Transthyretin as a Biomarker to Predict and Monitor Major Depressive Disorder Identified by Whole-Genome Transcriptomic Analysis in Mouse Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Chronic Mild Stress Procedures
2.2.1. Fear Stress
2.2.2. Cage Tilting
2.2.3. Cage Shaking
2.2.4. Wet Sawdust Bedding
2.2.5. Physical Restraint and Social Stress
2.2.6. Water Emergency
2.2.7. Tail Pinching
2.2.8. Continuous Overnight Illumination
2.3. Behavioral Testing Assessments
2.3.1. Running Wheel Test
2.3.2. Forced Swimming Test (FST)
2.3.3. Tail Suspension Test (TST)
2.4. Expression Microarray Experiments and Data Analysis
2.5. QPCR Validation
2.6. Immunohistochemical (IHC) Staining
2.7. TTR ELISA
2.8. Patient Samples
2.9. Statistical Analysis
3. Results
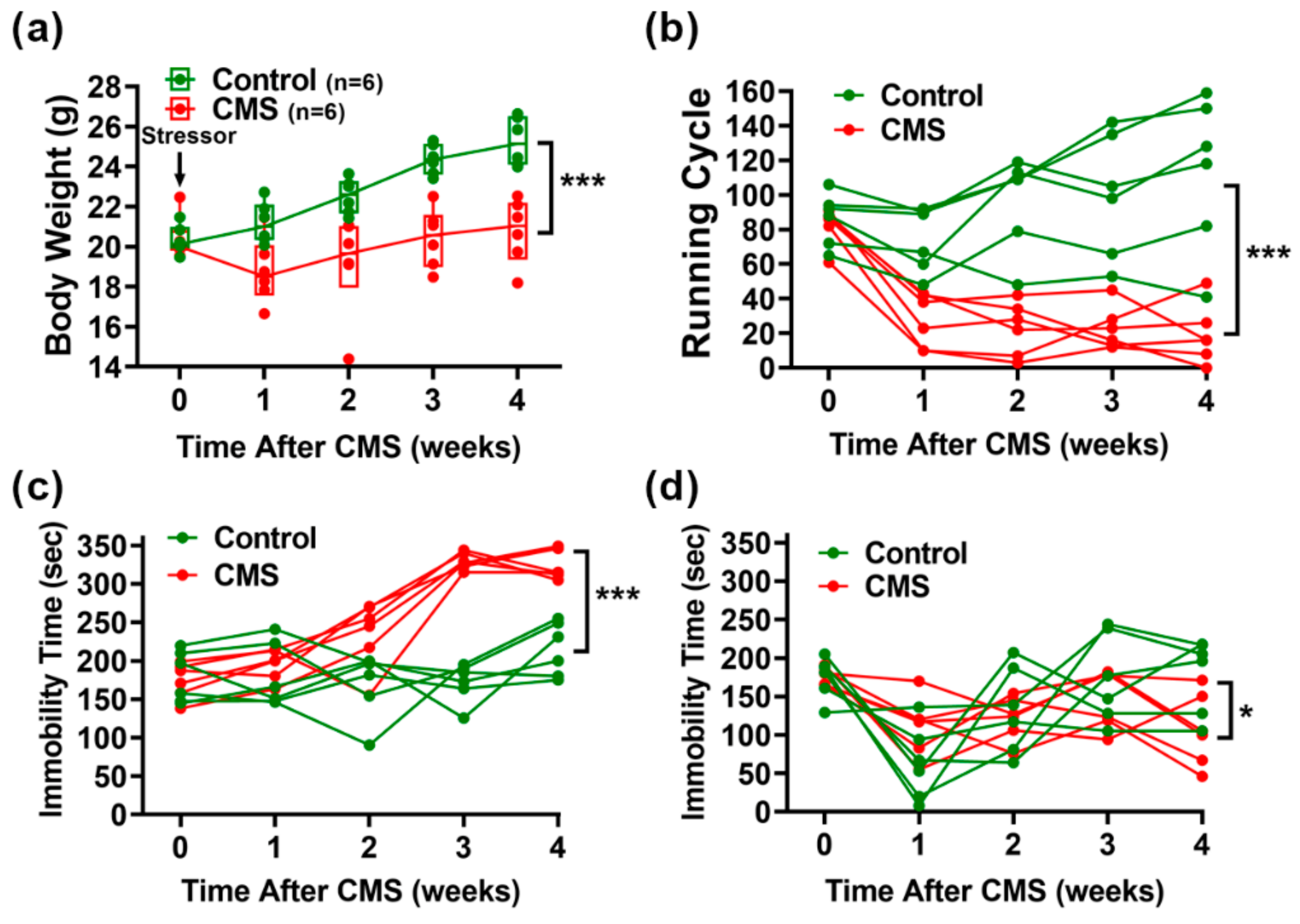
3.1. Mice That Underwent CMS Gained Less Body Weight and Exhibited Reduced Mobility
3.2. Histopathological Analysis of CMS-Treated Mouse Brains
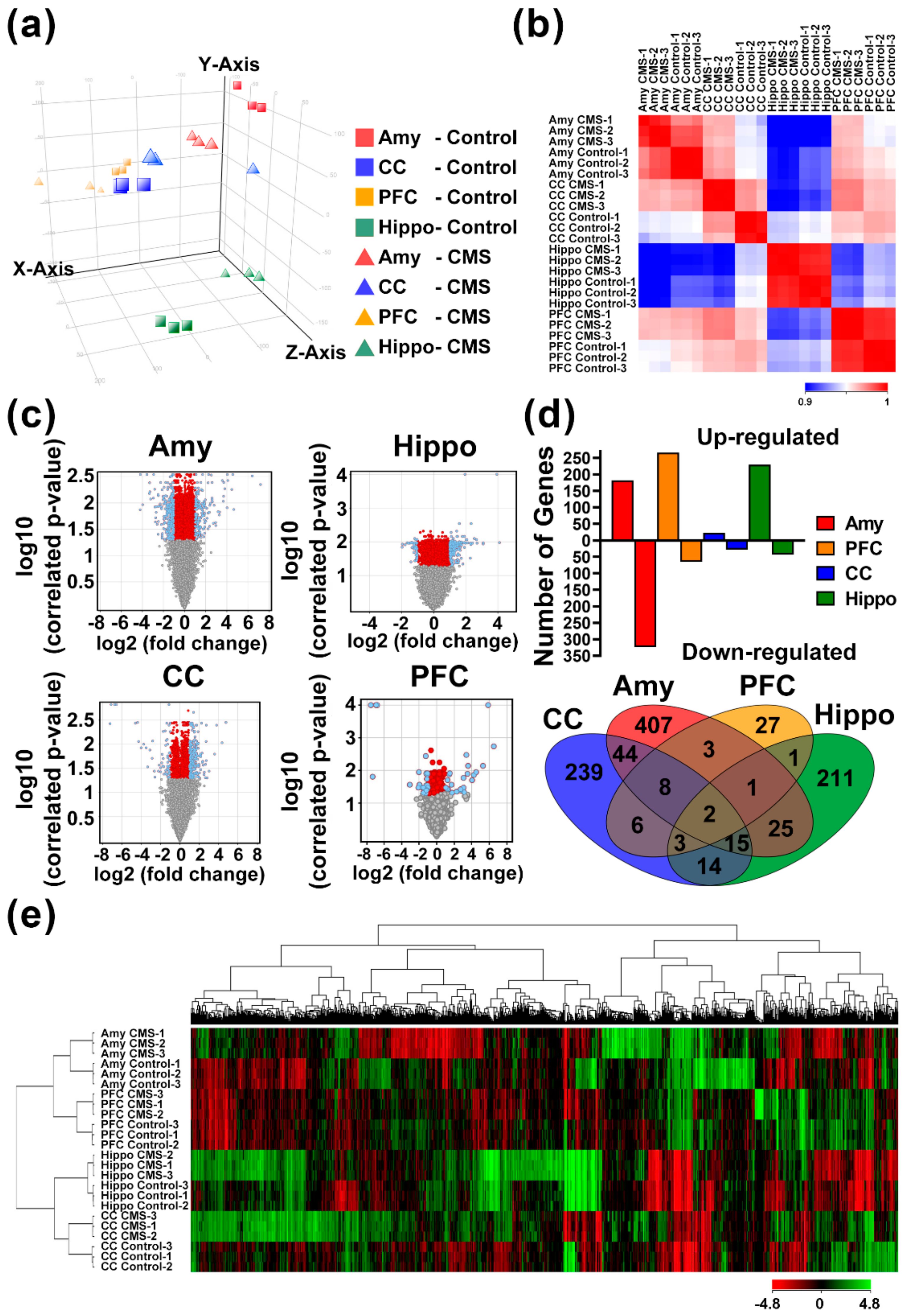
3.3. Identification of MDD-Related Gene Signatures Using Whole-Genome Transcriptomic Analysis
3.4. GSEA Predicts Potential Biological Function Related to CMS Induction
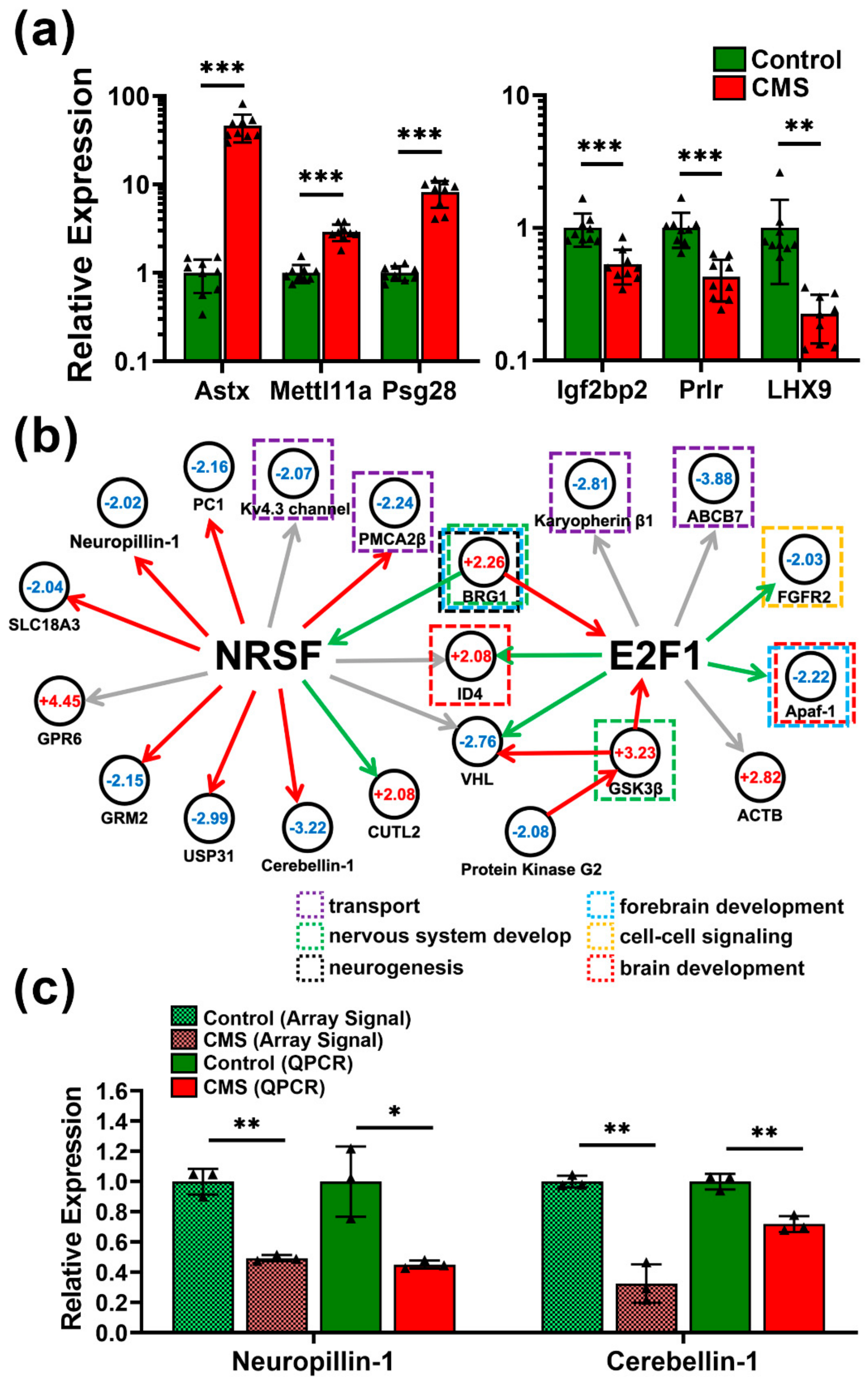
3.5. Pathway Prediction and Regulatory Network Construction in CMS-Treated Mice
3.6. Transthyretin (TTR) Is a Biomarker for MDD Monitoring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Assembly. Global Burden of Mental Disorders and the Need for a Comprehensive, Coordinated Response from Health and Social Sectors at the Country Level: Report by the Secretariat. 2012. Available online: https://apps.who.int/gb/ebwha/pdf_files/eb130/b130_9-en.pdf (accessed on 1 December 2011).
- Kessler, R.C.; Chiu, W.T.; Demler, O.; Merikangas, K.R.; Walters, E.E. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry 2005, 62, 617–627. [Google Scholar] [CrossRef] [Green Version]
- Tye, K.M.; Mirzabekov, J.J.; Warden, M.R.; Ferenczi, E.A.; Tsai, H.C.; Finkelstein, J.; Kim, S.Y.; Adhikari, A.; Thompson, K.R.; Andalman, A.S.; et al. Dopamine neurons modulate neural encoding and expression of depression-related behaviour. Nature 2013, 493, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Goetzel, R.Z.; Pei, X.; Tabrizi, M.J.; Henke, R.M.; Kowlessar, N.; Nelson, C.F.; Metz, R.D. Ten modifiable health risk factors are linked to more than one-fifth of employer-employee health care spending. Health Aff. 2012, 31, 2474–2484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duman, R.S.; Malberg, J.; Nakagawa, S.; D’Sa, C. Neuronal plasticity and survival in mood disorders. Biol. Psychiatry 2000, 48, 732–739. [Google Scholar] [CrossRef]
- Hannestad, J.; DellaGioia, N.; Bloch, M. The effect of antidepressant medication treatment on serum levels of inflammatory cytokines: A meta-analysis. Neuropsychopharmacology 2011, 36, 2452–2459. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lin, W.; Wang, W.; Qi, X.; Wang, D.; Tang, M. The effects of central pro-and anti-inflammatory immune challenges on depressive-like behavior induced by chronic forced swim stress in rats. Behav. Brain Res. 2013, 247, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Schiepers, O.J.; Wichers, M.C.; Maes, M. Cytokines and major depression. Prog. Neuropsychopharmacol. Biol Psychiatry 2005, 29, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.S.; Monteggia, L.M. A neurotrophic model for stress-related mood disorders. Biol. Psychiatry 2006, 59, 1116–1127. [Google Scholar] [CrossRef]
- Ulrich-Lai, Y.M.; Herman, J.P. Neural regulation of endocrine and autonomic stress responses. Nat. Rev. Neurosci. 2009, 10, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, M.H.; Rush, A.J.; Wisniewski, S.R.; Nierenberg, A.A.; Warden, D.; Ritz, L.; Norquist, G.; Howland, R.H.; Lebowitz, B.; McGrath, P.J.; et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: Implications for clinical practice. Am. J. Psychiatry 2006, 163, 28–40. [Google Scholar] [CrossRef]
- Hill, M.N.; Hellemans, K.G.; Verma, P.; Gorzalka, B.B.; Weinberg, J. Neurobiology of chronic mild stress: Parallels to major depression. Neurosci. Biobehav. Rev. 2012, 36, 2085–2117. [Google Scholar] [CrossRef] [Green Version]
- Alfarez, D.N.; Joels, M.; Krugers, H.J. Chronic unpredictable stress impairs long-term potentiation in rat hippocampal CA1 area and dentate gyrus in vitro. Eur. J. Neurosci. 2003, 17, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, N.; Zuo, P. cDNA microarray analysis of gene expression in the cerebral cortex and hippocampus of BALB/c mice subjected to chronic mild stress. Cell. Mol. Neurobiol. 2010, 30, 1035–1047. [Google Scholar] [CrossRef]
- Yamanishi, K.; Doe, N.; Sumida, M.; Watanabe, Y.; Yoshida, M.; Yamamoto, H.; Xu, Y.; Li, W.; Yamanishi, H.; Okamura, H.; et al. Hepatocyte nuclear factor 4 alpha is a key factor related to depression and physiological homeostasis in the mouse brain. PLoS ONE 2015, 10, e0119021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hines, L.M.; Hoffman, P.L.; Bhave, S.; Saba, L.; Kaiser, A.; Snell, L.; Goncharov, I.; LeGault, L.; Dongier, M.; Grant, B.; et al. A sex-specific role of type VII adenylyl cyclase in depression. J. Neurosci. 2006, 26, 12609–12619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, A.; Li, Q.; Koenig, E.A.; Gold, E.; Stephenson, D.; Yang, R.J.; Dreiling, J.; Sullivan, T.; Crawley, J.N. Phenotypic assessment of galanin overexpressing and galanin receptor R1 knockout mice in the tail suspension test for depression-related behavior. Psychopharmacology 2005, 178, 276–285. [Google Scholar] [CrossRef]
- Czeh, B.; Fuchs, E.; Wiborg, O.; Simon, M. Animal models of major depression and their clinical implications. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 64, 293–310. [Google Scholar] [CrossRef]
- Roozendaal, B.; McEwen, B.S.; Chattarji, S. Stress, memory and the amygdala. Nat. Rev. Neurosci. 2009, 10, 423–433. [Google Scholar] [CrossRef]
- Groenewold, N.A.; Opmeer, E.M.; de Jonge, P.; Aleman, A.; Costafreda, S.G. Emotional valence modulates brain functional abnormalities in depression: Evidence from a meta-analysis of fMRI studies. Neurosci. Biobehav. Rev. 2013, 37, 152–163. [Google Scholar] [CrossRef]
- Swartz, J.R.; Knodt, A.R.; Radtke, S.R.; Hariri, A.R. A neural biomarker of psychological vulnerability to future life stress. Neuron 2015, 85, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Murty, V.P.; Ritchey, M.; Adcock, R.A.; LaBar, K.S. fMRI studies of successful emotional memory encoding: A quantitative meta-analysis. Neuropsychologia 2010, 48, 3459–3469. [Google Scholar] [CrossRef] [Green Version]
- McEwen, B.S.; Akil, H. Revisiting the Stress Concept: Implications for Affective Disorders. J. Neurosci. 2020, 40, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; An, S.C.; Xu, C.; Ma, X.M. Role of proBDNF and BDNF in dendritic spine plasticity and depressive-like behaviors induced by an animal model of depression. Brain Res. 2017, 1663, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Sousa, N.; Lukoyanov, N.V.; Madeira, M.D.; Almeida, O.F.; Paula-Barbosa, M.M. Erratum to “Reorganization of the morphology of hippocampal neurites and synapses after stress-induced damage correlates with behavioral improvement”. Neuroscience 2000, 101, 483. [Google Scholar] [CrossRef]
- Zhuang, P.C.; Tan, Z.N.; Jia, Z.Y.; Wang, B.; Grady, J.J.; Ma, X.M. Treadmill Exercise Reverses Depression Model-Induced Alteration of Dendritic Spines in the Brain Areas of Mood Circuit. Front. Behav. Neurosci. 2019, 13, 93. [Google Scholar] [CrossRef]
- Duman, R.S.; Sanacora, G.; Krystal, J.H. Altered Connectivity in Depression: GABA and Glutamate Neurotransmitter Deficits and Reversal by Novel Treatments. Neuron 2019, 102, 75–90. [Google Scholar] [CrossRef]
- Kohler, C.A.; Freitas, T.H.; Maes, M.; de Andrade, N.Q.; Liu, C.S.; Fernandes, B.S.; Stubbs, B.; Solmi, M.; Veronese, N.; Herrmann, N.; et al. Peripheral cytokine and chemokine alterations in depression: A meta-analysis of 82 studies. Acta Psychiatr. Scand. 2017, 135, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.S.; Park, H.J.; Woo, J.; Lee, C.J.; Shim, I. Role of astrocytic GABAergic system on inflammatory cytokine-induced anxiety-like behavior. Neuropharmacology 2019, 160, 107776. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.J.; Petralia, R.S.; Mattson, M.P. Sonic Hedgehog Signaling and Hippocampal Neuroplasticity. Trends Neurosci. 2016, 39, 840–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, C.; Guo, Y.; Wang, C.; Liao, D.; Han, W.; Zhang, J.; Jiang, P. Systematic impacts of chronic unpredictable mild stress on metabolomics in rats. Sci. Rep. 2020, 10, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez Lopez, A.L.; Escobar Villanueva, M.C.; Brianza Padilla, M.; Bonilla Jaime, H.; Alarcon Aguilar, F.J. Chronic Unpredictable Mild Stress Progressively Disturbs Glucose Metabolism and Appetite Hormones in Rats. Acta Endocrinol. 2018, 14, 16–23. [Google Scholar] [CrossRef]
- Warburton, A.; Breen, G.; Rujescu, D.; Bubb, V.J.; Quinn, J.P. Characterization of a REST-Regulated Internal Promoter in the Schizophrenia Genome-Wide Associated Gene MIR137. Schizophr. Bull. 2015, 41, 698–707. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Aron, L.; Zullo, J.; Pan, Y.; Kim, H.; Chen, Y.; Yang, T.H.; Kim, H.M.; Drake, D.; Liu, X.S.; et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature 2014, 507, 448–454. [Google Scholar] [CrossRef] [Green Version]
- Warburton, A.; Savage, A.L.; Myers, P.; Peeney, D.; Bubb, V.J.; Quinn, J.P. Molecular signatures of mood stabilisers highlight the role of the transcription factor REST/NRSF. J. Affect. Disord. 2014, 172, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Zhao, D.; Zhao, H.; Yang, L. NRSF: An Angel or a Devil in Neurogenesis and Neurological Diseases. J. Mol. Neurosci. 2015, 56, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Hashimoto, E.; Ukai, W.; Tateno, M.; Yoshinaga, T.; Saito, S.; Sohma, H.; Saito, T. Lithium-induced suppression of transcription repressor NRSF/REST: Effects on the dysfunction of neuronal differentiation by ethanol. Eur. J. Pharmacol. 2008, 593, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Espada, L.; Udapudi, B.; Podlesniy, P.; Fabregat, I.; Espinet, C.; Tauler, A. Apoptotic action of E2F1 requires glycogen synthase kinase 3-beta activity in PC12 cells. J. Neurochem. 2007, 102, 2020–2028. [Google Scholar] [CrossRef]
- Zhu, Q.; Yang, J.; Han, S.; Liu, J.; Holzbeierlein, J.; Thrasher, J.B.; Li, B. Suppression of glycogen synthase kinase 3 activity reduces tumor growth of prostate cancer in vivo. Prostate 2011, 71, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Lessard, J.; Wu, J.I.; Ranish, J.A.; Wan, M.; Winslow, M.M.; Staahl, B.T.; Wu, H.; Aebersold, R.; Graef, I.A.; Crabtree, G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007, 55, 201–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manduca, J.D.; Theriault, R.K.; Perreault, M.L. Glycogen synthase kinase-3: The missing link to aberrant circuit function in disorders of cognitive dysfunction? Pharmacol. Res. 2020, 157, 104819. [Google Scholar] [CrossRef] [PubMed]
- Banagozar Mohammadi, A.; Torbati, M.; Farajdokht, F.; Sadigh-Eteghad, S.; Fazljou, S.M.B.; Vatandoust, S.M.; Golzari, S.E.J.; Mahmoudi, J. Sericin alleviates restraint stress induced depressive- and anxiety-like behaviors via modulation of oxidative stress, neuroinflammation and apoptosis in the prefrontal cortex and hippocampus. Brain Res. 2019, 1715, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Fatima, M.; Srivastav, S.; Ahmad, M.H.; Mondal, A.C. Effects of chronic unpredictable mild stress induced prenatal stress on neurodevelopment of neonates: Role of GSK-3beta. Sci. Rep. 2019, 9, 1305. [Google Scholar] [CrossRef] [Green Version]
- Maes, M.; Meltzer, H.Y.; Bosmans, E.; Bergmans, R.; Vandoolaeghe, E.; Ranjan, R.; Desnyder, R. Increased plasma concentrations of interleukin-6, soluble interleukin-6, soluble interleukin-2 and transferrin receptor in major depression. J. Affect. Disord. 1995, 34, 301–309. [Google Scholar] [CrossRef]
- Maes, M.; Delange, J.; Ranjan, R.; Meltzer, H.Y.; Desnyder, R.; Cooremans, W.; Scharpe, S. Acute phase proteins in schizophrenia, mania and major depression: Modulation by psychotropic drugs. Psychiatry Res. 1997, 66, 1–11. [Google Scholar] [CrossRef]
- Munkholm, K.; Brauner, J.V.; Kessing, L.V.; Vinberg, M. Cytokines in bipolar disorder vs. healthy control subjects: A systematic review and meta-analysis. J. Psychiatr. Res. 2013, 47, 1119–1133. [Google Scholar] [CrossRef]
- Modabbernia, A.; Taslimi, S.; Brietzke, E.; Ashrafi, M. Cytokine alterations in bipolar disorder: A meta-analysis of 30 studies. Biol. Psychiatry 2013, 74, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.M.; Cardoso, I.; Fernandes, R.; Guimaraes, A.; Saraiva, M.J. Deposition of transthyretin in early stages of familial amyloidotic polyneuropathy: Evidence for toxicity of nonfibrillar aggregates. Am. J. Pathol. 2001, 159, 1993–2000. [Google Scholar] [CrossRef]
- Judge, D.P.; Heitner, S.B.; Falk, R.H.; Maurer, M.S.; Shah, S.J.; Witteles, R.M.; Grogan, M.; Selby, V.N.; Jacoby, D.; Hanna, M.; et al. Transthyretin Stabilization by AG10 in Symptomatic Transthyretin Amyloid Cardiomyopathy. J. Am. Coll. Cardiol. 2019, 74, 285–295. [Google Scholar] [CrossRef]
- Maurer, M.S.; Schwartz, J.H.; Gundapaneni, B.; Elliott, P.M.; Merlini, G.; Waddington-Cruz, M.; Kristen, A.V.; Grogan, M.; Witteles, R.; Damy, T.; et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N. Engl. J. Med. 2018, 379, 1007–1016. [Google Scholar] [CrossRef]
- Dickson, P.W.; Howlett, G.J.; Schreiber, G. Metabolism of prealbumin in rats and changes induced by acute inflammation. Eur. J. Biochem. 1982, 129, 289–293. [Google Scholar] [CrossRef]
- Ingenbleek, Y.; Young, V. Transthyretin (prealbumin) in health and disease: Nutritional implications. Annu. Rev. Nutr. 1994, 14, 495–533. [Google Scholar] [CrossRef]
- Fava, M.; Labbate, L.A.; Abraham, M.E.; Rosenbaum, J.F. Hypothyroidism and hyperthyroidism in major depression revisited. J. Clin. Psychiatry 1995, 56, 186–192. [Google Scholar]
- Sullivan, G.M.; Mann, J.J.; Oquendo, M.A.; Lo, E.S.; Cooper, T.B.; Gorman, J.M. Low cerebrospinal fluid transthyretin levels in depression: Correlations with suicidal ideation and low serotonin function. Biol. Psychiatry 2006, 60, 500–506. [Google Scholar] [CrossRef]
- Bijur, G.N.; Jope, R.S. Opposing actions of phosphatidylinositol 3-kinase and glycogen synthase kinase-3beta in the regulation of HSF-1 activity. J. Neurochem. 2000, 75, 2401–2408. [Google Scholar] [CrossRef]
- Chu, B.; Zhong, R.; Soncin, F.; Stevenson, M.A.; Calderwood, S.K. Transcriptional activity of heat shock factor 1 at 37 degrees C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Calpha and Czeta. J. Biol. Chem. 1998, 273, 18640–18646. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Cattaneo, F.; Ryno, L.; Hulleman, J.; Reixach, N.; Buxbaum, J.N. The systemic amyloid precursor transthyretin (TTR) behaves as a neuronal stress protein regulated by HSF1 in SH-SY5Y human neuroblastoma cells and APP23 Alzheimer’s disease model mice. J. Neurosci. 2014, 34, 7253–7265. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.-L.; Chu, S.S.-T.; Chien, M.-H.; Kuo, P.-H.; Yang, P.-C.; Su, K.-Y. Transthyretin as a Biomarker to Predict and Monitor Major Depressive Disorder Identified by Whole-Genome Transcriptomic Analysis in Mouse Models. Biomedicines 2021, 9, 1124. https://doi.org/10.3390/biomedicines9091124
Yu S-L, Chu SS-T, Chien M-H, Kuo P-H, Yang P-C, Su K-Y. Transthyretin as a Biomarker to Predict and Monitor Major Depressive Disorder Identified by Whole-Genome Transcriptomic Analysis in Mouse Models. Biomedicines. 2021; 9(9):1124. https://doi.org/10.3390/biomedicines9091124
Chicago/Turabian StyleYu, Sung-Liang, Selina Shih-Ting Chu, Min-Hui Chien, Po-Hsiu Kuo, Pan-Chyr Yang, and Kang-Yi Su. 2021. "Transthyretin as a Biomarker to Predict and Monitor Major Depressive Disorder Identified by Whole-Genome Transcriptomic Analysis in Mouse Models" Biomedicines 9, no. 9: 1124. https://doi.org/10.3390/biomedicines9091124





