How Using Dedicated Software Can Improve RECIST Readings
Abstract
:1. Introduction
- -
- Reading times,
- -
- Accuracy of calculated responses (calculation errors),
- -
- Reliability of calculated responses (inter and intra-operator reproducibility).
2. Materials and Methodological Section
2.1. Materials
2.1.1. Study Population


2.1.2. Computer Material
Medical Images Acquisition
Images Post Treatment
- Readings were exclusively performed with the same software application with or without specific options:
- Standard reading: Manual determination of RECIST response using a standard viewer (Myrian®, Intrasense, France) without specific options dedicated to oncologic follow ups. This reading is called “manual reading” in the rest of the article.
- Reading with dedicated software: Determination of RECIST response with the dedicated application for oncologic follow ups, Myrian® XL-Onco, with specific functionalities:
- -
- Automated calculation of elements required for RECIST response determination (sum of diameters, percentage of tumour evolution, RECIST response (CR, PD, PR, and SD)),
- -
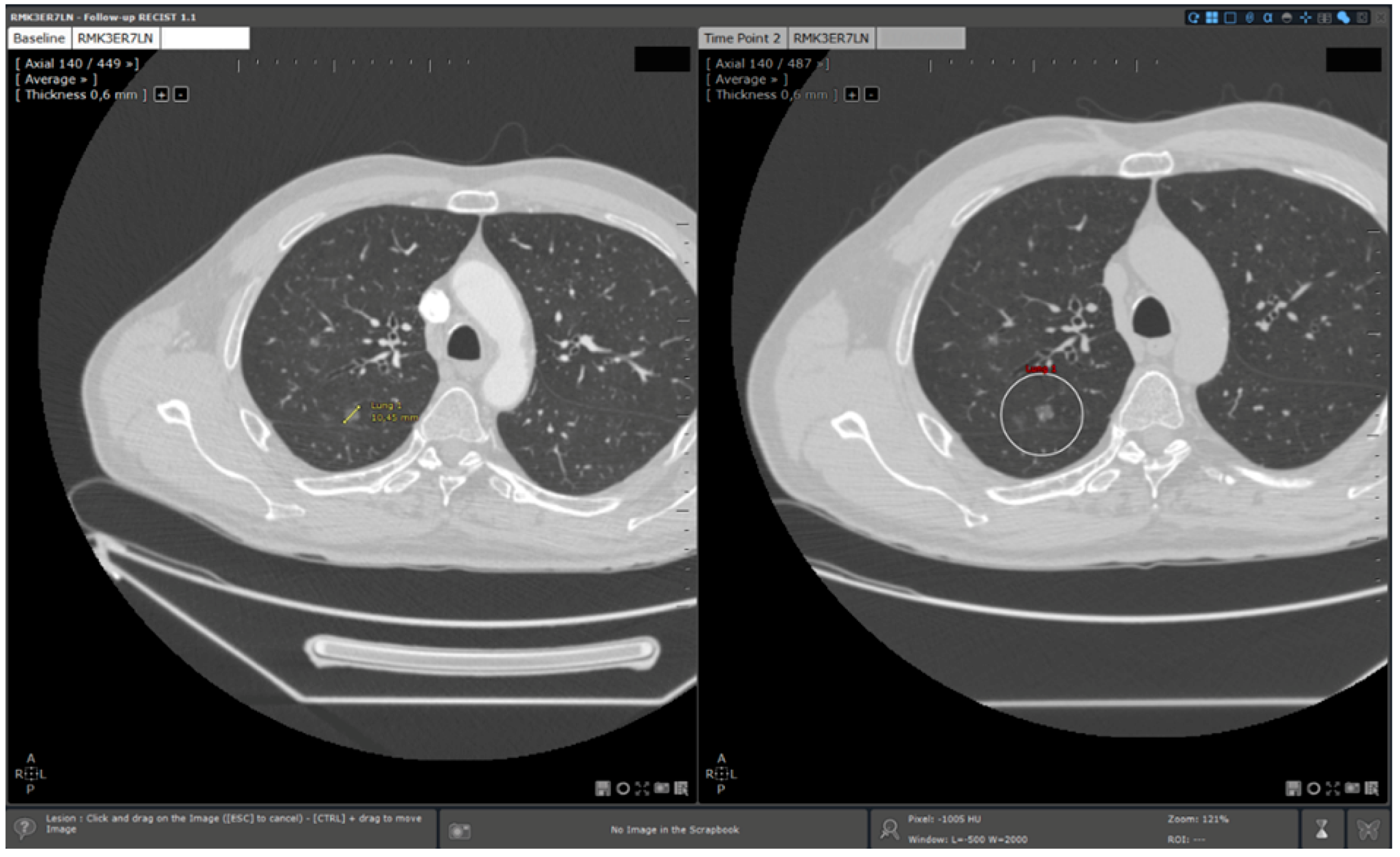
- 3D target matching technology to facilitate lesion localisation in the follow up study.
2.2. Method
2.2.1. Evaluation Criteria
Main Evaluation Criterion: Quantification of Saving in Reading Times Using Automated Reading
- ∆n%: Saving in reading time in % for a patient.
- Tm: Manual reading time for a patient (in seconds).
- Ta: Automated reading time for the same patient (in seconds).
- ∆m%: Mean saving in reading times for the whole database.
- n: Sample size.
Secondary Evaluation Criteria
- Inter and intra-operators reproducibility: For each reading method, it was evaluated comparing for the same patient, the real sums of measured diameters obtained during two distinct reading sessions.
- Manual calculation errors: Manual calculations only occurred during manual readings (without dedicated application). Errors were counted when a difference was detected between the RECIST response (CR, PD, SD, PR) recorded by a radiologist and the theoretical RECIST response based on the diameters measured.
2.2.2. Methodology of Analysis
Baseline Study Reading


Reading Protocol
- -
- Radiologist 1: senior radiologist: 10 years of experience
- -
- Radiologist 2: junior radiologist: 3 years of experience
| Manual Reading Standard Viewer | Automated Reading Dedicated Software | ||
|---|---|---|---|
| Step 1 | Follow up study opening | Manual | Manual |
| Step 2 | Baseline study opening | Manual | Automated |
| Step 3 | Target localisation on baseline study | Manual | Automated |
| Step 4 | Target localisation on follow up study | Manual | Automated |
| Step 5 | Target measurements on follow up study | Manual | Manual |
| Step 6 | Calculation of sums and percentage necessary to tumoral response evaluation (baseline/follow up) | Manual | Automated |
| Step 7 | RECIST response determination | Manual | Automated |
- -
- For manual reading: all the steps are manually performed by the reader
- -
- For automated reading: follow up opening and target measurements are the only steps manually performed by the reader.
Data Reporting
- Reading time evaluation: timekeeping from follow up study opening to follow up study closing and RECIST response determination (from step 1 to step 7, see Table 1),
- Intra and inter-operator reproducibility evaluation: target measurements values on follow up study,
- Calculation error quantification:
- -
- Results of diameter sums calculated by the radiologist on baseline and follow up studies,
- -
- Results of tumoral evolution percentage calculated by the radiologist,
- -
- RECIST score (CR, PD, SD and PR).
2.2.3. Statistical Evaluation
3. Results
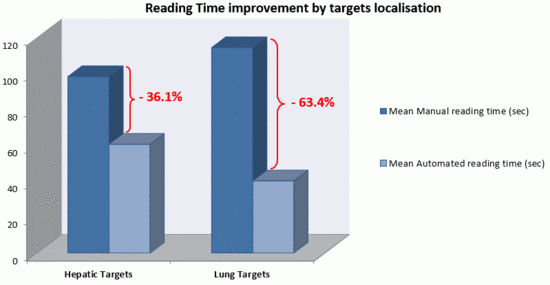
3.1. Saving in Reading Times

- -
- Mean Manual Reading times: senior radiologist: 107.2 s (±29.4)/junior radiologist: 104.6 (±34.4).
- -
- Mean Automated Reading times: senior radiologist: 55.2 s (±24.6)/junior radiologist: 40.3 (±10.7).
3.2. Inter and Intra-Operator Reproducibility
- -
- For manual readings: 0.97 [0.96–0.98]
- -
- For automated readings: 0.98 [0.96–0.99]
- -
- Radiologist 1: 0.97 [0.94–0.98] for manual reading/0.97 [0.95–0.99] for automated reading
- -
- Radiologist 2: 0.94 [0.87–0.98] for manual reading/0.93 [0.84–0.97] for automated reading.
3.3. Calculation Errors
4. Discussion
- -
- Calculation errors
- -
- Roundings by approximation: rounding off performed by physicians during calculation process in order to save time
- -
- Roundings by interpretation: rounding off voluntary performed by physicians in order to modify RECIST score
- -
- Direct impact on patient: when abnormalities are not detected and induce a modification of RECIST response. This can imply a modification of the therapeutic strategy.
- -
- Indirect impact on patient: when abnormalities are not detected but do not directly impact RECIST response. Indeed, conservation of these errors during follow up can potentially impact RECIST response if the concerned exam is baseline or NADIR.
- -
- Direct impact on trial results: when trial statistics can be impacted by the abnormality.
- -
- Correction: waste of time for physicians when error is detected and has to be corrected.

5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Eisenhauer, E.A.; Therasse, P. New response evaluation criteria in solid tumours: Revised RECIST guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Torkzad, M.R. Interobserver and intraobserver variability in the response evaluation of cancer therapy according to RECIST and WHO-criteria. Acta Oncol. 2010, 49, 509–514. [Google Scholar] [CrossRef]
- Thiesse, P.; Ollivier, L. Response rate accuracy in oncology trials: Reasons for interobserver variability. Groupe Français d’Immunothérapie of the Fédération Nationale des Centres de Lutte Contre le Cancer. J. Clin. Oncol. 1997, 15, 3507–3514. [Google Scholar] [PubMed]
- Levy, M.A.; Rubin, D.L. Tool support to enable evaluation of the clinical response to treatment. AMIA Ann. Symp. Proc. 2008, 2008, 399–403. [Google Scholar]
- Goo, J.M. A computer-aided diagnosis for evaluating lung nodules on chest CT: The current status and perspective. Korean J. Radiol. 2011, 12, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Folio, L.R.; Sandouk, A. Consistency and Efficiency of CT Analysis of Metastatic Disease: Semi-automated Lesion Management Application Within a PACS. Am. J. Roentgenol. 2013, 201, 618–625. [Google Scholar] [CrossRef]
- Williams, J.C. A data-based method for assessing and reducing human error to improve operational performance. In Proceedings of the Conference Record for 1988 IEEE Fourth Conference on Human Factors and Power Plants, Monterey, CA, USA, 5–9 June 1988; pp. 436–450.
- Reason, J. Understanding adverse events: Human factors. Qual. Health Care 1995, 4, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Therasse, P.; Arbuck, S. G. New guidelines to evaluate the response to treatment in solid tumors. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Chaudet, H.; Anceaux, F. Facteurs humains et ergonomie en informatique médicale. In Informatique médicale, e-Santé; Springer: Paris, France, 2013; pp. 495–520. [Google Scholar]
- Hopper, K.D.; Kasales, C.J. Analysis of interobserver and intraobserver variability in CT tumor measurements. Am. J. Roentgenol. 1996, 167, 851–854. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
René, A.; Aufort, S.; Mohamed, S.S.; Daures, J.P.; Chemouny, S.; Bonnel, C.; Gallix, B. How Using Dedicated Software Can Improve RECIST Readings. Informatics 2014, 1, 160-173. https://doi.org/10.3390/informatics1020160
René A, Aufort S, Mohamed SS, Daures JP, Chemouny S, Bonnel C, Gallix B. How Using Dedicated Software Can Improve RECIST Readings. Informatics. 2014; 1(2):160-173. https://doi.org/10.3390/informatics1020160
Chicago/Turabian StyleRené, Amandine, Sophie Aufort, Salim Si Mohamed, Jean Pierre Daures, Stéphane Chemouny, Christophe Bonnel, and Benoit Gallix. 2014. "How Using Dedicated Software Can Improve RECIST Readings" Informatics 1, no. 2: 160-173. https://doi.org/10.3390/informatics1020160