Toxic Effects of Two Representative Rare Earth Elements (La and Gd) on Danio rerio Based on Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Acute Toxicity Test
2.3. Bioaccumulation
2.4. Toxicity of Mixture
2.5. Transcriptome High-Throughput Sequencing
2.6. Quantitative Real-Time Polymerase Chain Reaction
2.7. Quality Control and Data Processing
3. Results
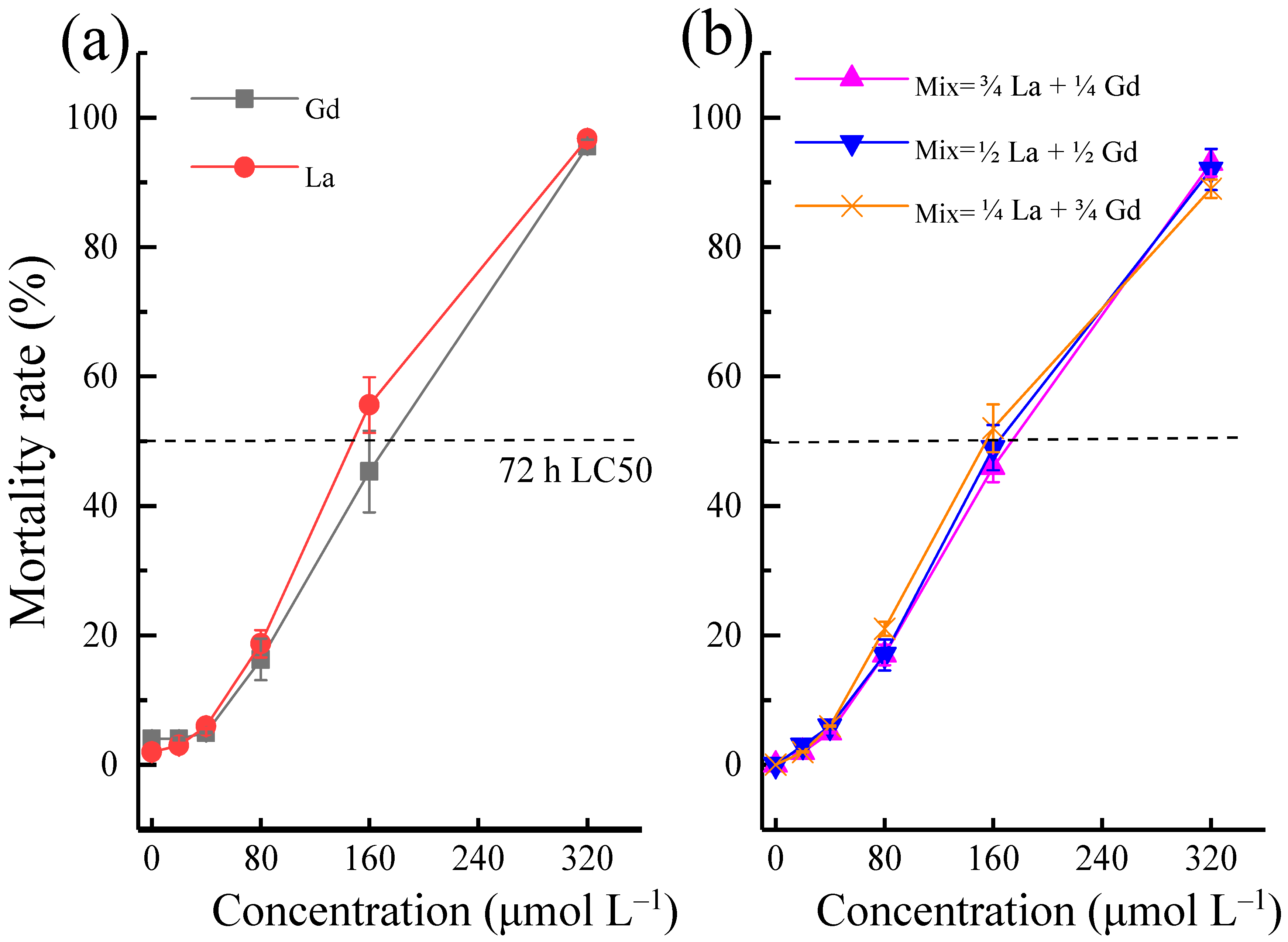
3.1. Toxic Effects and Associated Mechanisms of La and Gd on Danio rerio
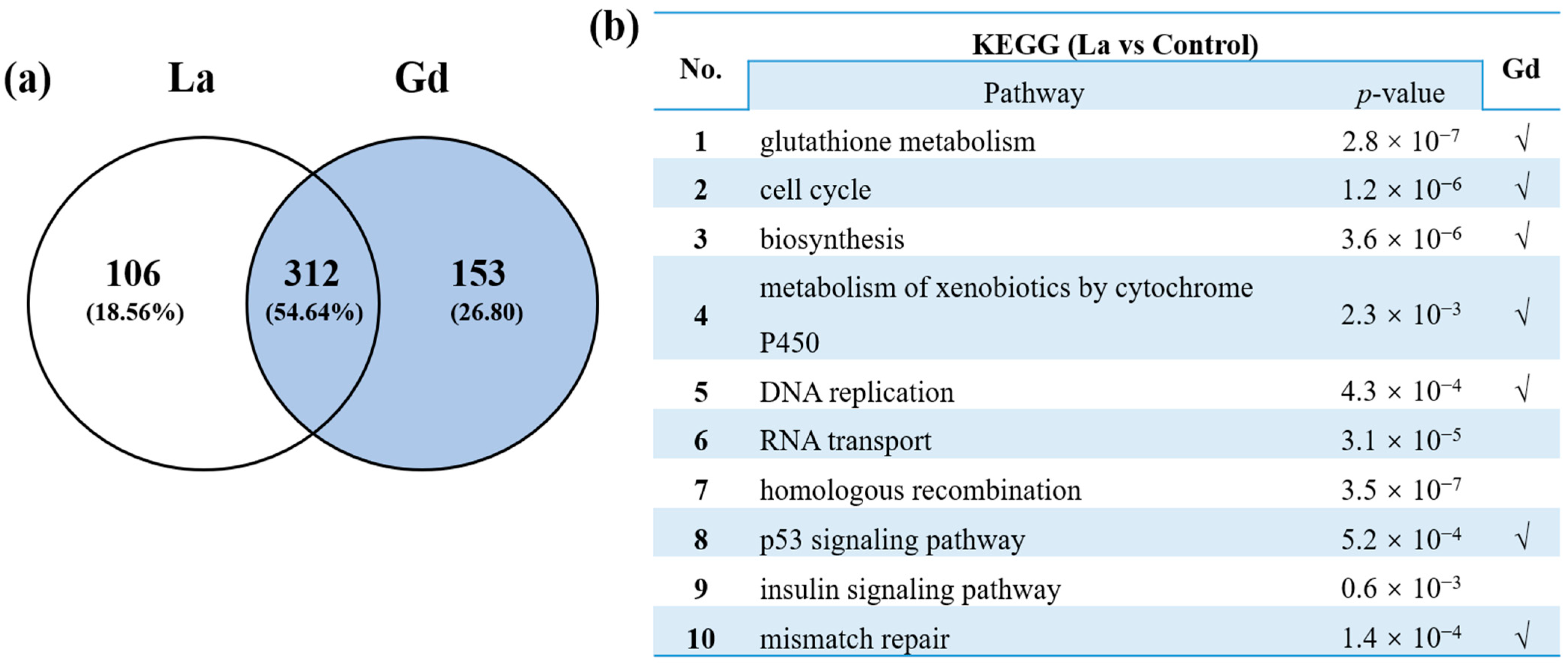
3.2. Transcriptome Analysis of Danio rerio under La and Gd Stress
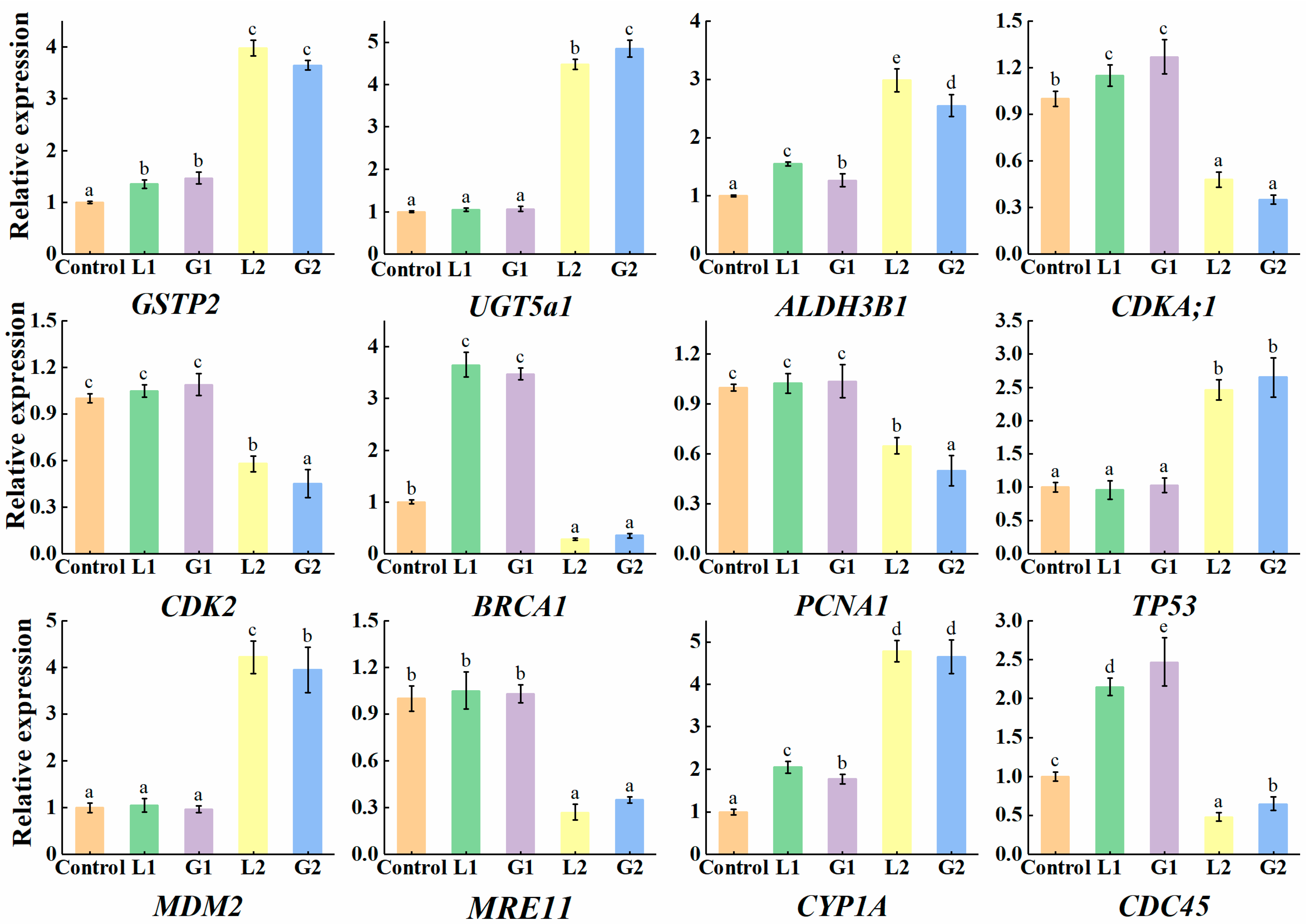
3.3. Validation of DEGs Induced by La and Gd
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Migaszewski, Z.M.; Galuszka, A. The characteristics, occurrence, and geochemical behavior of rare earth elements in the environment: A review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 429–471. [Google Scholar] [CrossRef]
- Sun, L.; Dong, H.; Zhang, P.; Yan, C. Up conversion of rare earth nanomaterials. Annu. Rev. Phys. Chem. 2015, 66, 619–642. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Chang, L.; Finkelman, R.B.; Wang, W.; Liu, J.; Li, J.; Xing, J.; Hou, C. Distribution of rare earth elements in PM10 emitted from burning coals and soil-mixed coal briquettes. J. Environ. Sci. 2020, 97, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Li, Z.; Liu, T.; Chen, J.; Wu, T.; Feng, X. Rare earth elements in street dust and associated health risk in a municipal industrial base of central China. Environ. Geochem. Health. 2017, 39, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Justyna, R.; Ewa, O.; Wojciech, R.; Lidia, W. Gadolinium as a New Emerging Contaminant of Aquatic Environments. Environ. Toxicol. Chem. 2018, 37, 1523–1534. [Google Scholar] [CrossRef]
- Pagano, G.; Guida, M.; Tommasi, F.; Oral, R. Health effects and toxicity mechanisms of rare earth elements-knowledge gaps and research prospects. Ecotoxicolo. Environ. Saf. 2015, 115, 40–48. [Google Scholar] [CrossRef]
- Willis, G.; Lynda, M.; Concilia, D.; Nhamo, C.; Nothando, D.; Edmond, S. Sources, behaviour, and environmental and human health risks of hightechnology rare earth elements as emerging contaminants. Sci. Total Environ. 2018, 636, 299–313. [Google Scholar] [CrossRef]
- Li, X.F.; Che, Z.B.; Chen, Z.Q.; Zhang, Y. A human health risk assessment of rare earth elements in soil and vegetables from a mining area in Fujian Province, Southeast China. Chemosphere 2013, 93, 1240–1246. [Google Scholar] [CrossRef]
- Herrmann, H.; Nolde, J.; Berger, S.; Heise, S. Aquatic ecotoxicity of lanthanum: A review and anattempt to derive water and sediment quality criteria. Ecotoxicol. Environ. Saf. 2016, 124, 213–238. [Google Scholar] [CrossRef]
- Marco, T.; Giovanni, P.; Marco, G.; Anna, P.; Antonietta, S.; Maria, G.; Daniel, M.L.; Petra, B.; Maja, L.; Philippe, J.T.; et al. Comparative toxicity of seven rare earth elements in sea urchin early life stages. Environ. Sci. Pollut. Res. 2017, 24, 20803–20810. [Google Scholar] [CrossRef] [Green Version]
- Dahle, J.T.; Arai, Y. Environmental geochemistry of cerium: Applications and toxicology of cerium oxide nanoparticles. Int. J. Environ. Res. Public Health 2015, 12, 1253–1278. [Google Scholar] [CrossRef] [PubMed]
- Giovanni, P.; Philippe, J.; Aldo, D.; Macro, T. Human exposures to rare earth elements: Present knowledge and research prospects. Environ. Res. 2019, 171, 493–500. [Google Scholar] [CrossRef]
- Kulaksız, S.; Bau, M. Rare earth elements in the Rhine River, Germany: First case of anthropogenic lanthanum as a dissolved microcontaminant in the hydrosphere. Environ. Int. 2011, 37, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.G. Detection of anthropogenic gadolinium in the Brisbane River plume in Moreton Bay, Queensland, Australia. Mar. Pollut. Bull. 2010, 60, 1113–1116. [Google Scholar] [CrossRef]
- Jacinto, J.; Henriques, B.; Duarte, A.C.; Vale, C.; Pereira, E. Removal and recovery of Critical Rare Elements from contaminated waters by living Gracilaria gracilis. J. Hazard. Mater. 2018, 344, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.B.; Liang, J.H.; Meng, H.Y.; Yin, Y.; Zhang, K. Rare Earth Elements Lanthanum and Praseodymium Adversely Affect Neural and Cardiovascular Development in Zebrafish (Danio rerio). Environ. Sci. Technol. 2021, 55, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Rim, K.T. Effects of rare earth elements on the environment and human health: A literature review. Toxicol. Environ. Health Sci. 2016, 8, 189–200. [Google Scholar] [CrossRef]
- Carpenter, D.; Boutin, C.; Allison, J.E.; Parsons, J.L.; Ellis, D.M. Uptake and effects of six rare earth elements (rees) on selected native and crop species growing in contaminated soils. PLoS ONE 2015, 10, e0129936. [Google Scholar] [CrossRef]
- Liu, Z.H.; Guo, C.; Tai, P.D.; Sun, L.Z.; Chen, Z.B. The exposure of gadolinium at environmental relevant levels induced genotoxic effects in Arabidopsis thaliana (L.). Ecotoxicol. Environ. Saf. 2021, 215, 112–138. [Google Scholar] [CrossRef]
- De Oliveira, C.; Ramos, S.J.; Siqueira, J.O.; Faquin, V.; De Castro, E.M.; Amaral, D.C.; Techio, V.H.; Coelho, L.C.; ESilva, P.H.; Schnug, E.; et al. Bioaccumulation and effects of lanthanum on growth and mitotic index in soybean plants. Ecotoxicological Environ. Saf. 2015, 122, 136–144. [Google Scholar] [CrossRef]
- Gonzalez, V.; Vignati, D.A.L.; Leyval, C.; Giamberini, L. Environmental fate and ecotoxicity of lanthanides: Are they a uniform group beyond chemistry? Environ. Int. 2014, 71, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Tai, P.D.; Zhao, Q.; Su, D.; Li, P.J.; Stagnitti, F. Biological toxicity of lanthanide elements on algae. Chemosphere 2010, 80, 1031–1035. [Google Scholar] [CrossRef]
- Blinova, I.; Lukjanova, A.; Muna, M.; Vija, H.; Kahru, A. Evaluation of the potential hazard of lanthanides to freshwater microcrustaceans. Sci. Total Environ. 2018, 642, 1100–1107. [Google Scholar] [CrossRef]
- Romero-Freire, A.; Joonas, E.; Muna, M.; Cossu-Leguille, C.; Vignati, D.A.L.; Giamberini, L. Assessment of the toxic effects of mixtures of three lanthanides (Ce, Gd, Lu) to aquatic biota. Sci. Total Environ. 2019, 661, 276–284. [Google Scholar] [CrossRef]
- Romero-Freire, A.; Minguez, L.; Pelletier, M.; Cayer, A.; Caillet, C.; Devin, S.; Gross, E.M.; Guérold, F.; Pain-Devin, S.; Vignati, D.A.L.; et al. Assessment of baseline ecotoxicity of sediments from a prospective mining area enriched in light rare earth elements. Sci. Total Environ. 2018, 612, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Bing, G.; Erkai, H.; Hao, Q.; Jianqiu, L.; Jie, J.; Ling, Z.; Xinde, C. Phytotoxicity of individual and binary mixtures of rare earth elements (Y, La, and Ce) in relation to bioavailability. Environ. Pollut. 2019, 246, 114–121. [Google Scholar] [CrossRef]
- Yang, G.; Kevin, J.W. Bio-uptake of a rare earth metal (Nd) by Chlamydomonas reinhardtii bioavailability of small organic complexes and role of hardness ions. Environ. Pollut. 2018, 243, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Webster, T.M.U.; Bury, N.; Van, A.R.; Santos, E.M. Global transcriptome profiling reveals molecular mechanisms of metal tolerance in a chronically exposed wild population of Brown Trout. Environ. Sci. Technol. 2013, 47, 8869–8877. [Google Scholar] [CrossRef]
- Loughery, J.R.; Marentette, J.R.; Frank, R.A.; Hewitt, L.M.; Martyniuk, C.J. Transcriptome profiling in larval fathead minnow exposed to commercial naphthenic acids and extracts from fresh and aged oil sands process-affected water. Environ. Sci. Technol. 2019, 53, 10435–10444. [Google Scholar] [CrossRef]
- Fuertes, I.; Campos, B.; Rivetti, C.; Pia, B.; Barata, C. Effects of single and combined low concentrations of neuroactive drugs on Daphnia magna reproduction and transcriptomic Responses. Environ. Sci. Technol. 2019, 53, 11979–11987. [Google Scholar] [CrossRef]
- Tu, Q.; Wang, X.R.; Tian, L.Q.; Dai, L.M. Bioaccumulation of the rare earth elements lanthanum, gadolinium and yttrium in carp (Cyprinus carpio). Environ. Pollut. 1994, 85, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, P.; Miramand, P. Subcellular and body distributions of 17 trace elements in the variegated scallop Chlamys varia from the French coast of the Bay of Biscay. Sci. Total Environ. 2005, 337, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.J.; Carpenter, D.; Boutin, C.; Allison, J.E. Rare earth elements (REEs): Effects on germination and growth of selected crop and native plant species. Chemosphere 2014, 96, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, M.; Chu, Y.; Chen, D.D.Y.; Huang, X.; Zhou, Q. Responses of plant calmodulin to endocytosis induced by rare earth elements. Chemosphere 2016, 154, 408–415. [Google Scholar] [CrossRef]
- Cheng, J.; Li, N.; Cheng, Z.; Hua, R.; Cai, J.; Si, W.; Hong, F. Splenocyte apopotic pathway in mice following exposure to cerium chloride. Chemosphere 2011, 83, 612–617. [Google Scholar] [CrossRef]
- Silvestre, F.; Boni, R.; Fissore, R.A.; Tosti, E. Ca2+ signaling during maturation of cumulus-oocyte complex in mammals. Mol. Reprod. Dev. 2011, 78, 744–756. [Google Scholar] [CrossRef]
- Boni, R.; Gualtieri, R.; Talevi, R.; Tosti, E. Calcium and other ion dynamics during gamete maturation and fertilization. Theriogenology 2007, 68, 156–164. [Google Scholar] [CrossRef]
- Martin, B.; Richardson, F.S. Lanthanides as probes for calcium in biological systems. Q. Rev. Biophys. 1979, 12, 181–209. [Google Scholar] [CrossRef]
- Hu, Z.; Cools, T.; De Veylder, L. Mechanisms used by plants to cope with DNA damage. Annu. Rev. Plant Biol. 2016, 67, 439–462. [Google Scholar] [CrossRef]
- Lario, S.; Casalots, A.; Sanfeliu, E.; Boix, L.; García-Iglesias, P.; Sánchez-Delgado, J.; Montserrat, A. Real-time PCR improves helicobacter pylori detection in patients with peptic ulcer bleeding. PLoS ONE 2011, 6, e20009. [Google Scholar] [CrossRef] [Green Version]
- Cools, T.; Veylder, D.L. DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 2009, 12, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Kawashima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef]
- Glisic, B.; Mihaljevic, I.; Popovic, M.; Zaja, R.; Loncar, J.; Fent, K.; Kovacevic, R.; Smital, T. Characterization of glutathione-S-transferases in zebrafish (Danio rerio). Aquat. Toxicol. 2015, 158, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, G.D.; Bridge, W.J. The glutathione system and the related thiol network in Caenorhabditis elegans. Redox Biol. 2019, 24, 101171. [Google Scholar] [CrossRef] [PubMed]
- Marchitti, S.A.; Brocker, C.; Orlicky, D.J.; Vasiliou, V. Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic. Biol. Med. 2010, 49, 1432–1443. [Google Scholar] [CrossRef]
- Li, Z.L.; Liu, Z.H.; Chen, R.J.; Li, X.J.; Tai, P.D.; Gong, Z.Q.; Jia, C.Y.; Liu, W. DNA damage and genetic methylation changes caused by cd in Arabidopsis thaliana seedlings. Environ. Toxicol. Chem. 2015, 34, 2095–2103. [Google Scholar] [CrossRef]
- Banin, A.; Navrot, J. Origin of life: Clues from relations between chemical compositions of living organisms and natural environments. Science 1975, 189, 550–551. [Google Scholar] [CrossRef]
- Pagano, G.; Guida, M.; Siciliano, A.; Oral, R.; Kocbas, F.; Palumbo, A.; Castellano, I.; Migliaccio, O.; Thomas, P.J.; Trifuoggi, M. Comparative toxicities of selected rare earth elements: Sea urchin embryogenesis and fertilization damage with redox and cytogenetic effects. Environ. Res. 2016, 147, 453–460. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Tai, P.; Sun, L.; Yang, X. Toxicity of ammonia, cadmium, and nitrobenzene to four local fishes in the Liao River, China and the derivation of site-specific water quality criteria. Ecotoxicol. Environ. Saf. 2017, 147, 656–663. [Google Scholar] [CrossRef]
- Liu, Z.; Tai, P.; Li, X.; Kong, L.; Matthews, T.G.; Lester, R.E.; Mondon, J.A. Deriving site-specific water quality criteria for ammonia from national versus international toxicity data. Ecotoxicol. Environ. Saf. 2019, 171, 665–676. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Ahmad, S.; Sun, M.; Zhang, Q. Selection of optimal reference genes for qRT-PCR analysis of shoot development and graviresponse in prostrate and erect chrysanthemums. PLoS ONE 2019, 14, e0225241. [Google Scholar] [CrossRef]
- Louren, A.P.; Mackert, A.; Cristino, A. Validation of reference genes for gene expression studies in the honey bee, apis mellifera, by quantitative real-time rt-pcr. Apidologie 2008, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]

| Concentration (μmol L−1) | Uptake (μg g−1, Fresh Weight) (BCF) | |||
|---|---|---|---|---|
| Muscle | Gills | Liver | ||
| 15 | La | 0.53 ± 0.14 b (0.25) | 9.36 ± 1.56 b (4.49) | 19.45 ± 3.21 b (9.33) |
| Gd | 0.41 ± 0.09 a (0.17) | 8.43 ± 1.22 a (3.58) | 18.21 ± 3.68 a (7.73) | |
| 30 | La | 1.38 ± 0.26 c (0.33) | 20.35 ± 3.72 d (4.88) | 35.41 ± 8.94 c (8.49) |
| Gd | 1.22 ± 0.35 c (0.26) | 15.21 ± 2.54 c (3.23) | 39.85 ± 7.62 d (8.46) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.; Guo, C.; Xue, C.; Ma, C.; Mu, H.; Sun, L. Toxic Effects of Two Representative Rare Earth Elements (La and Gd) on Danio rerio Based on Transcriptome Analysis. Toxics 2022, 10, 519. https://doi.org/10.3390/toxics10090519
Kang S, Guo C, Xue C, Ma C, Mu H, Sun L. Toxic Effects of Two Representative Rare Earth Elements (La and Gd) on Danio rerio Based on Transcriptome Analysis. Toxics. 2022; 10(9):519. https://doi.org/10.3390/toxics10090519
Chicago/Turabian StyleKang, Shu, Cheng Guo, Chenyang Xue, Chenshu Ma, Huaizhong Mu, and Lizong Sun. 2022. "Toxic Effects of Two Representative Rare Earth Elements (La and Gd) on Danio rerio Based on Transcriptome Analysis" Toxics 10, no. 9: 519. https://doi.org/10.3390/toxics10090519




