LRP-1 Matricellular Receptor Involvement in Triple Negative Breast Cancer Tumor Angiogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. Tumor- and HUVEC-Conditioned Media Preparation
2.3. In Vivo Studies
2.4. Orthotopic Xenograft Model
2.5. Matrigel® Plug
2.6. Optical Imaging
2.7. MRI Imaging
2.8. Chick Chorioallantoic Membrane (CAM) Assay
2.9. Histology
2.10. Immunohistochemistry and Immunofluorescence
2.11. Protein Extraction and Western Blot (WB) Analysis
2.12. RNA Isolation and Real-Time PCR
2.13. Tubule Formation
2.14. Endothelial Proliferation and Migration
2.15. Proteomics
2.16. S-2251TM Assay
2.17. Patient Tumor-Derived Breast Cancer Xenografts (PDX)
2.18. RNA Seq
2.19. Statistical Analysis
3. Results
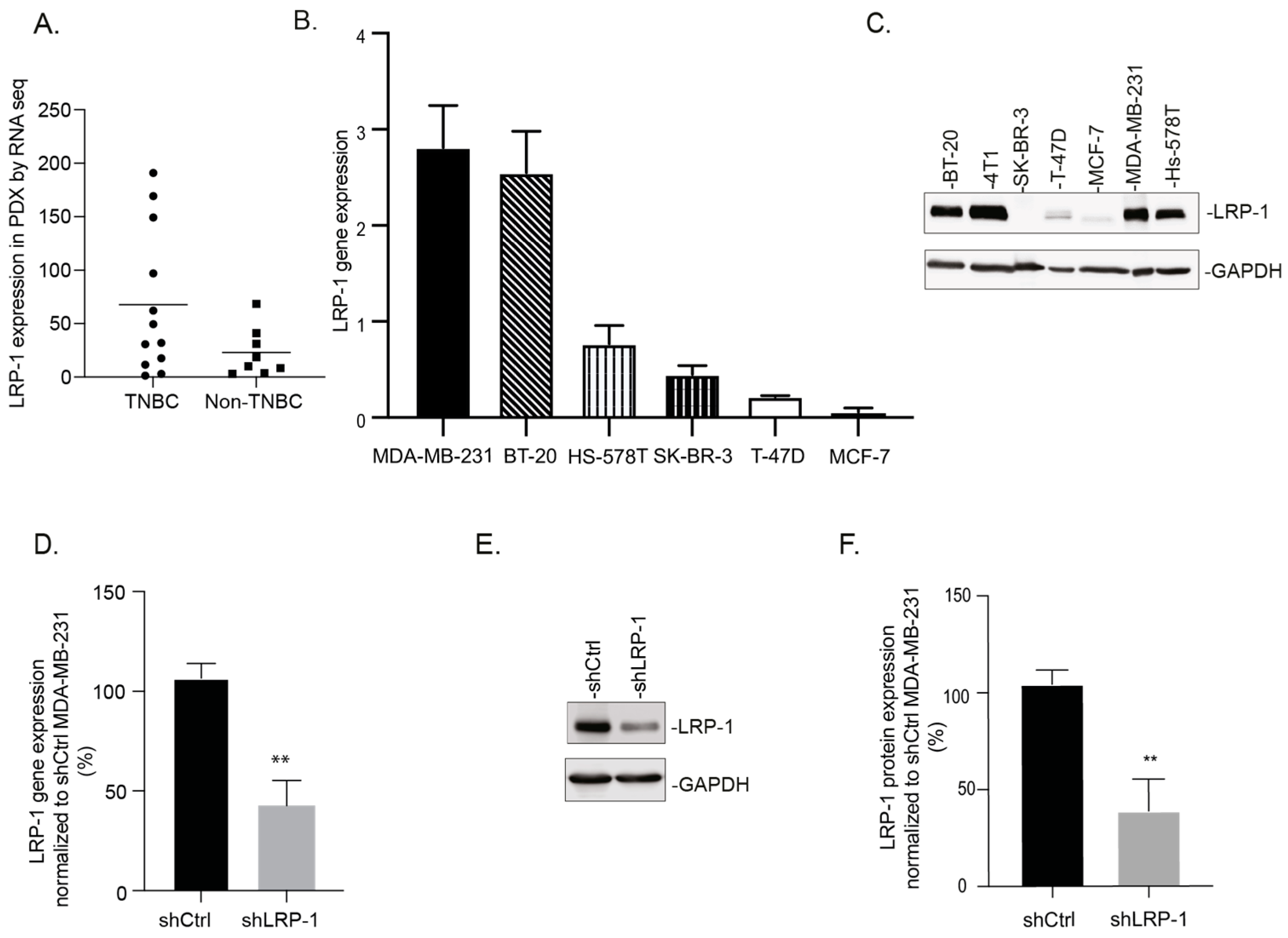
3.1. LRP-1 Is Preferentially Expressed in TNBC Cell Lines
3.2. LRP-1 Acts as a Pro-Tumorigenic Receptor, by Modulating Tumor Angiogenesis, in an Orthotopic Mammary Fat Pad TNBC Model
3.3. LRP-1 Repression Alters Angiogenesis in MDA-MB-231 Matrigel® Plugs and CAMs Assays
3.4. LRP-1-Down-Regulated MDA-MB-231 Secretome Modulates the Angiogenic Potential of Endothelial Cells
3.5. MDA-MB-231 Secretome Analysis Reveals That LRP-1 Angiogenic Effects Involved TGF-β and Plasminogen/Plasmin Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [Green Version]
- Metzger-Filho, O.; Tutt, A.; de Azambuja, E.; Saini, K.S.; Viale, G.; Loi, S.; Bradbury, I.; Bliss, J.M.; Azim, H.A., Jr.; Ellis, P.; et al. Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 2012, 30, 1879–1887. [Google Scholar] [CrossRef] [Green Version]
- Cleator, S.; Heller, W.; Coombes, R.C. Triple-negative breast cancer: Therapeutic options. Lancet Oncol. 2007, 8, 235–244. [Google Scholar] [CrossRef]
- Catalano, V.; Turdo, A.; Di Franco, S.; Dieli, F.; Todaro, M.; Stassi, G. Tumor and its microenvironment: A synergistic interplay. Semin. Cancer Biol. 2013, 23, 522–532. [Google Scholar] [CrossRef]
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: Zeb1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Alluri, P.; Newman, L.A. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef] [Green Version]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J.; Hanahan, D. Switch to the angiogenic phenotype during tumorigenesis. Princess Takamatsu Symp. 1991, 22, 339–347. [Google Scholar] [PubMed]
- Weis, S.M.; Cheresh, D.A. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat. Med. 2011, 17, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [Green Version]
- Lillis, A.P.; Van Duyn, L.B.; Murphy-Ullrich, J.E.; Strickland, D.K. Ldl receptor-related protein 1: Unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 2008, 88, 887–918. [Google Scholar] [CrossRef] [Green Version]
- Emonard, H.; Theret, L.; Bennasroune, A.H.; Dedieu, S. Regulation of lrp-1 expression: Make the point. Pathol. Biol. (Paris) 2014, 62, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Strickland, D.K. Lrp: A multifunctional scavenger and signaling receptor. J. Clin. Investig. 2001, 108, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Theret, L.; Jeanne, A.; Langlois, B.; Hachet, C.; David, M.; Khrestchatisky, M.; Devy, J.; Herve, E.; Almagro, S.; Dedieu, S. Identification of lrp-1 as an endocytosis and recycling receptor for beta1-integrin in thyroid cancer cells. Oncotarget 2017, 8, 78614–78632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langlois, B.; Perrot, G.; Schneider, C.; Henriet, P.; Emonard, H.; Martiny, L.; Dedieu, S. Lrp-1 promotes cancer cell invasion by supporting erk and inhibiting jnk signaling pathways. PLoS ONE 2010, 5, e11584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catasus, L.; Gallardo, A.; Llorente-Cortes, V.; Escuin, D.; Munoz, J.; Tibau, A.; Peiro, G.; Barnadas, A.; Lerma, E. Low-density lipoprotein receptor-related protein 1 is associated with proliferation and invasiveness in her-2/neu and triple-negative breast carcinomas. Hum. Pathol. 2011, 42, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Dedieu, S.; Langlois, B. Lrp-1: A new modulator of cytoskeleton dynamics and adhesive complex turnover in cancer cells. Cell Adh. Migr. 2008, 2, 77–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berquand, A.; Meunier, M.; Thevenard-Devy, J.; Ivaldi, C.; Campion, O.; Dedieu, S.; Molinari, M.; Devy, J. A gentle approach to investigate the influence of lrp-1 silencing on the migratory behavior of breast cancer cells by atomic force microscopy and dynamic cell studies. Nanomedicine 2019, 18, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Campion, O.; Al Khalifa, T.; Langlois, B.; Thevenard-Devy, J.; Salesse, S.; Savary, K.; Schneider, C.; Etique, N.; Dedieu, S.; Devy, J. Contribution of the low-density lipoprotein receptor family to breast cancer progression. Front. Oncol. 2020, 10, 882. [Google Scholar] [CrossRef]
- Bikfalvi, A. Tumoral angiogenesis: Models, targets and inhibition. J. Soc. Biol. 2009, 203, 167–170. [Google Scholar] [CrossRef] [PubMed]
- El Alaoui-Lasmaili, K.; Djermoune, E.H.; Tylcz, J.B.; Meng, D.; Plenat, F.; Thomas, N.; Faivre, B. A new algorithm for a better characterization and timing of the anti-vegf vascular effect named “normalization”. Angiogenesis 2017, 20, 149–162. [Google Scholar] [CrossRef]
- Jerman, T.; Pernus, F.; Likar, B.; Spiclin, Z. Enhancement of vascular structures in 3d and 2d angiographic images. IEEE Trans. Med. Imaging 2016, 35, 2107–2118. [Google Scholar] [CrossRef]
- Montoya-Zegarra, J.A.; Russo, E.; Runge, P.; Jadhav, M.; Willrodt, A.H.; Stoma, S.; Norrelykke, S.F.; Detmar, M.; Halin, C. Autotube: A novel software for the automated morphometric analysis of vascular networks in tissues. Angiogenesis 2019, 22, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Fuselier, C.; Quemener, S.; Dufay, E.; Bour, C.; Boulagnon-Rombi, C.; Bouland, N.; Djermoune, E.H.; Devy, J.; Martiny, L.; Schneider, C. Anti-tumoral and anti-angiogenic effects of low-diluted phenacetinum on melanoma. Front. Oncol. 2021, 11, 597503. [Google Scholar] [CrossRef]
- Alexander, R.A.; Prager, G.W.; Mihaly-Bison, J.; Uhrin, P.; Sunzenauer, S.; Binder, B.R.; Schutz, G.J.; Freissmuth, M.; Breuss, J.M. Vegf-induced endothelial cell migration requires urokinase receptor (upar)-dependent integrin redistribution. Cardiovasc. Res. 2012, 94, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Henriet, E.; Abou Hammoud, A.; Dupuy, J.W.; Dartigues, B.; Ezzoukry, Z.; Dugot-Senant, N.; Leste-Lasserre, T.; Pallares-Lupon, N.; Nikolski, M.; Le Bail, B.; et al. Argininosuccinate synthase 1 (ass1): A marker of unclassified hepatocellular adenoma and high bleeding risk. Hepatology 2017, 66, 2016–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; Garcia-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The proteomexchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef] [Green Version]
- Marangoni, E.; Vincent-Salomon, A.; Auger, N.; Degeorges, A.; Assayag, F.; de Cremoux, P.; de Plater, L.; Guyader, C.; De Pinieux, G.; Judde, J.G.; et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 2007, 13, 3989–3998. [Google Scholar] [CrossRef] [Green Version]
- de Gonzalo-Calvo, D.; Lopez-Vilaro, L.; Nasarre, L.; Perez-Olabarria, M.; Vazquez, T.; Escuin, D.; Badimon, L.; Barnadas, A.; Lerma, E.; Llorente-Cortes, V. Intratumor cholesteryl ester accumulation is associated with human breast cancer proliferation and aggressive potential: A molecular and clinicopathological study. BMC Cancer 2015, 15, 460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, L.A.; Hegg, R.; Freitas, F.R.; Tavares, E.R.; Almeida, C.P.; Baracat, E.C.; Maranhao, R.C. Effect of neoadjuvant chemotherapy on low-density lipoprotein (ldl) receptor and ldl receptor-related protein 1 (lrp-1) receptor in locally advanced breast cancer. Braz. J. Med. Biol. Res. 2012, 45, 557–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Santos, C.R.; Domingues, G.; Matias, I.; Matos, J.; Fonseca, I.; de Almeida, J.M.; Dias, S. Ldl-cholesterol signaling induces breast cancer proliferation and invasion. Lipids Health Dis. 2014, 13, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cedo, L.; Reddy, S.T.; Mato, E.; Blanco-Vaca, F.; Escola-Gil, J.C. Hdl and ldl: Potential new players in breast cancer development. J. Clin. Med. 2019, 8, 853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.S.; Kim, J.; Lee, H.J.; Kwon, B.M.; Lee, D.K.; Hong, S.H. LRP1-dependent pepsin clearance induced by 2′-hydroxycinnamaldehyde attenuates breast cancer cell invasion. Int. J. Biochem. Cell Biol. 2014, 53, 15–23. [Google Scholar] [CrossRef]
- Dv, S. A Concurrent Comparison of the Epidemiology and Clinical Presentation of Patients Hospitalized with Pandemic 2009 (H1N1) Influenza and Seasonal Influenza-A in Sub-himalayan Region of Himachal Pradesh. J. Assoc. Physicians India 2019, 67, 70–74. [Google Scholar]
- Strickland, D.K.; Muratoglu, S.C. Lrp in endothelial cells: A little goes a long way. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.; Xie, L.; Pi, X. Low-density lipoprotein receptor-related protein-1 signaling in angiogenesis. Front. Cardiovasc. Med. 2017, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- El Asmar, Z.; Terrand, J.; Jenty, M.; Host, L.; Mlih, M.; Zerr, A.; Justiniano, H.; Matz, R.L.; Boudier, C.; Scholler, E.; et al. Convergent signaling pathways controlled by lrp1 (receptor-related protein 1) cytoplasmic and extracellular domains limit cellular cholesterol accumulation. J. Biol. Chem. 2016, 291, 5116–5127. [Google Scholar] [CrossRef] [Green Version]
- Njah, K.; Chakraborty, S.; Qiu, B.; Arumugam, S.; Raju, A.; Pobbati, A.V.; Lakshmanan, M.; Tergaonkar, V.; Thibault, G.; Wang, X.; et al. A role of agrin in maintaining the stability of vascular endothelial growth factor receptor-2 during tumor angiogenesis. Cell Rep. 2019, 28, 949–965.e947. [Google Scholar] [CrossRef] [Green Version]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent multi-mirna targeting of apoe drives lrp1/lrp8-dependent melanoma metastasis and angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.W.; Lo, Y.H.; Chen, C.H.; Lin, C.Y.; Tsai, C.H.; Chen, P.J.; Yang, Y.F.; Wang, C.H.; Tan, C.H.; Hou, M.F.; et al. Vldl and ldl, but not hdl, promote breast cancer cell proliferation, metastasis and angiogenesis. Cancer Lett. 2017, 388, 130–138. [Google Scholar] [CrossRef]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Safina, D.; Schlitt, F.; Romeo, R.; Pflanzner, T.; Pietrzik, C.U.; Narayanaswami, V.; Edenhofer, F.; Faissner, A. Low-density lipoprotein receptor-related protein 1 is a novel modulator of radial glia stem cell proliferation, survival, and differentiation. Glia 2016, 64, 1363–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Türkbey, B.; Thomasson, D.; Pang, Y.; Bernardo, M.; Choyke, P.L. The role of dynamic contrast-enhanced mri in cancer diagnosis and treatment. Diagn. Interv. Radiol. 2010, 16, 186. [Google Scholar] [PubMed] [Green Version]
- Onda, N.; Kemmochi, S.; Morita, R.; Ishihara, Y.; Shibutani, M. In vivo imaging of tissue-remodeling activity involving infiltration of macrophages by a systemically administered protease-activatable probe in colon cancer tissues. Transl. Oncol. 2013, 6, 628–637. [Google Scholar] [CrossRef] [Green Version]
- Montel, V.; Gaultier, A.; Lester, R.D.; Campana, W.M.; Gonias, S.L. The low-density lipoprotein receptor-related protein regulates cancer cell survival and metastasis development. Cancer Res. 2007, 67, 9817–9824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Zhang, S.; Gong, Z.; Li, X.; Cao, K.; Deng, H.; He, Y.; et al. The role of microenvironment in tumor angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 204. [Google Scholar] [CrossRef]
- Qutub, A.A.; Popel, A.S. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst. Biol. 2009, 3, 13. [Google Scholar]
- Goulet, C.R.; Pouliot, F. Tgfbeta signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 2021, 1270, 89–105. [Google Scholar] [PubMed]
- Moore-Smith, L.; Pasche, B. Tgfbr1 signaling and breast cancer. J. Mammary Gland. Biol. Neoplasia 2011, 16, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albo, D.; Berger, D.H.; Wang, T.N.; Hu, X.; Rothman, V.; Tuszynski, G.P. Thrombospondin-1 and transforming growth factor-beta l promote breast tumor cell invasion through up-regulation of the plasminogen/plasmin system. Surgery 1997, 122, 493–499, discussion 499–500. [Google Scholar] [CrossRef]
- Ren, B.; Yee, K.O.; Lawler, J.; Khosravi-Far, R. Regulation of tumor angiogenesis by thrombospondin-1. Biochim. Biophys. Acta 2006, 1765, 178–188. [Google Scholar] [CrossRef]
- Gupta, K.; Kshirsagar, S.; Li, W.; Gui, L.; Ramakrishnan, S.; Gupta, P.; Law, P.Y.; Hebbel, R.P. Vegf prevents apoptosis of human microvascular endothelial cells via opposing effects on mapk/erk and sapk/jnk signaling. Exp. Cell Res. 1999, 247, 495–504. [Google Scholar] [CrossRef]
- Greenaway, J.; Lawler, J.; Moorehead, R.; Bornstein, P.; Lamarre, J.; Petrik, J. Thrombospondin-1 inhibits vegf levels in the ovary directly by binding and internalization via the low density lipoprotein receptor-related protein-1 (lrp-1). J. Cell Physiol. 2007, 210, 807–818. [Google Scholar] [CrossRef] [Green Version]
- Iruela-Arispe, M.L.; Luque, A.; Lee, N. Thrombospondin modules and angiogenesis. Int. J. Biochem. Cell. Biol. 2004, 36, 1070–1078. [Google Scholar] [CrossRef]
- Do, H.S.; Park, S.W.; Im, I.; Seo, D.; Yoo, H.W.; Go, H.; Kim, Y.H.; Koh, G.Y.; Lee, B.H.; Han, Y.M. Enhanced thrombospondin-1 causes dysfunction of vascular endothelial cells derived from fabry disease-induced pluripotent stem cells. EBioMedicine 2020, 52, 102633. [Google Scholar] [CrossRef]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [Green Version]
- Rakic, J.M.; Maillard, C.; Jost, M.; Bajou, K.; Masson, V.; Devy, L.; Lambert, V.; Foidart, J.M.; Noel, A. Role of plasminogen activator-plasmin system in tumor angiogenesis. Cell Mol. Life Sci. 2003, 60, 463–473. [Google Scholar] [CrossRef] [PubMed]
- He, Y.L.; Li, M.M.; Wu, L.Y.; Zhao, T.; Di, Y.; Huang, X.; Ding, X.F.; Wu, K.W.; Fan, M.; Zhu, L.L. Enhanced hypoxia-inducible factor (hif)-1alpha stability induced by 5-hydroxymethyl-2-furfural (5-hmf) contributes to protection against hypoxia. Mol. Med. 2015, 20, 590–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luoto, K.R.; Kumareswaran, R.; Bristow, R.G. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Zou, M.; Bhatia, A.; Jayaprakash, P.; Hofman, F.; Ying, Q.; Chen, M.; Woodley, D.T.; Li, W. Breast cancer mda-mb-231 cells use secreted heat shock protein-90alpha (hsp90alpha) to survive a hostile hypoxic environment. Sci. Rep. 2016, 6, 20605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Wang, C.; Chen, S.; Liu, J.; Fu, Y.; Luo, Y. Extracellular hsp90alpha and clusterin synergistically promote breast cancer epithelial-to-mesenchymal transition and metastasis via lrp1. J. Cell Sci. 2019, 132, jcs228213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anant, S.; Ramamoorthy, P.; Tawfik, O.; Jensen, R.A. Effects of hsp90 inhibitors on patient derived triple negative breast cancer (tnbc) cells: Brca1 as a therapeutic target for tnbc. J. Am. Coll. Surg. 2017, 225, e6. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campion, O.; Thevenard Devy, J.; Billottet, C.; Schneider, C.; Etique, N.; Dupuy, J.-W.; Raymond, A.-A.; Boulagnon Rombi, C.; Meunier, M.; Djermoune, E.-H.; et al. LRP-1 Matricellular Receptor Involvement in Triple Negative Breast Cancer Tumor Angiogenesis. Biomedicines 2021, 9, 1430. https://doi.org/10.3390/biomedicines9101430
Campion O, Thevenard Devy J, Billottet C, Schneider C, Etique N, Dupuy J-W, Raymond A-A, Boulagnon Rombi C, Meunier M, Djermoune E-H, et al. LRP-1 Matricellular Receptor Involvement in Triple Negative Breast Cancer Tumor Angiogenesis. Biomedicines. 2021; 9(10):1430. https://doi.org/10.3390/biomedicines9101430
Chicago/Turabian StyleCampion, Océane, Jessica Thevenard Devy, Clotilde Billottet, Christophe Schneider, Nicolas Etique, Jean-William Dupuy, Anne-Aurélie Raymond, Camille Boulagnon Rombi, Marie Meunier, El-Hadi Djermoune, and et al. 2021. "LRP-1 Matricellular Receptor Involvement in Triple Negative Breast Cancer Tumor Angiogenesis" Biomedicines 9, no. 10: 1430. https://doi.org/10.3390/biomedicines9101430






