Controlling Nuclear NF-κB Dynamics by β-TrCP—Insights from a Computational Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Modelling Canonical NF-κB Signaling
2.2. Measures of Nuclear NF-κB Dynamics
2.3. Bifurcation Analysis
3. Results
3.1. Description of the Computational Model of Canonical NF-κB Signaling
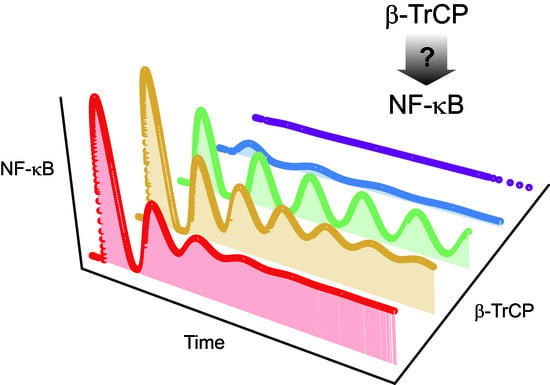
3.2. β-TrCP Abundance Influences the Transient Dynamics of Nuclear NF-κB upon TNF Stimulation
3.3. β-TrCP Abundance Affects Long-Term Dynamical Behavior of Nuclear NF-κB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hayden, M.S.; Ghosh, S. NF-kB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef]
- Bennett, J.; Capece, D.; Begalli, F.; Verzella, D.; D’Andrea, D.; Tornatore, L.; Franzoso, G. NF-kappaB in the crosshairs: Rethinking an old riddle. Int. J. Biochem. Cell Biol. 2018, 95, 108–112. [Google Scholar] [CrossRef]
- Begalli, F.; Bennett, J.; Capece, D.; Verzella, D.; D’Andrea, D.; Tornatore, L.; Franzoso, G. Unlocking the NF-kappaB Conundrum: Embracing Complexity to Achieve Specificity. Biomedicines 2017, 5, 50. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Carmody, R.J. NF-kappaB and the Transcriptional Control of Inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. [Google Scholar] [CrossRef]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef]
- Hinz, M.; Scheidereit, C. The IκB kinase complex in NF-κB regulation and beyond. EMBO Rep. 2014, 15, 46–61. [Google Scholar] [CrossRef]
- Staudt, L.M. Oncogenic Activation of NF-κB. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Müerköster, S.; Arlt, A.; Sipos, B.; Witt, M.; Großmann, M.; Klöppel, G.; Kalthoff, H.; Fölsch, U.R.; Schäfer, H. Increased Expression of the E3-Ubiquitin Ligase Receptor Subunit βTRCP1 Relates to Constitutive Nuclear Factor-κB Activation and Chemoresistance in Pancreatic Carcinoma Cells. Cancer Res. 2005, 65, 1316–1324. [Google Scholar] [CrossRef]
- Lau, A.W.; Fukushima, H.; Wei, W. The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front. Biosci. 2012, 17, 2197–2212. [Google Scholar] [CrossRef]
- Frankland-Searby, S.; Bhaumik, S.R. The 26S proteasome complex: An attractive target for cancer therapy. Biochim. Biophys. Acta 2012, 1825, 64–76. [Google Scholar] [CrossRef] [Green Version]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-kappaB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef]
- Wang, H.; Maitra, A.; Wang, H. The emerging roles of F-box proteins in pancreatic tumorigenesis. Semin. Cancer Biol. 2016, 36, 88–94. [Google Scholar] [CrossRef]
- Uddin, S.; Bhat, A.A.; Krishnankutty, R.; Mir, F.; Kulinski, M.; Mohammad, R.M. Involvement of F-BOX proteins in progression and development of human malignancies. Semin. Cancer Biol. 2016, 36, 18–32. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta 2010, 1799, 775–787. [Google Scholar] [CrossRef]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef]
- Gadina, M.; Gazaniga, N.; Vian, L.; Furumoto, Y. Small molecules to the rescue: Inhibition of cytokine signaling in immune-mediated diseases. J. Autoimmun. 2017, 85, 20–31. [Google Scholar] [CrossRef]
- Fuchs, S.Y.; Spiegelman, V.S.; Kumar, K.G. The many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene 2004, 23, 2028–2036. [Google Scholar] [CrossRef]
- Tang, W.; Li, Y.; Yu, D.; Thomas-Tikhonenko, A.; Spiegelman, V.S.; Fuchs, S.Y. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005, 65, 1904–1908. [Google Scholar] [CrossRef]
- Bhatia, N.; Herter, J.R.; Slaga, T.J.; Fuchs, S.Y.; Spiegelman, V.S. Mouse homologue of HOS (mHOS) is overexpressed in skin tumors and implicated in constitutive activation of NF-kappaB. Oncogene 2002, 21, 1501–1509. [Google Scholar] [CrossRef]
- Seo, E.; Kim, H.; Kim, R.; Yun, S.; Kim, M.; Han, J.K.; Costantini, F.; Jho, E.H. Multiple isoforms of beta-TrCP display differential activities in the regulation of Wnt signaling. Cell. Signal. 2009, 21, 43–51. [Google Scholar] [CrossRef]
- Fuchs, S.Y.; Chen, A.; Xiong, Y.; Pan, Z.Q.; Ronai, Z. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IkappaB and beta-catenin. Oncogene 1999, 18, 2039–2046. [Google Scholar] [CrossRef]
- Shi, M.; Cho, H.; Inn, K.S.; Yang, A.; Zhao, Z.; Liang, Q.; Versteeg, G.A.; Amini-Bavil-Olyaee, S.; Wong, L.Y.; Zlokovic, B.V.; et al. Negative regulation of NF-kappaB activity by brain-specific TRIpartite Motif protein 9. Nat. Commun. 2014, 5, 4820. [Google Scholar] [CrossRef]
- Guardavaccaro, D.; Kudo, Y.; Boulaire, J.; Barchi, M.; Busino, L.; Donzelli, M.; Margottin-Goguet, F.; Jackson, P.K.; Yamasaki, L.; Pagano, M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev. Cell 2003, 4, 799–812. [Google Scholar] [CrossRef]
- Nakayama, K.; Hatakeyama, S.; Maruyama, S.; Kikuchi, A.; Onoe, K.; Good, R.A.; Nakayama, K.I. Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 8752–8757. [Google Scholar] [CrossRef]
- Spiegelman, V.S.; Slaga, T.J.; Pagano, M.; Minamoto, T.; Ronai, Z.; Fuchs, S.Y. Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol. Cell 2000, 5, 877–882. [Google Scholar] [CrossRef]
- Benary, U.; Kofahl, B.; Hecht, A.; Wolf, J. Mathematical modelling suggests a differential impact of beta-transducin repeat-containing protein paralogues on Wnt/beta-catenin signalling dynamics. FEBS J. 2015, 282, 1080–1096. [Google Scholar] [CrossRef]
- Zhang, M.; Yan, Y.; Lim, Y.B.; Tang, D.; Xie, R.; Chen, A.; Tai, P.; Harris, S.E.; Xing, L.; Qin, Y.X.; et al. BMP-2 modulates beta-catenin signaling through stimulation of Lrp5 expression and inhibition of beta-TrCP expression in osteoblasts. J. Cell Biochem. 2009, 108, 896–905. [Google Scholar] [CrossRef]
- Spiegelman, V.S.; Tang, W.; Chan, A.M.; Igarashi, M.; Aaronson, S.A.; Sassoon, D.A.; Katoh, M.; Slaga, T.J.; Fuchs, S.Y. Induction of homologue of Slimb ubiquitin ligase receptor by mitogen signaling. J. Biol. Chem. 2002, 277, 36624–36630. [Google Scholar] [CrossRef]
- Shanzer, M.; Adler, J.; Ricardo-Lax, I.; Reuven, N.; Shaul, Y. The nonreceptor tyrosine kinase c-Src attenuates SCF(beta-TrCP) E3-ligase activity abrogating Taz proteasomal degradation. Proc. Natl. Acad. Sci. USA 2017, 114, 1678–1683. [Google Scholar] [CrossRef]
- Spiegelman, V.S.; Stavropoulos, P.; Latres, E.; Pagano, M.; Ronai, Z.; Slaga, T.J.; Fuchs, S.Y. Induction of beta-transducin repeat-containing protein by JNK signaling and its role in the activation of NF-kappaB. J. Biol. Chem. 2001, 276, 27152–27158. [Google Scholar] [CrossRef]
- Schmidt, M.L.; Donninger, H.; Clark, G.J. Ras regulates SCF(beta-TrCP) protein activity and specificity via its effector protein NORE1A. J. Biol. Chem. 2014, 289, 31102–31110. [Google Scholar] [CrossRef]
- Besnard-Guerin, C.; Belaidouni, N.; Lassot, I.; Segeral, E.; Jobart, A.; Marchal, C.; Benarous, R. HIV-1 Vpu sequesters beta-transducin repeat-containing protein (betaTrCP) in the cytoplasm and provokes the accumulation of beta-catenin and other SCFbetaTrCP substrates. J. Biol. Chem. 2004, 279, 788–795. [Google Scholar] [CrossRef]
- Hoffmann, A.; Levchenko, A.; Scott, M.L.; Baltimore, D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science 2002, 298, 1241–1245. [Google Scholar] [CrossRef]
- Lipniacki, T.; Paszek, P.; Brasier, A.R.; Luxon, B.; Kimmel, M. Mathematical model of NF-kappaB regulatory module. J. Theor. Biol. 2004, 228, 195–215. [Google Scholar] [CrossRef]
- Lipniacki, T.; Kimmel, M. Deterministic and stochastic models of NFkappaB pathway. Cardiovasc. Toxicol. 2007, 7, 215–234. [Google Scholar] [CrossRef]
- Cheong, R.; Hoffmann, A.; Levchenko, A. Understanding NF-kappaB signaling via mathematical modeling. Mol. Syst. Biol. 2008, 4, 192. [Google Scholar] [CrossRef]
- Werner, S.L.; Kearns, J.D.; Zadorozhnaya, V.; Lynch, C.; O’Dea, E.; Boldin, M.P.; Ma, A.; Baltimore, D.; Hoffmann, A. Encoding NF-kappaB temporal control in response to TNF: Distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008, 22, 2093–2101. [Google Scholar] [CrossRef]
- Basak, S.; Behar, M.; Hoffmann, A. Lessons from mathematically modeling the NF-kappaB pathway. Immunol. Rev. 2012, 246, 221–238. [Google Scholar] [CrossRef]
- Longo, D.M.; Selimkhanov, J.; Kearns, J.D.; Hasty, J.; Hoffmann, A.; Tsimring, L.S. Dual delayed feedback provides sensitivity and robustness to the NF-kappaB signaling module. PLoS Comput. Biol. 2013, 9, e1003112. [Google Scholar] [CrossRef]
- Zambrano, S.; Bianchi, M.E.; Agresti, A. A simple model of NF-kappaB dynamics reproduces experimental observations. J. Theor. Biol. 2014, 347C, 44–53. [Google Scholar] [CrossRef]
- Williams, R.; Timmis, J.; Qwarnstrom, E. Computational Models of the NF-KB Signalling Pathway. Computation 2014, 2, 131–158. [Google Scholar] [CrossRef] [Green Version]
- Fagerlund, R.; Behar, M.; Fortmann, K.T.; Lin, Y.E.; Vargas, J.D.; Hoffmann, A. Anatomy of a negative feedback loop: The case of IkappaBalpha. J. R. Soc. Interface 2015, 12, 0262. [Google Scholar] [CrossRef]
- Mothes, J.; Busse, D.; Kofahl, B.; Wolf, J. Sources of dynamic variability in NF-κB signal transduction: A mechanistic model. Bioessays 2015, 37, 452–462. [Google Scholar] [CrossRef]
- Inoue, K.; Shinohara, H.; Behar, M.; Yumoto, N.; Tanaka, G.; Hoffmann, A.; Aihara, K.; Okada-Hatakeyama, M. Oscillation dynamics underlie functional switching of NF-kappaB for B-cell activation. NPJ Syst. Biol. Appl. 2016, 2, 16024. [Google Scholar] [CrossRef]
- Adamson, A.; Boddington, C.; Downton, P.; Rowe, W.; Bagnall, J.; Lam, C.; Maya-Mendoza, A.; Schmidt, L.; Harper, C.V.; Spiller, D.G.; et al. Signal transduction controls heterogeneous NF-kappaB dynamics and target gene expression through cytokine-specific refractory states. Nat. Commun. 2016, 7, 12057. [Google Scholar] [CrossRef]
- Harper, C.V.; Woodcock, D.J.; Lam, C.; Garcia-Albornoz, M.; Adamson, A.; Ashall, L.; Rowe, W.; Downton, P.; Schmidt, L.; West, S.; et al. Temperature regulates NF-kappaB dynamics and function through timing of A20 transcription. Proc. Natl. Acad. Sci. USA 2018, 115, E5243–E5249. [Google Scholar] [CrossRef]
- Yilmaz, Z.B.; Kofahl, B.; Beaudette, P.; Baum, K.; Ipenberg, I.; Weih, F.; Wolf, J.; Dittmar, G.; Scheidereit, C. Quantitative dissection and modeling of the NF-kappaB p100-p105 module reveals interdependent precursor proteolysis. Cell Rep. 2014, 9, 1756–1769. [Google Scholar] [CrossRef]
- Sung, M.H.; Simon, R. In silico simulation of inhibitor drug effects on nuclear factor-kappaB pathway dynamics. Mol. Pharmacol. 2004, 66, 70–75. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, B.; Li, S.; Zhao, Q. A formal model for analyzing drug combination effects and its application in TNF-alpha-induced NFkappaB pathway. BMC Syst. Biol. 2010, 4, 50. [Google Scholar] [CrossRef]
- Peng, H.; Wen, J.; Li, H.; Chang, J.; Zhou, X. Drug inhibition profile prediction for NFkappaB pathway in multiple myeloma. PLoS ONE 2011, 6, e14750. [Google Scholar] [CrossRef]
- Behar, M.; Barken, D.; Werner, S.L.; Hoffmann, A. The dynamics of signaling as a pharmacological target. Cell 2013, 155, 448–461. [Google Scholar] [CrossRef]
- Klipp, E. Timing matters. FEBS Lett. 2009, 583, 4013–4018. [Google Scholar] [CrossRef] [Green Version]
- Llorens, M.; Nuno, J.C.; Rodriguez, Y.; Melendez-Hevia, E.; Montero, F. Generalization of the theory of transition times in metabolic pathways: A geometrical approach. Biophys. J. 1999, 77, 23–36. [Google Scholar] [CrossRef]
- Heinrich, R.; Neel, B.G.; Rapoport, T.A. Mathematical models of protein kinase signal transduction. Mol. Cell 2002, 9, 957–970. [Google Scholar] [CrossRef]
- Kofahl, B.; Klipp, E. Modelling the dynamics of the yeast pheromone pathway. Yeast 2004, 21, 831–850. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.E.; Walker, S.R.; Savery, K.; Frank, D.A.; Gaudet, S. Fold Change of Nuclear NF-kappaB Determines TNF-Induced Transcription in Single Cells. Mol. Cell 2014. [Google Scholar] [CrossRef]
- Heinrich, R.; Schuster, S. The Regulation of Cellular Systems; Chapman & Hall: New York, NY, USA, 1996. [Google Scholar]
- Kroll, M.; Margottin, F.; Kohl, A.; Renard, P.; Durand, H.; Concordet, J.P.; Bachelerie, F.; Arenzana-Seisdedos, F.; Benarous, R. Inducible degradation of IkappaBalpha by the proteasome requires interaction with the F-box protein h-betaTrCP. J. Biol. Chem. 1999, 274, 7941–7945. [Google Scholar] [CrossRef]
- Spencer, E.; Jiang, J.; Chen, Z.J. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999, 13, 284–294. [Google Scholar] [CrossRef]
- Wang, X.; Adhikari, N.; Li, Q.; Guan, Z.; Hall, J.L. The role of [beta]-transducin repeat-containing protein ([beta]-TrCP) in the regulation of NF-[kappa]B in vascular smooth muscle cells. Arterioscler. Thromb Vasc. Biol. 2004, 24, 85–90. [Google Scholar] [CrossRef]
- Colombo, F.; Zambrano, S.; Agresti, A. NF-kappaB, the Importance of Being Dynamic: Role and Insights in Cancer. Biomedicines 2018, 6, 45. [Google Scholar] [CrossRef]
- Nelson, D.E.; Ihekwaba, A.E.C.; Elliott, M.; Johnson, J.R.; Gibney, C.A.; Foreman, B.E.; Nelson, G.; See, V.; Horton, C.A.; Spiller, D.G.; et al. Oscillations in NF-kB Signaling Control the Dynamics of Gene Expression. Science 2004, 306, 704–708. [Google Scholar] [CrossRef]
- Tay, S.; Hughey, J.J.; Lee, T.K.; Lipniacki, T.; Quake, S.R.; Covert, M.W. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 2010. [Google Scholar] [CrossRef]
- Kardynska, M.; Paszek, A.; Smieja, J.; Spiller, D.; Widlak, W.; White, M.R.H.; Paszek, P.; Kimmel, M. Quantitative analysis reveals crosstalk mechanisms of heat shock-induced attenuation of NF-kappaB signaling at the single cell level. PLoS Comput. Biol. 2018, 14, e1006130. [Google Scholar] [CrossRef]
- Ashall, L.; Horton, C.A.; Nelson, D.E.; Paszek, P.; Harper, C.V.; Sillitoe, K.; Ryan, S.; Spiller, D.G.; Unitt, J.F.; Broomhead, D.S.; et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science 2009, 324, 242–246. [Google Scholar] [CrossRef]
- Joo, J.; Plimpton, S.J.; Faulon, J.L. Statistical ensemble analysis for simulating extrinsic noise-driven response in NF-kappaB signaling networks. BMC Syst. Biol. 2013, 7, 45. [Google Scholar] [CrossRef]
- Sung, M.H.; Li, N.; Lao, Q.; Gottschalk, R.A.; Hager, G.L.; Fraser, I.D. Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-kappaB signaling. Sci. Signal. 2014, 7, ra6. [Google Scholar] [CrossRef]
- Zhang, Q.; Gupta, S.; Schipper, D.L.; Kowalczyk, G.J.; Mancini, A.E.; Faeder, J.R.; Lee, R.E.C. NF-kappaB Dynamics Discriminate between TNF Doses in Single Cells. Cell Syst. 2017, 5, 638–645 e635. [Google Scholar] [CrossRef]
- Wong, V.C.; Bass, V.L.; Bullock, M.E.; Chavali, A.K.; Lee, R.E.C.; Mothes, W.; Gaudet, S.; Miller-Jensen, K. NF-kappaB-Chromatin Interactions Drive Diverse Phenotypes by Modulating Transcriptional Noise. Cell Rep. 2018, 22, 585–599. [Google Scholar] [CrossRef]
- Novak, B.; Tyson, J.J. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell. Biol. 2008, 9, 981–991. [Google Scholar] [CrossRef] [Green Version]
- Ananthasubramaniam, B.; Herzel, H. Positive feedback promotes oscillations in negative feedback loops. PLoS ONE 2014, 9, e104761. [Google Scholar] [CrossRef]
- Baum, K.; Politi, A.Z.; Kofahl, B.; Steuer, R.; Wolf, J. Feedback, Mass Conservation and Reaction Kinetics Impact the Robustness of Cellular Oscillations. PLoS Comput. Biol. 2016, 12, e1005298. [Google Scholar] [CrossRef]
- Sung, M.H.; Salvatore, L.; De Lorenzi, R.; Indrawan, A.; Pasparakis, M.; Hager, G.L.; Bianchi, M.E.; Agresti, A. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS ONE 2009, 4, e7163. [Google Scholar] [CrossRef]
- Zambrano, S.; Bianchi, M.E.; Agresti, A. High-throughput analysis of NF-kappaB dynamics in single cells reveals basal nuclear localization of NF-kappaB and spontaneous activation of oscillations. PLoS ONE 2014, 9, e90104. [Google Scholar] [CrossRef]
- Hughey, J.J.; Gutschow, M.V.; Bajar, B.T.; Covert, M.W. Single-cell variation leads to population invariance in NF-kappaB signaling dynamics. Mol. Biol. Cell 2015, 26, 583–590. [Google Scholar] [CrossRef]
- Barken, D.; Wang, C.J.; Kearns, J.; Cheong, R.; Hoffmann, A.; Levchenko, A. Comment on "Oscillations in NF-kappaB Signaling Control the Dynamics of Gene Expression". Science 2005, 308, 52. [Google Scholar] [CrossRef]
- Nelson, D.E.; Horton, C.A.; See, V.; Johnson, J.R.; Nelson, G.; Spiller, D.G.; Kell, D.B.; White, M.R.H. Response to Comment on "Oscillations in NF-κB Signaling Control the Dynamics of Gene Expression". Science 2005, 308, 52. [Google Scholar] [CrossRef]
- Turner, D.A.; Paszek, P.; Woodcock, D.J.; Nelson, D.E.; Horton, C.A.; Wang, Y.; Spiller, D.G.; Rand, D.A.; White, M.R.; Harper, C.V. Physiological levels of TNFalpha stimulation induce stochastic dynamics of NF-kappaB responses in single living cells. J. Cell Sci. 2010, 123, 2834–2843. [Google Scholar] [CrossRef]
- Kellogg, R.A.; Tay, S. Noise facilitates transcriptional control under dynamic inputs. Cell 2015, 160, 381–392. [Google Scholar] [CrossRef]
- Zambrano, S.; De Toma, I.; Piffer, A.; Bianchi, M.E.; Agresti, A. NF-kappaB oscillations translate into functionally related patterns of gene expression. Elife 2016, 5, e09100. [Google Scholar] [CrossRef]
- Paszek, P.; Jackson, D.A.; White, M.R. Oscillatory control of signalling molecules. Curr. Opin. Genet. Dev. 2010, 20, 670–676. [Google Scholar] [CrossRef]
- Winston, J.T.; Strack, P.; Beer-Romero, P.; Chu, C.Y.; Elledge, S.J.; Harper, J.W. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999, 13, 270–283. [Google Scholar] [CrossRef]
- Low, T.Y.; Peng, M.; Magliozzi, R.; Mohammed, S.; Guardavaccaro, D.; R. Heck, A.J. A systems-wide screen identifies substrates of the SCFβTrCP ubiquitin ligase. Sci. Signal. 2014, 7, rs8. [Google Scholar] [CrossRef]
- Deng, J.; Miller, S.A.; Wang, H.-Y.; Xia, W.; Wen, Y.; Zhou, B.P.; Li, Y.; Lin, S.-Y.; Hung, M.-C. beta-catenin interacts with and inhibits NF-kappaB in human colon and breast cancer. Cancer Cell 2002, 2, 323–334. [Google Scholar] [CrossRef]
- Sun, J.; Hobert, M.E.; Duan, Y.; Rao, A.S.; He, T.C.; Chang, E.B.; Madara, J.L. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am. J. Physiol. Gastrointest Liver Physiol. 2005, 289, G129–G137. [Google Scholar] [CrossRef]
- Duan, Y.; Liao, A.P.; Kuppireddi, S.; Ye, Z.; Ciancio, M.J.; Sun, J. beta-Catenin activity negatively regulates bacteria-induced inflammation. Lab. Investig. 2007, 87, 613–624. [Google Scholar] [CrossRef]
- Nejak-Bowen, K.; Kikuchi, A.; Monga, S.P.S. Beta-catenin-NF-κB interactions in murine hepatocytes: A complex to die for. Hepatology 2012. [Google Scholar] [CrossRef]
- Nakajima, H.; Fujiwara, H.; Furuichi, Y.; Tanaka, K.; Shimbara, N. A novel small-molecule inhibitor of NF-κB signaling. Biochem. Biophys. Res. Commun. 2008, 368, 1007–1013. [Google Scholar] [CrossRef]
- Liu, Y.; Mallampalli, R.K. Small molecule therapeutics targeting F-box proteins in cancer. Semin. Cancer Biol. 2016, 36, 105–119. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benary, U.; Wolf, J. Controlling Nuclear NF-κB Dynamics by β-TrCP—Insights from a Computational Model. Biomedicines 2019, 7, 40. https://doi.org/10.3390/biomedicines7020040
Benary U, Wolf J. Controlling Nuclear NF-κB Dynamics by β-TrCP—Insights from a Computational Model. Biomedicines. 2019; 7(2):40. https://doi.org/10.3390/biomedicines7020040
Chicago/Turabian StyleBenary, Uwe, and Jana Wolf. 2019. "Controlling Nuclear NF-κB Dynamics by β-TrCP—Insights from a Computational Model" Biomedicines 7, no. 2: 40. https://doi.org/10.3390/biomedicines7020040