Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. SH-SY5Y Cell Culture
2.3. Cell Viability Assay
2.4. Oxidative Stress Sensor
2.5. Reverse Transcription Polymerase—Chain Reaction (RT-PCR)
2.6. RNA Sequencing
2.7. Differential Expression Analysis
2.8. Statistics
2.9. Data Availability
3. Results
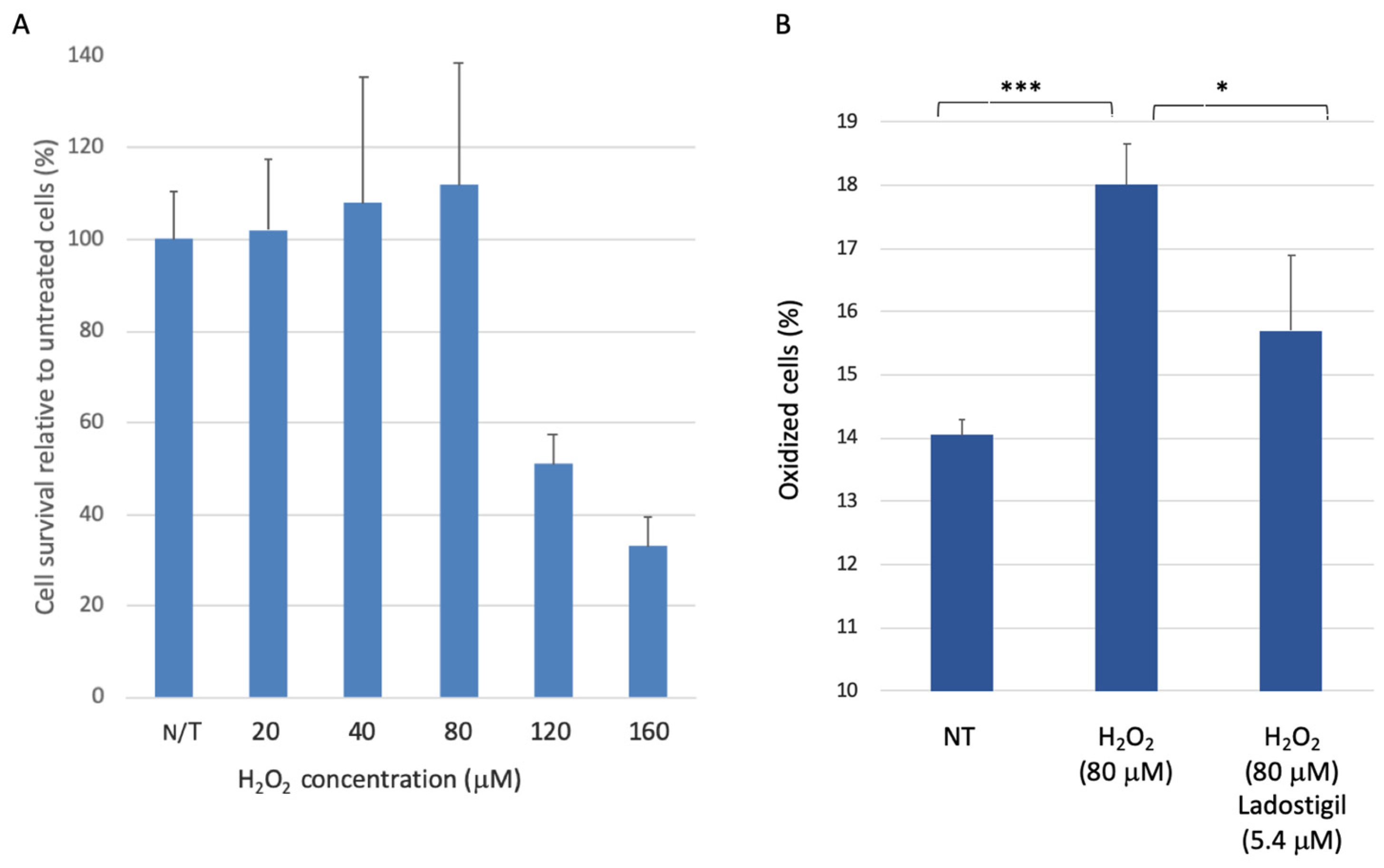
3.1. SH-SY5Y Cell Survival upon Acute Oxidative Stress
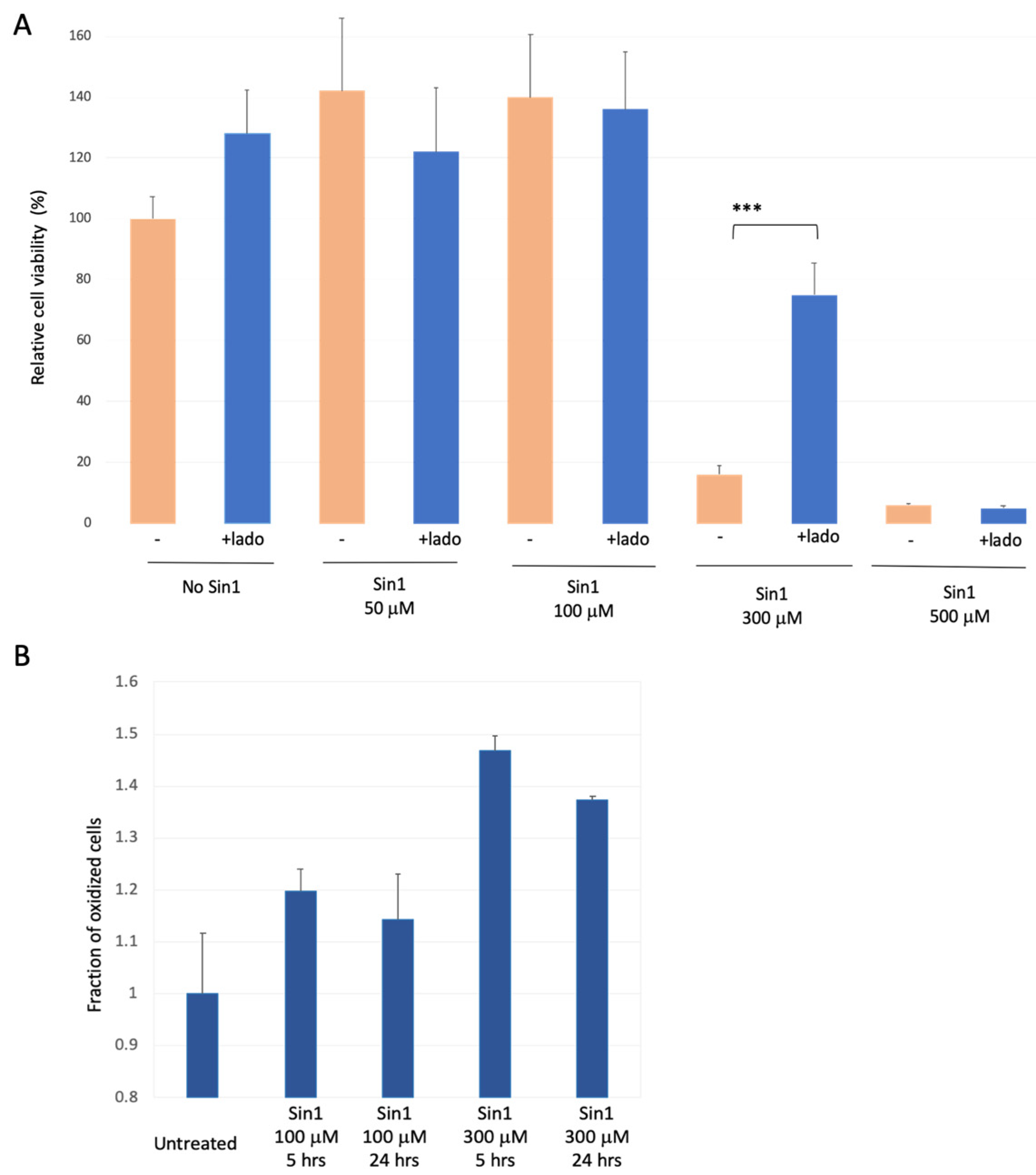
3.2. SH-SYS5 Redox State under Induced Long-Term Oxidation Stress
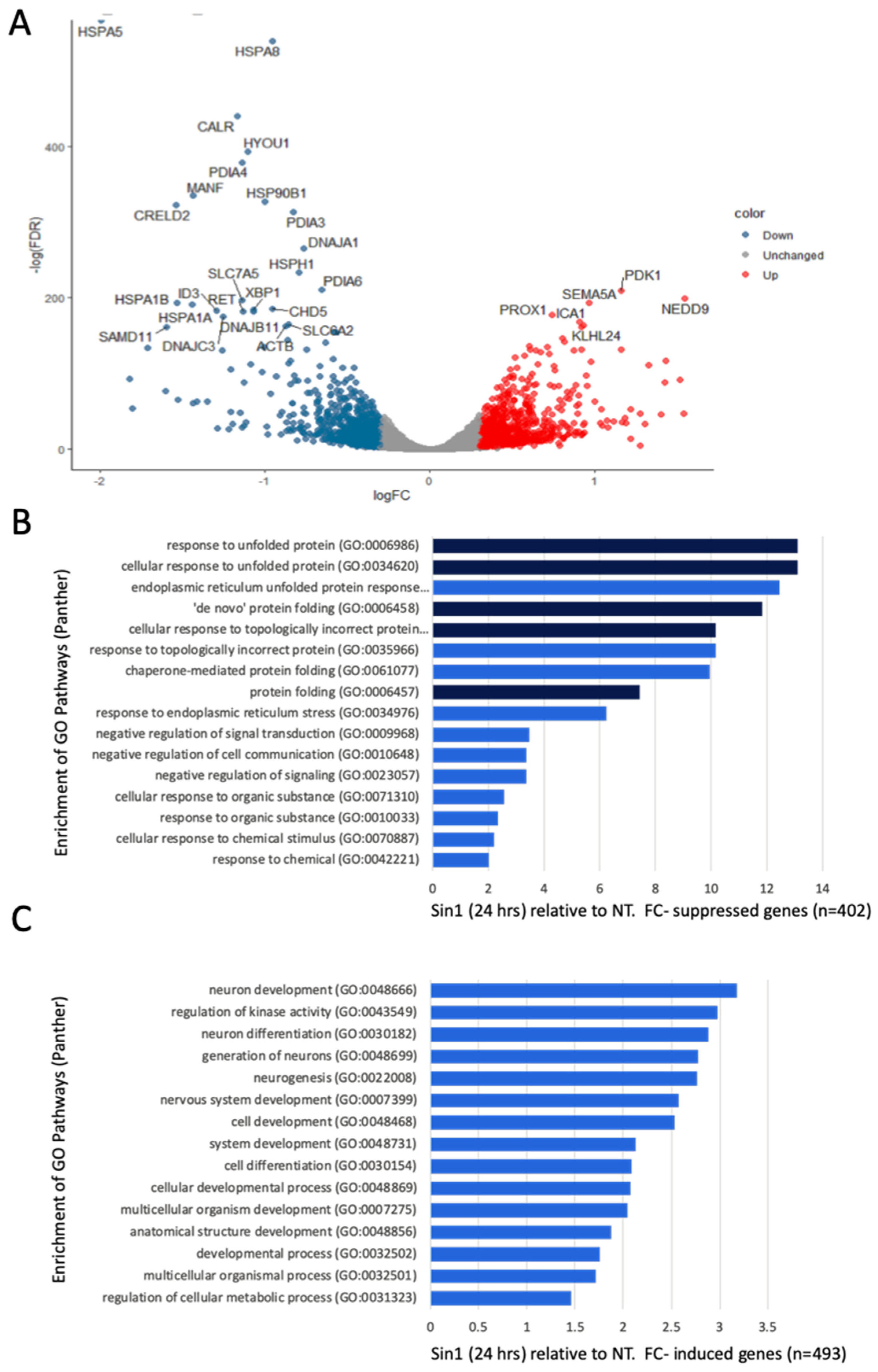
3.3. A Simultaneous Suppression of ER Chaperones and Folding Enzymes by a Chronic Oxidative Stress
3.4. Ladostigil Alters the Expression of Genes Act in Dissipating Oxidative Damage
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | central nerve system |
| DE | differentially expressed |
| ER | endoplasmic reticulum |
| FACS | fluorescence-activated cell sorting |
| FC | fold change |
| FCS | fetal calf serum |
| FDR | false discovery rate |
| GO | gene ontology |
| LPS | lipopolysaccharide |
| ROS | reactive oxygen species |
| TMM | trimmed mean of M-values |
References
- Rodríguez, J.J.; Yeh, C.-Y.; Terzieva, S.; Olabarria, M.; Kulijewicz-Nawrot, M.; Verkhratsky, A. Complex and region-specific changes in astroglial markers in the aging brain. Neurobiol. Aging 2014, 35, 15–23. [Google Scholar] [CrossRef]
- Ferreira, L.K.; Busatto, G.F. Resting-state functional connectivity in normal brain aging. Neurosci. Biobehav. Rev. 2013, 37, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, M.; Mansuy, I.M. Dendritic spine loss and synaptic alterations in Alzheimer’s disease. Mol. Neurobiol. 2008, 37, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorszewska, J. Cell biology of normal brain aging: Synaptic plasticity–cell death. Aging Clin. Exp. Res. 2013, 25, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Bertoni-Freddari, C.; Fattoretti, P.; Giorgetti, B.; Solazzi, M.; Balietti, M.; Meier-Ruge, W. Role of mitochondrial deterioration in physiological and pathological brain aging. Gerontology 2004, 50, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Proteostasis and aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Elobeid, A.; Libard, S.; Leino, M.; Popova, S.N.; Alafuzoff, I. Altered proteins in the aging brain. J. Neuropathol. Exp. Neurol. 2016, 75, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Chinta, S.J.; Woods, G.; Rane, A.; Demaria, M.; Campisi, J.; Andersen, J.K. Cellular senescence and the aging brain. Exp. Gerontol. 2015, 68, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Stefanatos, R.; Sanz, A. The role of mitochondrial ROS in the aging brain. FEBS Lett. 2018, 592, 743–758. [Google Scholar] [CrossRef] [Green Version]
- Albarracin, S.L.; Stab, B.; Casas, Z.; Sutachan, J.J.; Samudio, I.; Gonzalez, J.; Gonzalo, L.; Capani, F.; Morales, L.; Barreto, G.E. Effects of natural antioxidants in neurodegenerative disease. Nutr. Neurosci. 2012, 15, 1–9. [Google Scholar] [CrossRef]
- Wojsiat, J.; Zoltowska, K.M.; Laskowska-Kaszub, K.; Wojda, U. Oxidant/antioxidant imbalance in Alzheimer’s disease: Therapeutic and diagnostic prospects. Oxidative Med. Cell. Longev. 2018, 2018, 6435861. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Tammineni, P. Mitochondrial aspects of synaptic dysfunction in Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 57, 1087–1103. [Google Scholar] [CrossRef] [Green Version]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of brain aging: Adaptive and pathological modification by metabolic states. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Voisine, C.; Rolland, T.; Wachi, S.; Soper, J.H.; Zhu, Y.; Orton, K.; Villella, A.; Garza, D.; Vidal, M. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014, 9, 1135–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esopenko, C.; Levine, B. Aging, neurodegenerative disease, and traumatic brain injury: The role of neuroimaging. J. Neurotrauma 2015, 32, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Moradov, D.; Finkin-Groner, E.; Bejar, C.; Sunita, P.; Schorer-Apelbaum, D.; Barasch, D.; Nemirovski, A.; Cohen, M.; Weinstock, M. Dose-Limiting inhibition of acetylcholinesterase by ladostigil results from the rapid formation and fast hydrolysis of the drug–enzyme complex formed by its major metabolite, R-MCPAI. Biochem. Pharmacol. 2015, 94, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, M.; Luques, L.; Poltyrev, T.; Bejar, C.; Shoham, S. Ladostigil prevents age-related glial activation and spatial memory deficits in rats. Neurobiol. Aging 2011, 32, 1069–1078. [Google Scholar] [CrossRef]
- Panarsky, R.; Luques, L.; Weinstock, M. Anti-inflammatory effects of ladostigil and its metabolites in aged rat brain and in microglial cells. J. Neuroimmune Pharmacol. 2012, 7, 488–498. [Google Scholar] [CrossRef]
- Shoham, S.; Bejar, C.; Kovalev, E.; Schorer-Apelbaum, D.; Weinstock, M. Ladostigil prevents gliosis, oxidative–nitrative stress and memory deficits induced by intracerebroventricular injection of streptozotocin in rats. Neuropharmacology 2007, 52, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Linial, M.; Stern, A.; Weinstock, M. Effect of ladostigil treatment of aging rats on gene expression in four brain areas associated with regulation of memory. Neuropharmacology 2020, 177, 108229. [Google Scholar] [CrossRef] [PubMed]
- Shoham, S.; Linial, M.; Weinstock, M. Age-Induced spatial memory deficits in rats are correlated with specific brain region alterations in microglial morphology and gene expression. J. Neuroimmune Pharmacol. 2019, 14, 251–262. [Google Scholar] [CrossRef]
- Timmerman, R.; Burm, S.M.; Bajramovic, J.J. An overview of in vitro methods to study microglia. Front. Cell. Neurosci. 2018, 12, 242. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.T. Microglial aging in the healthy CNS: Phenotypes, drivers, and rejuvenation. Front. Cell. Neurosci. 2013, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-Y.; Tan, M.-S.; Yu, J.-T.; Tan, L. Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar]
- Camandola, S.; Mattson, M.P. NF-κB as a therapeutic target in neurodegenerative diseases. Expert Opin. Ther. Targets 2007, 11, 123–132. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Kaltschmidt, C. NF-kB: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997, 20, 252–258. [Google Scholar] [CrossRef]
- Singh, S.; Singh, T.G. Role of Nuclear Factor Kappa B (NF-kappaB) signalling in neurodegenerative diseases: An mechanistic approach. Curr. Neuropharmacol. 2020, 18, 918–935. [Google Scholar] [CrossRef]
- Block, M.L.; Zecca, L.; Hong, J.S. Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nat. Rev. Neurosci. 2007, 8, 57–69. [Google Scholar] [CrossRef]
- Wang, C.; He, J.; Xu, T.; Han, H.; Zhu, Z.; Meng, L.; Pang, Q.; Fan, R. Bisphenol A(BPA), BPS and BPB-induced oxidative stress and apoptosis mediated by mitochondria in human neuroblastoma cell lines. Ecotoxicol. Environ. Saf. 2021, 207, 111299. [Google Scholar] [CrossRef] [PubMed]
- Shefa, U.; Jeong, N.Y.; Song, I.O.; Chung, H.-J.; Kim, D.; Jung, J.; Huh, Y. Mitophagy links oxidative stress conditions and neurodegenerative diseases. Neural Regen. Res. 2019, 14, 749. [Google Scholar] [PubMed]
- Kamat, P.K.; Kalani, A.; Kyles, P.; Tyagi, S.C.; Tyagi, N. Autophagy of mitochondria: A promising therapeutic target for neurodegenerative disease. Cell Biochem. Biophys. 2014, 70, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Mecocci, P.; Boccardi, V.; Cecchetti, R.; Bastiani, P.; Scamosci, M.; Ruggiero, C.; Baroni, M. A long journey into aging, brain aging, and Alzheimer’s disease following the oxidative stress tracks. J. Alzheimer’s Dis. 2018, 62, 1319–1335. [Google Scholar]
- Sousa, S.R.; Vetter, I.; Ragnarsson, L.; Lewis, R.J. Expression and pharmacology of endogenous Cav channels in SH-SY5Y human neuroblastoma cells. PLoS ONE 2013, 8, e59293. [Google Scholar] [CrossRef] [Green Version]
- Ou, X.M.; Partoens, P.M.; Wang, J.M.; Walker, J.H.; Danks, K.; Vaughan, P.F.; De Potter, W.P. The storage of noradrenaline, neuropeptide Y and chromogranins in and stoichiometric release from large dense cored vesicles of the undifferentiated human neuroblastoma cell line SH-SY5Y. Int. J. Mol. Med. 1998, 1, 105–112. [Google Scholar] [CrossRef]
- Forster, J.I.; Koglsberger, S.; Trefois, C.; Boyd, O.; Baumuratov, A.S.; Buck, L.; Balling, R.; Antony, P.M. Characterization of differentiated SH-SY5Y as neuronal screening model reveals increased oxidative vulnerability. J. Biomol. Screen 2016, 21, 496–509. [Google Scholar] [CrossRef] [Green Version]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y human neuroblastoma cell line. J. Vis. Exp. 2016, 108, 53193. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef]
- Denn, E.R.; Schober, J.M. A single-wavelength flow cytometric approach using redox-sensitive green fluorescent protein probes for measuring redox stress in live cells. Biotechniques 2021, 70, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Hung, L.H.; Lloyd, W.; Yeung, K.Y. Hot-starting software containers for STAR aligner. Gigascience 2018, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. Gencode reference annotation for the human and mouse genomes. Nucleic. Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. Panther version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic. Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Ayer, A.; Gourlay, C.W.; Dawes, I.W. Cellular redox homeostasis, reactive oxygen species and replicative ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 60–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogg, N.; Darley-Usmar, V.M.; Wilson, M.T.; Moncada, S. Production of hydroxyl radicals from the simultaneous generation of superoxide and nitric oxide. Biochem. J. 1992, 281, 419–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, I.N.; Sullivan, P.G.; Hall, E.D. Peroxynitrite-mediated oxidative damage to brain mitochondria: Protective effects of peroxynitrite scavengers. J. Neurosci. Res. 2007, 85, 2216–2223. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [Green Version]
- Lue, H.; Kleemann, R.; Calandra, T.; Roger, T.; Bernhagen, J. Macrophage migration inhibitory factor (MIF): Mechanisms of action and role in disease. Microbes Infect. 2002, 4, 449–460. [Google Scholar] [CrossRef]
- Israelson, A.; Ditsworth, D.; Sun, S.; Song, S.; Liang, J.; Hruska-Plochan, M.; McAlonis-Downes, M.; Abu-Hamad, S.; Zoltsman, G.; Shani, T.; et al. Macrophage migration inhibitory factor as a chaperone inhibiting accumulation of misfolded SOD1. Neuron 2015, 86, 218–232. [Google Scholar] [CrossRef] [Green Version]
- Mungrue, I.N.; Pagnon, J.; Kohannim, O.; Gargalovic, P.S.; Lusis, A.J. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 2009, 182, 466–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Merianda, T.T.; Twiss, J.L.; Gallo, G. Mechanism and role of the intra-axonal Calreticulin translation in response to axonal injury. Exp. Neurol. 2020, 323, 113072. [Google Scholar] [CrossRef] [PubMed]
- Eesmaa, A.; Yu, L.Y.; Goos, H.; Noges, K.; Kovaleva, V.; Hellman, M.; Zimmermann, R.; Jung, M.; Permi, P.; Varjosalo, M.; et al. The cytoprotective protein MANF promotes neuronal survival independently from its role as a GRP78 cofactor. J. Biol. Chem. 2021, 296, 100295. [Google Scholar] [CrossRef]
- Newington, J.T.; Rappon, T.; Albers, S.; Wong, D.Y.; Rylett, R.J.; Cumming, R.C. Overexpression of pyruvate dehydrogenase kinase 1 and lactate dehydrogenase A in nerve cells confers resistance to amyloid beta and other toxins by decreasing mitochondrial respiration and reactive oxygen species production. J. Biol. Chem. 2012, 287, 37245–37258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Zhuang, Y.; Zhang, L.; Gao, H.; Neavin, D.; Carrillo-Roa, T.; Wang, Y.; Yu, J.; Qin, S.; Kim, D.C. ERICH3: Vesicular association and antidepressant treatment response. Mol. Psychiatry 2020, 26, 2415–2428. [Google Scholar] [CrossRef]
- Chao, C.C.; Huang, C.C.; Lu, D.Y.; Wong, K.L.; Chen, Y.R.; Cheng, T.H.; Leung, Y.M. Ca2+ store depletion and endoplasmic reticulum stress are involved in P2X7 receptor-mediated neurotoxicity in differentiated NG108-15 cells. J. Cell. Biochem. 2012, 113, 1377–1385. [Google Scholar] [CrossRef]
- Hirata, T.; Fujita, M.; Nakamura, S.; Gotoh, K.; Motooka, D.; Murakami, Y.; Maeda, Y.; Kinoshita, T. Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell. 2015, 26, 3071–3084. [Google Scholar] [CrossRef]
- Umezawa, Y. Detecting mitochondrial RNA and other cellular events in living cells. Anal. Bioanal. Chem. 2008, 391, 1591–1598. [Google Scholar] [CrossRef]
- Alfonso, J.; Pollevick, G.D.; Van Der Hart, M.G.; Flugge, G.; Fuchs, E.; Frasch, A.C. Identification of genes regulated by chronic psychosocial stress and antidepressant treatment in the hippocampus. Eur. J. Neurosci. 2004, 19, 659–666. [Google Scholar] [CrossRef]
- Jain, P.; Karthikeyan, C.; Moorthy, N.S.; Waiker, D.K.; Jain, A.K.; Trivedi, P. Human CDC2-like kinase 1 (CLK1): A novel target for Alzheimer’s disease. Curr. Drug Targets 2014, 15, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.Y.; Zhu, J.; Rizvi, A.; Zhu, X.; Tanaka, H.; Dreyfus, C.F. Synaptojanin1 deficiency upregulates basal autophagosome formation in astrocytes. J. Biol. Chem. 2021, 297, 100873. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhong, M.; Zhao, J.; Rhee, H.; Caesar, I.; Knight, E.M.; Volpicelli-Daley, L.; Bustos, V.; Netzer, W.; Liu, L. Reduction of synaptojanin 1 accelerates Aβ clearance and attenuates cognitive deterioration in an Alzheimer mouse model. J. Biol. Chem. 2013, 288, 32050–32063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell 2018, 44, 217–232 e211. [Google Scholar] [CrossRef]
- Liu, D.; Ke, Z.; Luo, J. Thiamine deficiency and neurodegeneration: The interplay among oxidative stress, endoplasmic reticulum stress, and autophagy. Mol. Neurobiol. 2017, 54, 5440–5448. [Google Scholar] [CrossRef]
- Bar-Am, O.; Amit, T.; Youdim, M.B.; Weinreb, O. Neuroprotective and neurorestorative potential of propargylamine derivatives in ageing: Focus on mitochondrial targets. J. Neural. Transm. 2016, 123, 125–135. [Google Scholar] [CrossRef]
- Rohrdanz, E.; Kahl, R. Alterations of antioxidant enzyme expression in response to hydrogen peroxide. Free Radic. Biol. Med. 1998, 24, 27–38. [Google Scholar] [CrossRef]
- Radi, R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA 2004, 101, 4003–4008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.J.; Hogg, N.; Joseph, J.; Konorev, E.; Kalyanaraman, B. The peroxynitrite generator, SIN-1, becomes a nitric oxide donor in the presence of electron acceptors. Arch. Biochem. Biophys. 1999, 361, 331–339. [Google Scholar] [CrossRef]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; Lefebvre d’Hellencourt, C.; Ravanan, P. A molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative stress: Harms and benefits for human health. Oxidative Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.R.; Brasil, F.B.; Furstenau, C.R. Evaluation of the Mitochondria-related redox and bioenergetics effects of gastrodin in SH-SY5Y cells exposed to hydrogen peroxide. J. Mol. Neurosci. 2018, 64, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Schneider, L.; Giordano, S.; Zelickson, B.R.; Johnson, M.S.; Benavides, G.A.; Ouyang, X.; Fineberg, N.; Darley-Usmar, V.M.; Zhang, J. Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic. Biol. Med. 2011, 51, 2007–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene Symbol (Synonym) | Gene Name | TaqMan ID Real Time-PCR | TaqMan Amplicon Length (nt) | Forward (F), Reverse (R) PCR Primers | PCR Amplicon Length (nt) a |
|---|---|---|---|---|---|
| SOD1 | Superoxide Dismutase 1 | Hs00533490_m1 | 60 | F: TTTGCGTCGTAGTCTCCTGC R: CTTTGGCCCACCGTGTTTTC | 308 |
| SOD2 | Superoxide Dismutase 2 | Hs00167309_m1 | 67 | F: CTGCTCCCCGCGCTTTCTTA R: CACGTTTGATGGCTTCCAGC | 373/490 |
| GPX1 | Glutathione Peroxidase 1 | Hs00829989_gH | 76 | F: TTACAGTGCTTGTTCGGGGC R: TCTTGGCGTTCTCCTGATGC | 314 |
| NFE2L2 (NRF2) | Nuclear Factor, Erythroid 2 Like 2 | Hs00975961_g1 | 74 | F: TTATAGCGTGCAAACCTCGC R: TGTGGGCAACCTGTCTCTTCA | 373 |
| MIF | Macrophage Migration Inhibitory Factor | Hs00236988_g1 | 56 | F: GTGGTGTCCGAGAAGTCAGG R: TTGCTGTAGGAGCGGTTCTG | 324 |
| ACTB | Actin Beta | Hs99999903_m1 | 171 | F: ACAGAGCCTCGCCTTTGCCGA R: CATGCCCACCATCAGCCCTGG | 196 |
| Symbol | Gene Name | FDR | Ratio | CPM | Label |
|---|---|---|---|---|---|
| AC006042.2 | peptidylprolyl isomerase (cyclophilin)-like 4 (PPIL4) pseudogene | 1.2 × 10−6 | 2.32 | 5.62 | pseudo. |
| BX005019.1 | novel transcript | 1.8 × 10−23 | 2.05 | 11.43 | lncRNA |
| ERICH3 | glutamate rich 3 | 7.6 × 10−49 | 2.50 | 21.34 | coding |
| IQCN | IQ motif containing N | 1.4 × 10−16 | 2.16 | 10.17 | coding |
| LINC02575 | long intergenic non-protein coding RNA 2575 | 3.5 × 10−39 | 2.68 | 8.82 | lncRNA |
| LOX | lysyl oxidase | 3.3 × 10−14 | 2.06 | 5.36 | coding |
| MTUS1 | microtubule associated scaffold protein 1 | 4.5 × 10−21 | 2.41 | 5.83 | coding |
| MYO15B | myosin XVB | 1.7 × 10−15 | 2.47 | 5.26 | coding |
| NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 | 4.0 × 10−87 | 2.91 | 16.74 | coding |
| NEGR1 | neuronal growth regulator 1 | 1.1 × 10−57 | 2.23 | 29.69 | coding |
| NYAP2 | neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 | 2.3 × 10−17 | 2.11 | 6.44 | coding |
| PDK1 | pyruvate dehydrogenase kinase 1 | 8.5 × 10−92 | 2.23 | 53.06 | coding |
| PLCL1 | phospholipase C like 1 (inactive) | 8.9 × 10−16 | 2.05 | 6.90 | coding |
| RN7SL1 | RNA component of signal recognition particle 7SL1 | 0.011 | 2.42 | 6.43 | ncRNA |
| SERTM2 | serine rich and transmembrane domain containing 2 | 6.4 × 10−24 | 2.32 | 6.68 | coding |
| SLIT2 | slit guidance ligand 2 | 3.5 × 10−16 | 2.24 | 5.24 | coding |
| TSPEAR-AS1 | TSPEAR antisense RNA 1 | 2.4 × 10−40 | 2.86 | 11.06 | lncRNA |
| TSPEAR-AS2 | TSPEAR antisense RNA 2 | 1.4 × 10−51 | 2.69 | 13.02 | lncRNA |
| ZBTB20 | zinc finger and BTB domain containing 20 | 6.3 × 10−19 | 2.27 | 21.96 | coding |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohar, K.; Lezmi, E.; Eliyahu, T.; Linial, M. Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells. Biomedicines 2021, 9, 1251. https://doi.org/10.3390/biomedicines9091251
Zohar K, Lezmi E, Eliyahu T, Linial M. Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells. Biomedicines. 2021; 9(9):1251. https://doi.org/10.3390/biomedicines9091251
Chicago/Turabian StyleZohar, Keren, Elyad Lezmi, Tsiona Eliyahu, and Michal Linial. 2021. "Ladostigil Attenuates Induced Oxidative Stress in Human Neuroblast-like SH-SY5Y Cells" Biomedicines 9, no. 9: 1251. https://doi.org/10.3390/biomedicines9091251







