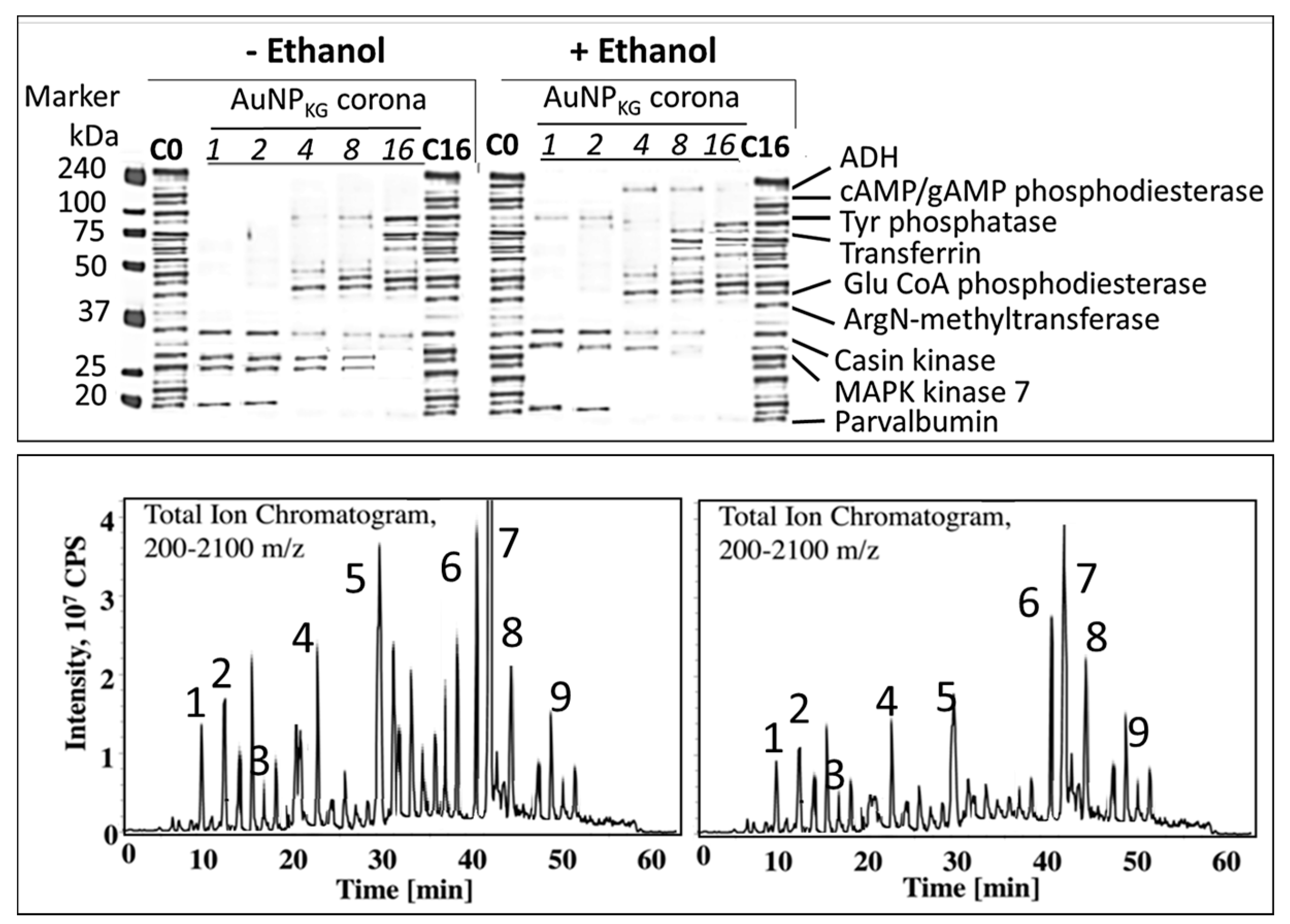
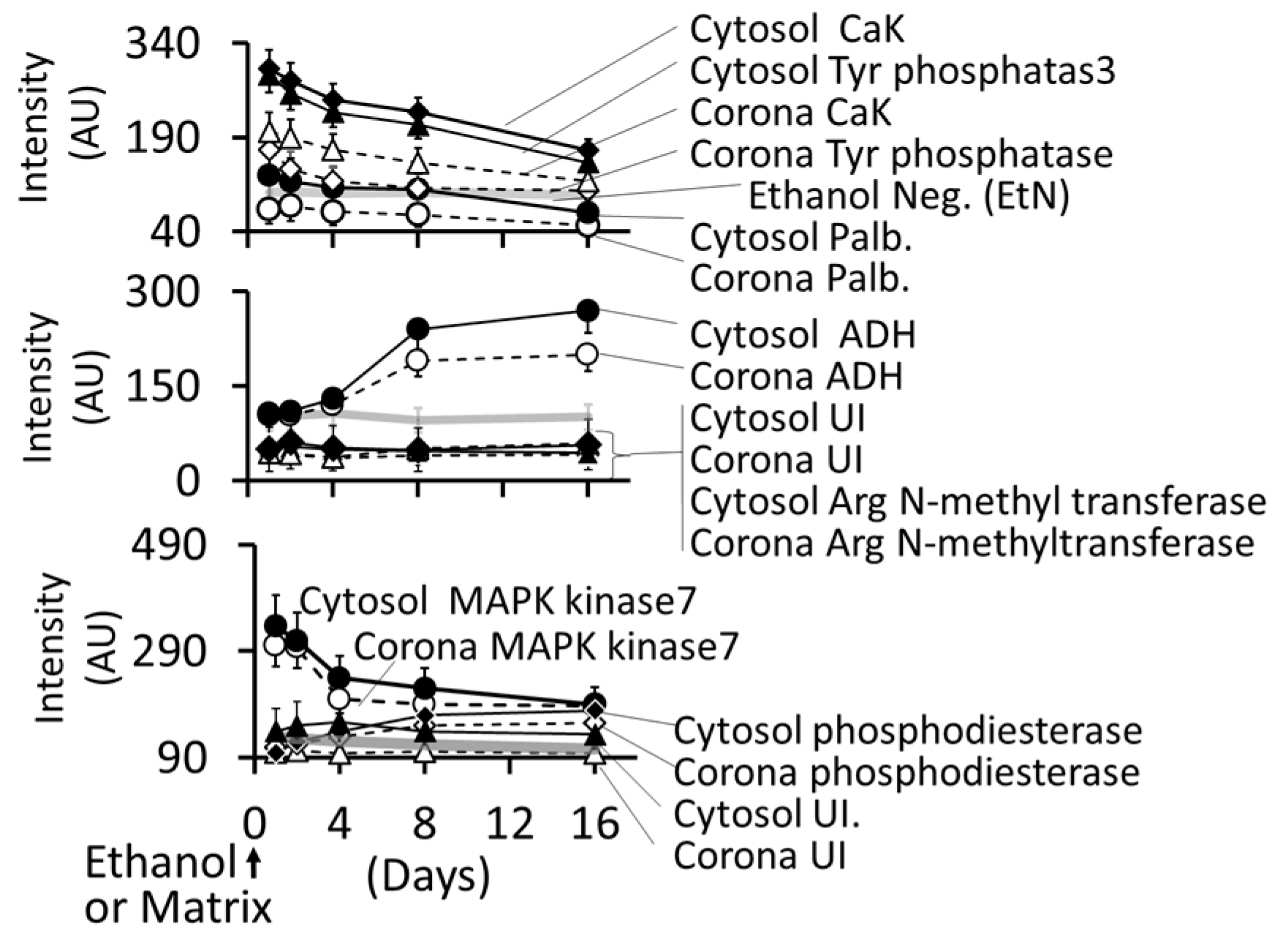
3.4.5. Formation of Intracellular Protein–Corona
- i
- Out of 35 proteins detected in control (An/En) cells, 9 cytosolic proteins were confirmed. Tyr phosphorylase, Arg N-methyl transferase, and MAPK kinase-7 yielded stronger bands (scan density 2.2 to 2.6) than parvalbumin, ADH, Glu-CoA dehydrogenase, cAMP/gAMP phosphodiesterase, transferrin, and casein kinase 1γ2 (scan density 0.3 to 1.4). Day 1 to day 16 values did not differ significantly.
- ii
- The AuNP corona protein/cytosol protein ratio for Tyr phosphorylase, Arg N-methyl transferase, and MAPK kinase-7 increased from <10 at day 1 to >20 at day 16. The ratio for other proteins either did not change or increased slightly.
- iii
- Ethanol exposure did not significantly alter the protein scan values, but significantly suppressed the AuNP corona protein/cytosol protein ratio. Ethanol specifically altered binding of MAPK kinase-7, casein kinase, Tyr phosphorylase and cAMP/gAMP phosphodiesterase to AuNPs, possibly by altering their concentrations in the cytosol. This suggests that ethanol exposure may have directly affected the AuNP–protein interaction.
- iv
- The composition of protein corona changed temporally. AuNPs extracted from day 1 and day 2 An/En or An/Ep cells interacted with smaller (<37 kDa) proteins, while AuNPs extracted from day 4 to day 16 cells interacted with proteins ranging from <37 kDa to 150 kDa. The band intensity increases with increasing incubation time.
4. Discussion
4.1. Synthesis and Characterization of AuNPs
4.1.1. AuNP Size, UV Absorption and FTIR Spectroscopy
4.1.2. Characterization of Surface Ligands
4.2. Cellular Uptake and Extrusion of AuNPs
4.3. Toxicity of Plant-Extract Synthesized AuNPs
4.4. Therapeutic Potency of Plant-Extract Synthesized AuNPs against Ethanol Toxicity
4.4.1. Ethanol Cytotoxicity
4.4.2. Protective Effects of AuNPs against Ethanol Toxicity
4.4.3. Possible Mechanisms for Protection against Ethanol
- (i)
- Gum ligands, such as arabinose, galactose, and fructose, that are part of a highly branched polysaccharide (MW 3 × 105) consisting of b-(1-3) galactose backbone with linked branches of arabinose and rhamnose [87]. Ali et al. [35] have provided direct evidence of anti-inflammatory and anti-oxidative capacities of edible gum Arabic (GA) that ameliorates superoxide production and DNA double strand breakage. Cuesta et al. [88] and Faggio et al. [89] have reported high immune-stimulatory, anti-inflammatory and anti-oxidation properties of GA in mammals and aquatic animals, respectively. Studies [90,91,92,93] have also shown that edible gum, in addition to being anti-oxidative, enhance the biocompatibility and bioavailability of AuNPs and other NPs, such as AgNPs and magnetic iron oxide nanoparticles (MNP). The GA coating offers two major benefits: it enhances colloidal stability and provides reactive functional groups suitable for coupling of bioactive compounds.
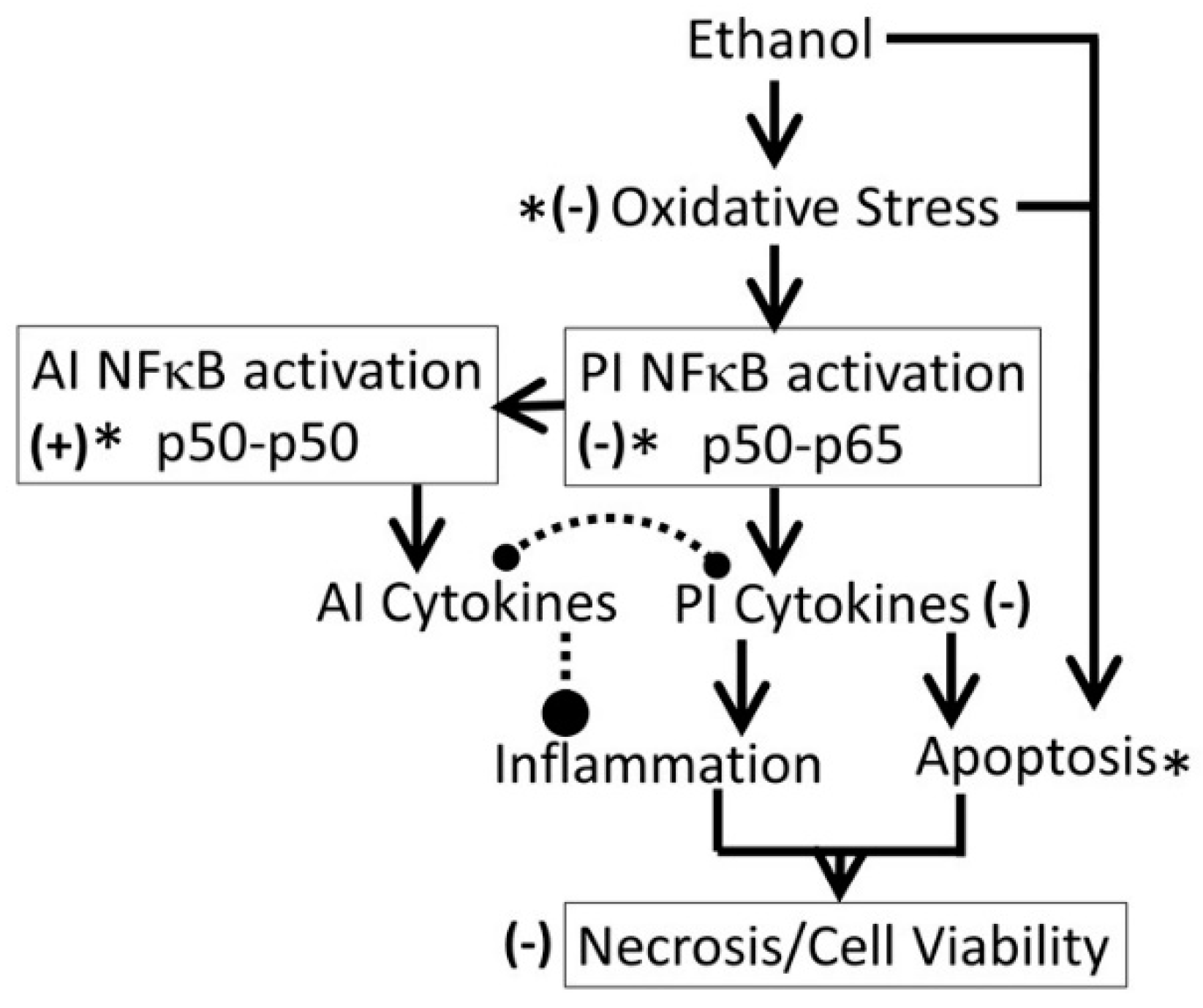
- (ii)
- Kudzu ligands, such as daidzein and puerarin possess potent anti-inflammatory and anti-oxidation properties, thus protecting against alcohol-mediated disorders [28,29,94,95,96,97]. As shown in Figure 10, ethanol induces oxidative stress and ensuing downstream activation of p50–p65 (a pro-inflammatory dimer of NFκB) and suppression of p50–p50 (an anti-inflammatory dimer of NFκB), resulting in dysregulation of inflammation and activation of pro-apoptosis signaling [32]. The addictive effects of ethanol are mediated by the brain addiction pathways including dopamine, GABA, NMDA, and serotonin neurons [5,98,99]. Earlier studies have shown that kudzu root extracts modulate the brain addiction circuits, especially the dopamine, GABA, NMDA, and Glu neurons/receptors in protecting the brain against the adverse effects of ethanol and suppressing the severity of the withdrawal symptoms [100,101,102].
4.4.4. AuNP–Protein Interaction
5. Conclusions
Conflicts of Interest
References
- Sacks, J.J.; Roeber, J.; Bouchery, E.E. State costs of excessive alcohol consumption, 2006. Am. J. Prev. Med. 2013, 45, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.F. Are societies paying unnecessarily for an otherwise free lunch? Final musings on the research on Alcoholics Anonymous and its mechanisms of behavior change. Addiction 2017, 112, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, R.; Agardh, E.; Coates, M.M.; Allebeck, P.; Danielsson, A.K. Alcohol-attributed disease burden and alcohol policies in the BRICS-countries during the years 1990–2013. J. Glob. Health 2017, 7, 010404. [Google Scholar] [CrossRef] [PubMed]
- Stahre, M. Contribution of excessive alcohol consumption to deaths and years of potential life lost in the United States. Prev. Chronic Dis. 2014, 11, E109. [Google Scholar] [CrossRef] [PubMed]
- Koob, G.F. A role for brain stress systems in addiction. Neuron 2008, 59, 11–34. [Google Scholar] [CrossRef] [PubMed]
- Koob, G.F.; Volkow, N.D. Neurocircuitry of addiction. Neuropsychopharm 2010, 35, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K. Critical Review of Alcohol, Alcoholism and the Withdrawal Symptoms I. Mechanisms of Addiction and Withdrawal Syndrome. Biomedicines 2017. submitted. [Google Scholar]
- Ducharme, L.J.; Chandler, R.K.; Harris, A.H. Implementing Effective Substance Abuse Treatments in General Medical Settings: Mapping the Research Terrain. J. Subst. Abuse Treat. 2016, 60, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K. Critical Review of Alcohol, Alcoholism and the Withdrawal Symptoms II. Treatment Strategies. Biomedicines 2017. submitted. [Google Scholar]
- Litten, R.Z.; Bradley, A.M.; Moss, H.B. Alcohol biomarkers in applied settings: Recent advances and future research opportunities. Alcohol. Clin. Exp. Res. 2010, 34, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Carai, M.A.; Agabio, R.; Bombardelli, E.; Bourov, I.; Gessa, G.L.; Lobina, C.; Morazzoni, P.; Pani, M.; Reali, R.; Vacca, G.; et al. Potential use of medicinal plants in the treatment of alcoholism. Fitoterapia 2000, 71 (Suppl. 1), S38–S42. [Google Scholar] [CrossRef]
- Taritatu, D.V.; Bagade, V.B.; Karambelkar, P.J.; Jadhav, V.M.; Kadam, V. Natural Bioenhancers: An overview. J. Pharmacogn. Phytochem. 2013, 2, 55–60. [Google Scholar]
- Benlhabib, E.; Baker, J.I.; Keyler, J.E.; Singh, A.K. Kudzu root extract suppresses voluntary alcohol intake and alcohol withdrawal symptoms in P rats receiving free access to water and alcohol. J. Med. Food 2004, 7, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Benlhabib, E.; Baker, J.I.; Keyler, D.E.; Singh, A.K. Effects of purified puerarin on voluntary alcohol intake and alcohol withdrawal symptoms in P rats receiving free access to water and alcohol. J. Med. Food 2004, 7, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K. An Introduction to Nanoparticles and their Applications in Alcoholism Treatment. Biomedicines 2017. submitted. [Google Scholar]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV−Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Jiang, Y.; Gupta, S.; Younus, M.; Ramzan, M. Anti-inflammatory potency of nano-formulated puerarin and curcumin in rats subjected to the lipopolysaccharide-induced inflammation. J. Med. Food 2013, 16, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.K.; Paul, W.; Sharma, C.P. Green synthesis of gold nanoparticles with Zingiber officinale extract: Characterization and blood compatibility. Process Biochem. 2011, 46, 2007–2013. [Google Scholar] [CrossRef]
- Nellore, J.; Pauline, C.; Amarnath, K.; Shilpa, P.N. Antioxidant effect of gold nanoparticles synthesised from curcuma longa restrains 1-methyl-2-phenyl pyridinium ion induced stress in PC 12 cells. J. Nanoneurosci. 2012, 2, 63–74. [Google Scholar] [CrossRef]
- Banse, V.; Ledwani, L. Green synthesis of nanoparticles: Their advantages and disadvantages. AIP Conf. Proc. 2016, 1724, 020048. [Google Scholar]
- Patra, J.K.; Baek, K.-H. Green Nanobiotechnology: Factors Affecting Synthesis and Characterization Techniques. J. Nanomater. 2014, 2014, 417305. [Google Scholar] [CrossRef]
- Elia, R.; Zach, R.; Hazan, S.; Kolusheva, S.; Porat, Z.; Zeiri, Y. Green synthesis of gold nanoparticles using plant extracts as reducing agents. Int. J. Nanomed. 2014, 9, 4007–4021. [Google Scholar]
- Khan, A.U.; Wei, Y.; Yuan, Q.; Tahir, K.; Khan, Z.U.H.; Khan, F.U.; Khan, S.U.; Khan, M.J.; Khan, F.A.; Ahmad, A. Photocatalytic and antibacterial response of biosynthesized Gold nanoparticles. J. Photochem. Photobiol. B Biol. 2016, 162, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Tahir, K.; Nazir, S.; Ahmad, A.; Li, B.; Khan, A.U.; Khan, Z.U.H.; Khan, F.U.; Khan, Q.U.; Khan, A.; Rahman, A.U. Facile and green synthesis of phytochemicals capped platinum nanoparticles and in vitro their superior antibacterial activity. J. Photochem. Photobiol. B Biol. 2017, 166, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Tahir, K.; Nazir, S.; Li, B.; Khan, A.U.; Khan, Z.U.H.; Ahmad, A.U.; Khan, Q.U.; Zhao, Y. Enhanced visible light photocatalytic inactivation of Escherichia coli using silver nanoparticles as photocatalyst. J. Photochem. Photobiol. B Biol. 2015, 153, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Tahir, K.; Ahmad, A.; Li, B.; Nazir, B.; Khan, A.U.; Nasir, T.; Khan, Z.U.A.; Naz, R.; Raza, M. Visible light photocatalytic inactivation of bacteria and photo degradation of methylene blue with Ag/TiO2 nanoompsite prepared by a novel method. J. Photochem. Photobiol. B Biol. 2016, 162, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Tahir, K.; Nazir, S.; Li, B.; Ahmad, A.; Nasir, T.; Khan, A.U.; Shah, S.A.A.; Khan, Z.U.H.; Yasin, G.; Hameed, M.U. Sapium sebiferum leaf extract mediated synthesis of palladium nanoparticles and in vitro investigation of their bacterial and photocatalytic activities. J. Photochem. Photobiol. B Biol. 2016, 164, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Tahir, K.; Ahmad, A.; Li, B.; Khan, A.U.; Nazir, S.; Khan, S.; Khan, Z.U.H.; Khan, S.U. Preparation, characterization and an efficient photocatalytic activity of Au/TiO2 nanocomposite prepared by green deposition method. Mater. Lett. 2016, 178, 56–59. [Google Scholar] [CrossRef]
- Ahmad, A.; Wei, Y.; Syed, F.; Khan, S.; Khan, G.M.; Tahir, K.; Khan, A.U.; Raza, M.; Khan, F.U.; Yuan, Q. Isatis tinctoria mediated synthesis of amphotericin B-bound silver nanoparticles with enhanced photoinduced antileishmanial activity: A novel green approach. J. Photochem. Photobiol. B Biol. 2016, 161, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.U.; Chen, Y.; Khan, N.U.; Ahmad, A.; Tahir, K.; Khan, Z.U.; Khan, A.U.; Khan, S.U.; Raza, M.; Wan, P. Visible light inactivation of E. coli, Cytotoxicity and ROS determination of biochemically capped gold nanoparticles. Microb. Pathog. 2017, 107, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Yuan, Q.; Wei, Y.; Khan, S.U.; Tahir, K.; Khan, Z.U.H.; Ahmad, A.; Ali, F.; Ali, S.; Nazir, S. Longan fruit juice mediated synthesis of uniformly dispersed spherical AuNPs: Cytotoxicity against human breast cancer cell line MCF-7, antioxidant and fluorescent properties. RSC Adv. 2016, 6, 23775–23782. [Google Scholar] [CrossRef]
- Khan, A.U.; Yuan, Q.; Wei, Y.; Tahir, K.; Khan, S.U.; Ahmad, A.; Khan, S.; Nazir, S.; Khan, F.U. Ultra-efficient photocatalytic deprivation of methylene blue and biological activities of biogenic silver nanoparticles. J. Photochem. Photobiol. B Biol. 2016, 159, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.U.H.; Khan, A.; Shah, A.; Wan, P.; Chen, Y.; Khan, G.M.; Khan, A.U.; Tahir, K.; Muhammad, N.; Khan, H.U. Enhanced photocatalytic and electrocatalytic applications of green synthesized silver nanoparticles. J. Mol. Liq. 2016, 220, 248–257. [Google Scholar] [CrossRef]
- Khan, F.U.; Chen, Y.; Khan, N.U.; Khan, Z.U.H.; Khan, A.U.; Ahmad, A.; Tahir, K.; Wang, L.; Khan, M.R.; Wan, P. Antioxidant and catalytic applications of silver nanoparticles using Dimocarpus longan seed extract as a reducing and stabilizing agent. J. Photochem. Photobiol. B Biol. 2016, 164, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Ali, B.H.; Al-Husseni, I.; Beegam, S.; Al-Shukaili, A.; Nemmar, A.; Schierling, S.; Queisser, N.; Schupp, N. Effect of Gum Arabic on Oxidative Stress and Inflammation in Adenine–Induced Chronic Renal Failure in Rats. PLoS ONE 2013, 8, e55242. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.M.; Graabaek, P.M.A. Unified method for the determination of gold in biological fluids by flameless atomic absorption spectroscopy. Scand. J. Clin. Lab. Investig. 1977, 37, 91–94. [Google Scholar] [CrossRef]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. Methods Enzymol. 1978, 52, 302–310. [Google Scholar] [PubMed]
- Chan, F.K.M.; Moriwaki, K.; De Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar] [PubMed]
- Singh, A.K.; Jiang, Y. Differential activation of NFκB/RelA-p50 and NFκB/p50-p50 in control and alcohol-drinking rats subjected to carrageenin-induced pleurisy. Mediat. Inflamm. 2004, 13, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Gupta, S. Analysis of recombinant human erythropoietin and darbepoietin in spiked plasma. Proteom. Clin. Appl. 2007, 1, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Amendola, V.; Meneghetti, M. Laser ablation synthesis in solution and size manipulation of noble metal nanoparticles. Phys. Chem. Chem. Phys. 2009, 11, 3805–3821. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; Pal, T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem. Rev. 2007, 107, 4797–4862. [Google Scholar] [CrossRef] [PubMed]
- Noguez, C. Surface plasmons on metal nanoparticles: The influence of shape and physical environment. J. Phys. Chem. C 2007, 111, 3806–3819. [Google Scholar] [CrossRef]
- Kizil, R.; Irudayaraj, J.; Seetharaman, K. Characterization of Irradiated Starches by Using FT-Raman and FTIR Spectroscopy. J. Agric. Food Chem. 2002, 50, 3912–3918. [Google Scholar] [CrossRef] [PubMed]
- Kora, A.J.; Beedu, S.R.; Jayaraman, A. Size-controlled green synthesis of silver nanoparticles mediated by gum ghatti (Anogeissus latifolia) and its biological activity. Org. Med. Chem. Lett. 2012, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Penttinen, P.; Guan, T.; Xiao, J.; Chen, Q.; Xu, J.; Lindström, K.; Zhang, L.; Zhang, X.; Strobel, G.A. The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr. Microbiol. 2011, 62, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Chan, W.C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Alkilany, A.M.; Murphy, C.J. Toxicity and cellular uptake of gold nanoparticles: What we have learned so far? J. Nanopart. Res. 2010, 12, 2313–2333. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wu, Y.; Jin, S.; Tian, Y.; Zhang, X.; Zhao, Y.; Yu, L.; Liang, X.J. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano 2011, 5, 8629–8639. [Google Scholar] [CrossRef] [PubMed]
- Worley, S.L.; Vaughn, B.J.; Terry, A.I.; Gardiner, C.S.; DeKrey, G.K. Time- and dose-dependent effects of ethanol on mouse embryonic stem cells. Reprod. Toxicol. 2015, 57, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Ghosh, R.N.; Maxfield, F.R. Endocytosis. Physiol. Rev. 1997, 77, 759–803. [Google Scholar] [PubMed]
- Chithrani, D.B.; Jelveh, S.; Jalali, F.; van Prooijen, M.; Allen, C.; Bristow, R.G.; Hill, R.P.; Jaffray, D.A. Gold nanoparticles as radiation sensitizers in cancer therapy. Radiat. Res. 2010, 173, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Kam, N.W.S.; Liu, Z.; Dai, H. Carbon nanotubes as intracellular transporters for proteins and DNA: An investigation of the uptake mechanism and pathway. Angew. Chem. 2006, 118, 591–595. [Google Scholar] [CrossRef]
- Wilhelm, C.; Gazeau, F.; Roger, J.; Pons, J.N.; Bacri, J.C. Interaction of anionic superparamagnetic nanoparticles with cells: Kinetic analyses of membrane adsorption and subsequent internalization. Langmuir 2002, 18, 8148–8155. [Google Scholar] [CrossRef]
- Oh, N.; Park, J.H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed. 2014, 9 (Suppl. 1), 51–63. [Google Scholar]
- Sakhtianchi, R.; Minchin, R.F.; Lee, K.B.; Alkilany, A.M.; Serpooshan, V.; Mahmoudi, M. Exocytosis of nanoparticles from cells: Role in cellular retention and toxicity. Adv. Colloid Interface Sci. 2013, 201, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.J.; Gole, A.M.; Stone, J.W.; Sisco, P.N.; Alkilany, A.M.; Goldsmith, E.C.; Baxter, S.C. Gold nanoparticles in biology: Beyond toxicity to cellular imaging. Acc. Chem. Res. 2008, 41, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, N.; Dykman, L. Biodistribution and toxicity of engineered gold nanoparticles: A review of in vitro and in vivo studies. Chem. Soc. Rev. 2011, 40, 1647–1671. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Lin, S.J.; Lin, C.H.; Tsai, C.S.; Lu, Y.J. Size effect of Ag nanoparticles on surface plasmon resonance. Surf. Coat. Technol. 2008, 202, 5339–5342. [Google Scholar] [CrossRef]
- Chompoosor, A.; Saha, K.; Ghosh, P.S.; Macarthy, D.J.; Miranda, O.R.; Zhu, Z.J.; Arcaro, K.F.; Rotello, V.M. The role of surface functionality on acute cytotoxicity, ROS generation and DNA damage by cationic gold nanoparticles. Small 2010, 6, 2246–2249. [Google Scholar] [CrossRef] [PubMed]
- Aratono, M.; Toyomasu, T.; Villeneuve, M.; Uchizono, Y.; Takiue, T.; Motomura, K.; Ikeda, N. Thermodynamic study on the surface formation of the mixture of water and ethanol. J. Colloid Interface Sci. 1997, 191, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Holte, L.L.; Gawrisch, K. Determining ethanol distribution in phospholipid multilayers with MAS−NOESY spectra. Biochemistry 1997, 36, 4669–4674. [Google Scholar] [CrossRef] [PubMed]
- Patra, M.; Salonen, E.; Terama, E.; Vattulainen, I.; Faller, R.; Lee, B.W.; Holopainen, J.; Karttunen, M. Under the influence of alcohol: The effect of ethanol and methanol on lipid bilayers. Biophys. J. 2006, 90, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Pernodet, N.; Fang, X.; Sun, Y.; Bakhtina, A.; Ramakrishnan, A.; Sokolov, J.; Ulman, A.; Rafailovich, M. Adverse effects of citrate/gold nanoparticles on human dermal fibroblasts. Small 2006, 2, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Connor, E.E.; Mwamuka, J.; Gole, A.; Murphy, C.J.; Wyatt, M.D. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small 2005, 1, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Ganesan, S. In vitro cytotoxicity assay on gold nanoparticles with different stabilizing agents. J. Nanomater. 2012, 2012, 14. [Google Scholar] [CrossRef]
- Choi, S.Y.; Jeong, S.; Jang, S.H.; Park, J.; Park, J.H.; Ock, K.S.; Lee, S.Y.; Joo, S.W. In vitro toxicity of serum protein-adsorbed citrate-reduced gold nanoparticles in human lung adenocarcinoma cells. Toxicol. In Vitro 2012, 26, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Neuman, M.G.; Shear, N.H.; Bellentani, S.; Tiribelli, C. Role of cytokines in ethanol-induced cytotoxicity in vitro in Hep G2 cells. Gastroenterology 1998, 115, 157–166. [Google Scholar] [CrossRef]
- Busby, W.F.; Ackermann, J.M.; Crespi, C.L. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab. Dispos. 1999, 27, 246–249. [Google Scholar] [PubMed]
- Prince, P.S.M.; Menon, V.P.; Gunasekaran, G. Hypolipidaemic action of Tinospora cordifolia roots in alloxan diabetic rats. J. Ethnopharmacol. 1998, 64, 53–57. [Google Scholar] [CrossRef]
- Prince, S.M.P.; Menon, V.P. Hypoglycaemic and hypolipidaemic action of alcohol extract of Tinospora cordifolia roots in chemical induced diabetes in rats. Phytother. Res. 2003, 17, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Perfumi, M.; Ciccocioppo, R.; Angeletti, S.; Cucculelli, M.; Massi, M. Effects of Hypericum perforatum extract on alcohol intake in Marchigian Sardinian alcohol-preferring rats. Alcohol Alcohol. 1999, 34, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Overstreet, D.H.; Keung, W.M.; Rezvani, A.H.; Massi, M.; Lee, D.Y. Herbal remedies for alcoholism: Promises and possible pitfalls. Alcohol. Clin. Exp. Res. 2003, 27, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Shati, A.A.; Elsaid, F.G. Effects of water extracts of thyme (Thymus vulgaris) and ginger (Zingiber officinale Roscoe) on alcohol abuse. Food Chem. Toxicol. 2009, 47, 1945–1949. [Google Scholar] [CrossRef] [PubMed]
- Tomczyk, M.; Zovko-Koncic, M.; Chrostek, L. Phytotherapy of alcoholism. Nat. Prod. Commun. 2012, 7, 273–280. [Google Scholar] [PubMed]
- Ozarowski, M.; Mikolajczak, P.L.; Thiem, B. Medicinal plants in the phytotherapy of alcohol or nicotine addiction. Implication for plants in vitro cultures. Prz. Lek. 2013, 70, 869–874. [Google Scholar] [PubMed]
- Otunola, G.A.; Afolayan, A.J. Therapeutic effect of aqueous extracts of three dietary spices and their mixture on lipid metabolism and oxidative stress in a rat model of chronic alcohol consumption. Pak. J. Pharm. Sci. 2016, 29, 1151–1155. [Google Scholar]
- Sharma, B.; Dabur, R. Protective Effects of Tinospora cordifolia on Hepatic and Gastrointestinal Toxicity Induced by Chronic and Moderate Alcoholism. Alcohol Alcohol. 2016, 51, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shirolkar, A.; Sharma, B.; Lata, S.; Dabur, R. Guduchi Sawras (Tinospora cordifolia): An Ayurvedic drug treatment modulates the impaired lipid metabolism in alcoholics through dopaminergic neurotransmission and anti-oxidant defense system. Biomed. Pharmacother. 2016, 83, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, C.; Padma, M.; Mareeswaran, R.; Suyavaran, A.; Kuma, M.S.; Premkumar, K.; Thirunavukkarasu, C. The extra cellular synthesis of gold and silver nanoparticles and their free radical scavenging and antibacterial properties. Colloids Surf. B Biointerfaces 2013, 102, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, A.; Ganesh, M.; Jayaprakash, J.; Jang, H.T. Biosynthesis of silver nanoparticles using Cassia tora leaf extract and its antioxidant and antibacterial activities. J. Ind. Eng. Chem. 2015, 28, 277–281. [Google Scholar] [CrossRef]
- Ravichandran, V.; Vasanthi, S.; Shalini, S.; Shah, S.A.A.; Harish, R. Green synthesis of silver nanoparticles using Atrocarpus altilis leaf extract and the study of their antimicrobial and antioxidant activity. Mater. Lett. 2016, 180, 264–267. [Google Scholar] [CrossRef]
- Wang, C.; Mathiyalagan, R.; Kin, Y.J.; Castro-Aceituno, V.; Singh, P.; Ahn, S.; Wang, D.; Yang, D.C. Rapid green synthesis of silver and gold nanoparticles using Dendropanax morbifera leaf extract and their anticancer activities. Int. J. Nanomed. 2016, 11, 3691–3701. [Google Scholar]
- Markus, J.; Wang, D.; Kim, Y.-J.; Ahn, S.; Mathiyalagan, R.; Wang, C.; Yang, D.C. Biosynthesis, Characterization, and Bioactivities Evaluation of Silver and Gold Nanoparticles Mediated by the Roots of Chinese Herbal Angelica pubescens Maxim. Nanoscale Res. Lett. 2017, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Amid, B.T.; Mirhosseini, H.; Kostadinović, S. Chemical composition and molecular structure of polysaccharide-protein biopolymer from Durio Zibethinus seed: extraction and purification process. Chem. Central J. 2012, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Dror, Y.; Cohen, Y.; Yerushalmi-Rozen, R. Structure of gum arabic in aqueous solution. J. Polym. Sci. Part B Polym. Phys. 2006, 44, 3265–3271. [Google Scholar] [CrossRef]
- Cuesta, A.; Rodríguez, A.; Esteban, MA.; Meseguer, J. In vivo effects of propolis, a honeybee product, on gilthead seabream innate immune responses. Fish Shellfish Immunol. 2005, 18, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Faggio, C.; Fazio, F.; Marafioti, S.; Arfuso, F.; Piccione, G. Oral administration of Gum Arabic: Effects on haematological parameters and oxidative stress markers in Mugil cephalus. Iran. J. Fish. Sci. 2015, 14, 60–72. [Google Scholar]
- Williams, D.N.; Gold, K.A.; Holoman, T.R.P.; Ehrman, S.H.; Wilson, O.C. Surface modification of magnetic nanoparticles using gum arabic. J. Nanopart. Res. 2006, 8, 749–753. [Google Scholar] [CrossRef]
- Kattumuri, V.; Katti, K.; Bhaskaran, S.; Boote, E.J.; Casteel, S.W.; Fent, G.M.; Robertson, D.J.; Chandrasekhar, M.; Kannan, R.; Katti, K.V. Gum Arabic as a Phytochemical Construct for the Stabilization of Gold Nanoparticles: In Vivo Pharmacokinetics and X-ray-Contrast-Imaging Studies. Small 2007, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Roque, A.C.; Bicho, A.; Batalha, I.L.; Cardoso, A.S.; Hussain, A. Biocompatible and bioactive gum Arabic coated iron oxide magnetic nanoparticles. J. Biotechnol. 2009, 144, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Chen, D.H. Facile green synthesis of gold nanoparticles with gum arabic as a stabilizing agent and reducing agent. Gold Bull. 2010, 43, 234–240. [Google Scholar] [CrossRef]
- Chen, X.; Li, R.; Liang, T.; Zhang, K.; Gao, Y.; Xu, L. Puerarin improves metabolic function leading to hepatoprotective effects in chronic alcohol-induced liver injury in rats. Phytomedicine 2013, 20, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zheng, N.; Liang, T.; He, Q.; Xu, L. Puerarin attenuates neuronal degeneration and blocks oxidative stress to elicit a neuroprotective effect on substantia nigra injury in 6-OHDA-lesioned rats. Brain Res. 2013, 1517, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shangguan, Z.; Liu, Y.; Wang, J.; Li, X.; Yang, S.; Liu, S. Puerarin protects pancreatic β-cell survival via PI3K/Akt signaling pathway. J. Mol. Endocrinol. 2014, 53, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Du, Y.Q.; Yuan, L.; Wang, N.N. Protective effect of puerarin on acute alcoholic liver injury. Am. J. Chin. Med. 2010, 38, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Leshner, A.I. Addiction is a brain disease, and it matters. Science 1997, 278, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Le Moal, M.; Koob, G.F. Drug addiction: Pathways to the disease and pathophysiological perspectives. Eur. Neuropsychopharmacol. 2007, 17, 377–393. [Google Scholar] [CrossRef] [PubMed]
- Terenius, L. Rational treatment of addiction. Cur. Opin. Chem. Biol. 1998, 2, 541–547. [Google Scholar] [CrossRef]
- Meletis, C.D.; Zabriskie, N. Natural approaches to treating addiction. Altern. Complement. Ther. 2008, 14, 275–281. [Google Scholar] [CrossRef]
- Ma, Z.Z.; Xu, W.; Jensen, N.H.; Roth, B.L.; Liu-Chen, L.Y.; Lee, D.Y. Isoquinoline alkaloids isolated from Corydalis yanhusuo and their binding affinities at the dopamine D1 receptor. Molecules 2008, 13, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Casals, E.; Pfaller, T.; Duschl, A.; Oostingh, G.J.; Puntes, V. Time evolution of the nanoparticle protein corona. ACS Nano 2010, 4, 3623–3632. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Lynch, I.; Ejtehadi, M.R.; Monopoli, M.P.; Bombelli, F.B.; Laurent, S. Protein-nanoparticle interactions: Opportunities and challenges. Chem. Rev. 2011, 111, 5610–5637. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, M.; Saffar, S.; Abd Emamy, B.; Peirovi, A.; Shokrgozar, M.A.; Serpooshan, V.; Mahmoudi, M. Plasma concentration gradient influences the protein corona decoration on nanoparticles. RSC Adv. 2013, 3, 1119–1126. [Google Scholar] [CrossRef]
- Nel, A.E.; Madler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Treuel, L.; Nienhaus, G.U. Toward a molecular understanding of nanoparticle-protein interactions. Biophys. Rev. 2012, 4, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Chan, W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggard, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Foy, M.; Berggad, T.; Donnelly, S.; Cagney, G.; Linse, S.; Dawson, K. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. 2007, 46, 5754–5756. [Google Scholar] [CrossRef] [PubMed]
- Landsiedel, R.; Ma-Hock, L.; Kroll, A.; Hahn, D.; Schnekenburger, J.; Wiench, K.; Wohlleben, W. Testing Metal-Oxide Nanomaterials for Human Safety. Adv. Mater. 2010, 22, 2601–2627. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Sethson, I.; Jonsson, B.-H. Protein Adsorption onto Silica Nanoparticles: Conformational Changes Depend on the Particles’ Curvature and the Protein Stability. Langmuir 2004, 20, 10639–10647. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Sethson, I.; Jonsson, B.-H. Transient. Interaction with Nanoparticles “Freezes” a Protein in an Ensemble of Metastable Near-Native Conformations. Biochemistry 2005, 44, 10093–10099. [Google Scholar] [CrossRef] [PubMed]



| Protein | Test: IS or MS: Sequence | m/z (+1) | m/z (+2) |
|---|---|---|---|
| Bin1 protein | MS: AEEELIK | 832.2 | 418.1 |
| Casein Kinase 1 γ2 | MS: MEYVHTK | 908.0 | 456.3 |
| cAMP/cGMP phosphodiesterase | MS: VOYHNWK | 944.4 | - |
| Glutaryl-Co-A dehydrogenase | MS: CEDNCIR | 853.3 | 439.1 |
| Arginine N-methyltransferase | MS: QYKDYK | 845.5 | 424.3 |
| Lactate dehydrogenase | MS: MVSGESR | 766.52 | 384.3 |
| MAPK kinase 7 | MS: LCDFGISGR | 968.7 | - |
| Parvalbumin | MS: FFQMVGLK | 970.5 | 486.4 |
| Protein Tyr Phosphatase | MS: MPVIVSR | 803.1 | - |
| Transferrin | MS: EDPQTFYY | 815.5 | 209.3 |
| Triose phosphate isomerase | MS: AISDNVK | 731.4 | - |
| NP | 0 | 5 | 10 | 20 | 40 | 60 | 100 |
|---|---|---|---|---|---|---|---|
| An/En cells (no AuNPs or ethanol) | |||||||
| - | An−PI− | - | - | - | - | - | - |
| Ap/En cells (AuNPs positive but ethanol negative) | |||||||
| AuNPKG-90,50,37 AuNPK-90,50,37 | - | An−PI− | An−PI− | An−PI− | An−PI− | An++PI− | An−PI− An++PI+ An+++PI+ |
| AuNPG-50,90 | - | An+PI− | An+PI− | An+PI− | An+PI− | An+PI− | An−PI− An+++PI− An+PI++ |
| AuNPPEG, PEG+K+G | - | An+PI− | An+PI− | Anl+PI− | An++PI− | An+++PI− An++PI− An+PI+ | An+PI− An+++PI+ An−PI++ |
| An/Ep cells (AuNPs negative, but ethanol positive) | |||||||
| An−PI+++ An++PI− An+PI++ An+++PI− | − | - | - | - | - | - | |
| Ap/Ep cells (AuNPs and ethanol positive) | |||||||
| AuNPKG-90,-50,-37 | - | An−PI− An++PI− An++PI+ | An++PI− | An++PI− | An++PI− | An+PI− An+PI+ An++PI+ | An−PI− An++PI+ An−PI++ |
| AuNPK-90,-50,-37 | - | An−PI− An++PI− An++PI+ | An−PI− An+PI+ | An+PI− An+PI++ | An+PI− An+PI++ | An−PI− An++PI+ | An−PI+ An++PI− An++PI++ |
| AuNPG-50,-90 | - | An−PI− An+PI++ | An+PI− An+PI++ | An+PI− An+PI+ | An+PI− An++PI+ An+PI+ | An+PI− An++PI+ An−PI+ | An−PI+ An++PI− An++PI+ |
| AuNPPEG,PEG+K+G | - | An++PI− | An++PI− An++PI+ | An++PI− An+PI++ | An++PI− An++PI+ An−PI++ | An++PI− An++PI+ An−PI++ | An−PI− An++PI− An++PI+ An−PI+++ |
| Peak cm−1 | Functional Group/Vibration Mode |
|---|---|
| 750 | Aromatic protons |
| 927 | C–O–C vibration modes of α-1,4 glycosidic linkage |
| 1040/1162 | Carbonyl (C=O) group, C–O stretching alcohols (primary, secondary and tertiary), carboxylic acids, esters, and ethers |
| 1020/1079 1116/1156 | C–O/C stretching in exo/endo-cyclic bonds and C–O–H/C deformation modes of oligo/polysaccharides and absorption bands for carbohydrates |
| 1383/1456 | The spectral bands for C–O–H stretching in pyranose and C–H deformation in esters |
| 1621/1637 | Aromatic C=C/C–OH stretching in exocyclic bond |
| 2923 | Glucoside ring due to the stretching vibration of O–H [44] |
| 3420 | O–H, as also the H-bonded alcohols and phenols, carboxylic acid |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.K. Comparative Therapeutic Effects of Plant-Extract Synthesized and Traditionally Synthesized Gold Nanoparticles on Alcohol-Induced Inflammatory Activity in SH-SY5Y Cells In Vitro. Biomedicines 2017, 5, 70. https://doi.org/10.3390/biomedicines5040070
Singh AK. Comparative Therapeutic Effects of Plant-Extract Synthesized and Traditionally Synthesized Gold Nanoparticles on Alcohol-Induced Inflammatory Activity in SH-SY5Y Cells In Vitro. Biomedicines. 2017; 5(4):70. https://doi.org/10.3390/biomedicines5040070
Chicago/Turabian StyleSingh, Ashok K. 2017. "Comparative Therapeutic Effects of Plant-Extract Synthesized and Traditionally Synthesized Gold Nanoparticles on Alcohol-Induced Inflammatory Activity in SH-SY5Y Cells In Vitro" Biomedicines 5, no. 4: 70. https://doi.org/10.3390/biomedicines5040070





