Toxicity of Three Optical Brighteners: Potential Pharmacological Targets and Effects on Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Molecular Docking of OBs
2.2.1. Optical Brighteners Structure
2.2.2. Preparation of Protein Structures
2.2.3. Virtual Screening Protocol
2.2.4. Refinement Docking Experiments
2.2.5. Conformational Analysis
2.2.6. Molecular Dynamics Simulations
2.3. Toxicity of OBs in Experiments with Caenorhabditis elegans
2.3.1. Optical Brightener
2.3.2. Strains and Nematodes: Maintenance and Treatments
2.3.3. Lethality Assay
2.3.4. Body Length Assay
2.3.5. Locomotion Assay
2.3.6. Reproduction Assay
2.3.7. Gene Expression Using Fluorescence Measuring
2.4. Statistical Analysis
3. Results
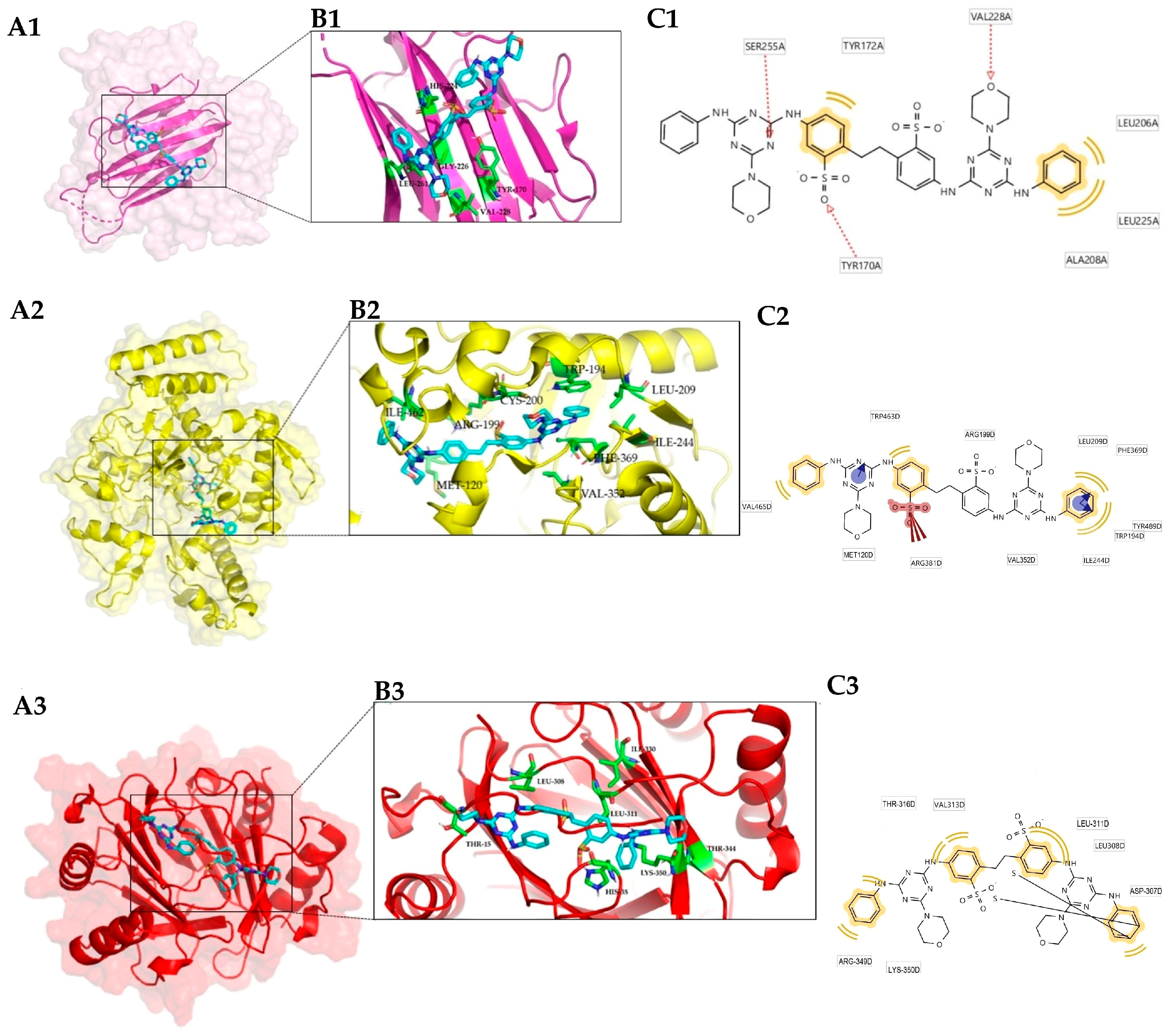
3.1. Protein Target Selection
3.2. Conformational Analysis
3.3. Molecular Dynamics Simulation
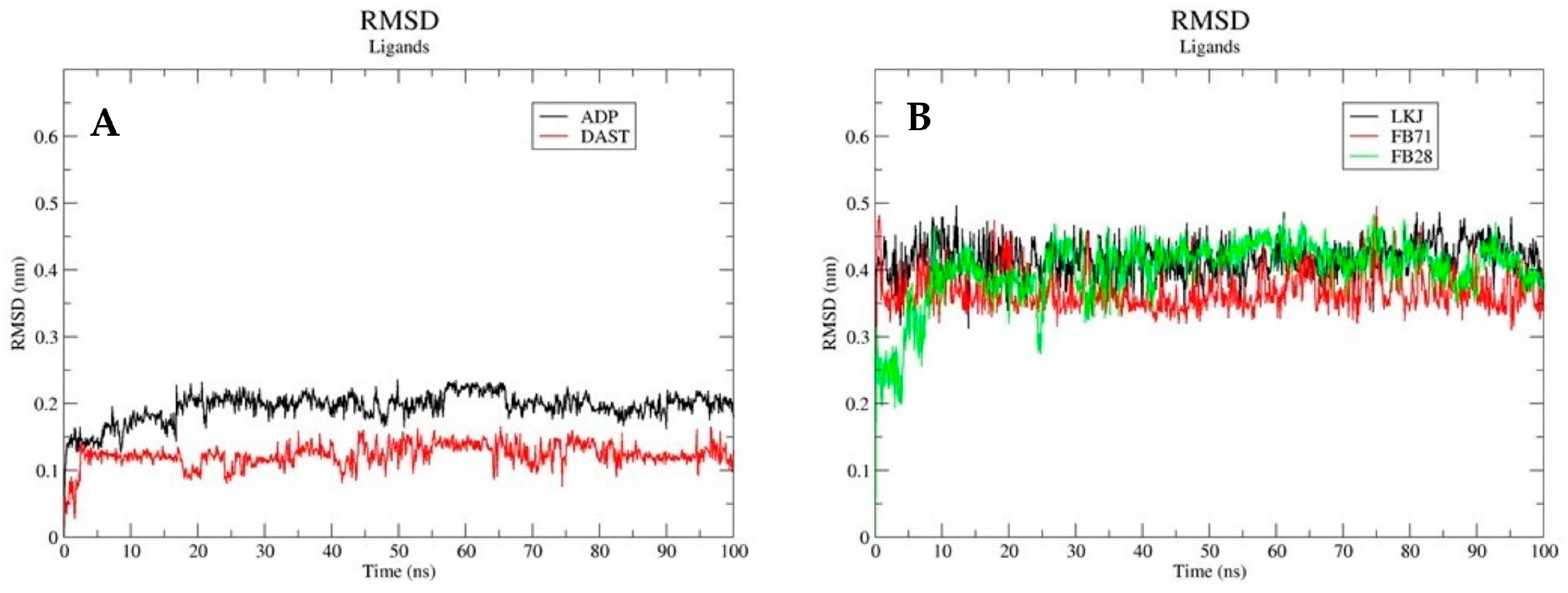
3.3.1. Root Mean Square Deviations (RSMD)
3.3.2. Root Mean Square Fluctuations (RSMF)
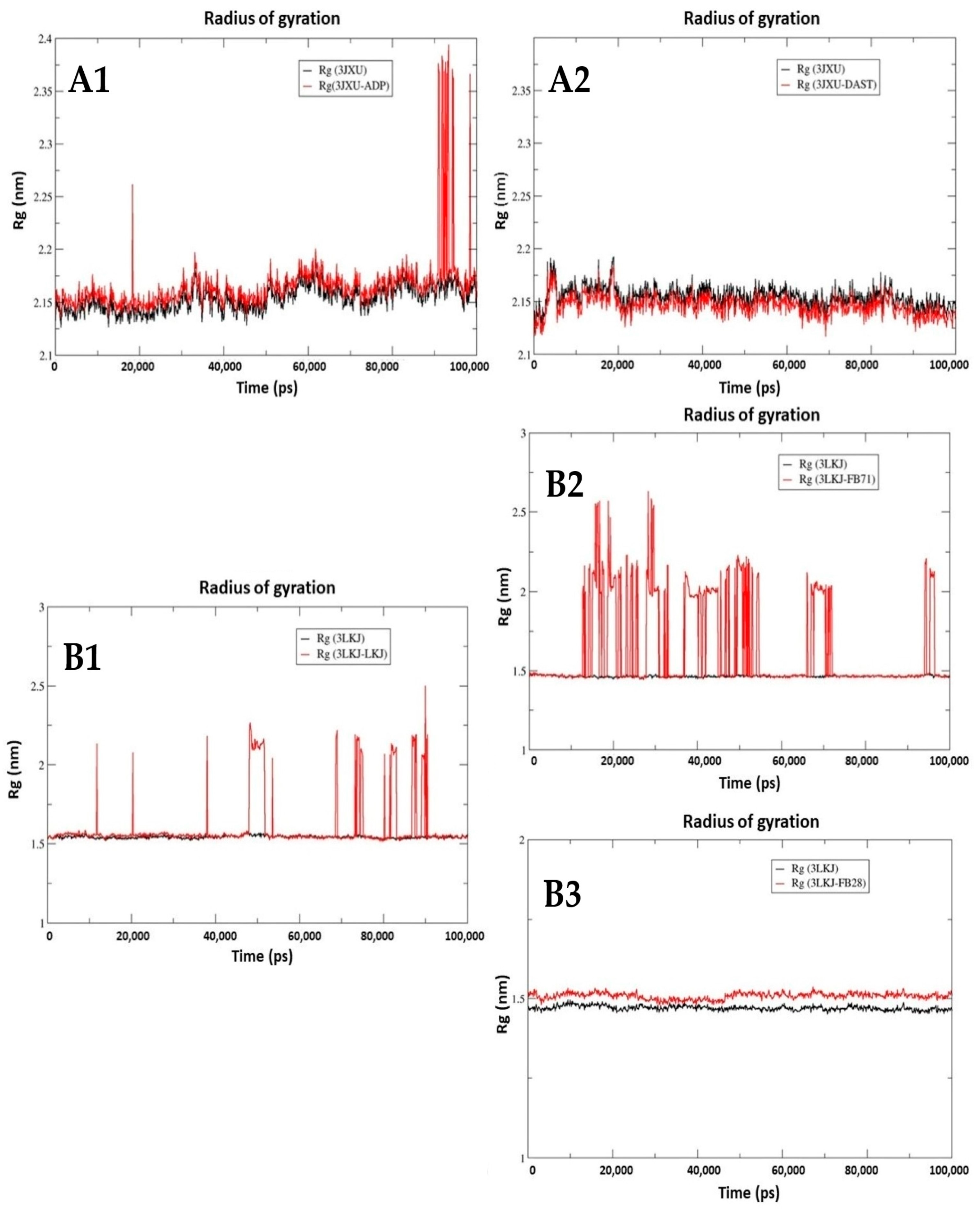
3.3.3. Radius of Gyration (Rg)
3.3.4. Molecular Mechanics Energies Combined with Surface Area Continuum Solvation (MMGBSA)
3.4. Toxicity of OBs in Caenorhabditis elegans
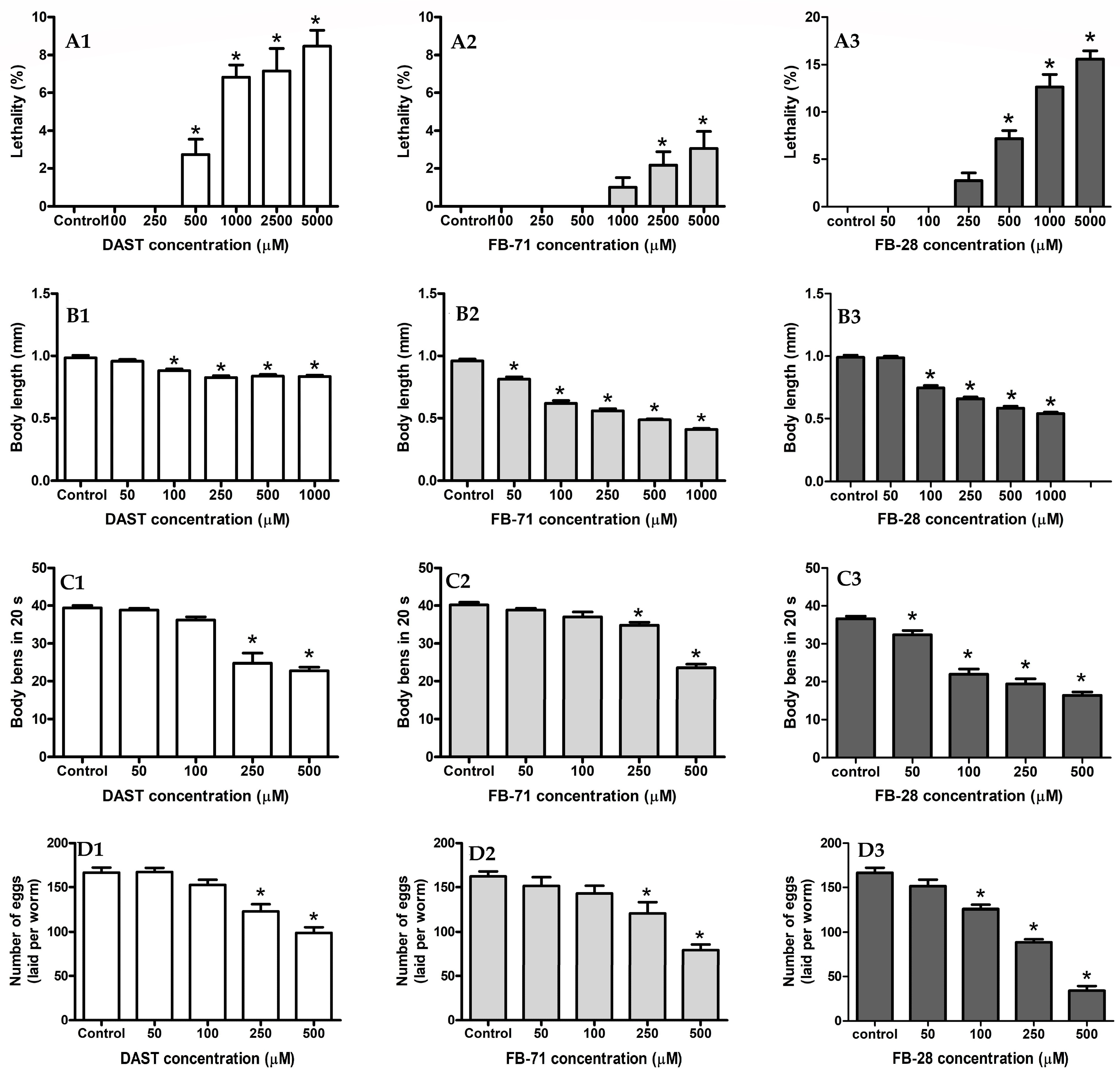
3.4.1. Lethality Assay
3.4.2. Body Length Assay
3.4.3. Locomotion Assay
3.4.4. Reproduction Assay
3.4.5. Transgenic Reporter Assays
4. Discussion
4.1. In Silico Behavior of OBs
4.2. Effects of OBs on the Physiology and Gene Expression of C. elegans
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Álvarez, M. La industria de los surfactantes: Tendencias mundiales y perspectivas para Colombia. Palmas 2004, 25, 340–353. [Google Scholar]
- Azizullah, A.; Richter, P.; Häder, D. Toxicity assessment of a common laundry detergent using the freshwater flagellate Euglena gracilis. Chemosphere 2011, 84, 1392–1400. [Google Scholar] [CrossRef]
- Grand View Research Inc. Optical brighteners market worth $1.11bn by 2024. Focus Surfactants 2018, 2018, 4. [Google Scholar] [CrossRef]
- Giagnorio, M.; Amelio, A.; Grüttner, H.; Tiraferri, A. Environmental impacts of detergents and benefits of their recovery in the laundering industry. J. Clean. Prod. 2017, 154, 593–601. [Google Scholar] [CrossRef]
- Bajpai, D.; Tyagi, V.K. Laundry Detergents: An Overview. J. Oleo Sci. 2007, 56, 327–340. [Google Scholar] [CrossRef]
- Ojo, O.A.; Oso, B.A. Biodegradation of synthetic detergents in wastewater. Afr. J. Biotechnol. 2009, 8, 1090–1109. [Google Scholar]
- Hollender, J. Unanswered Questions: The Health and Environmental Hazards Hidden in Traditional Household Cleaning Products; Household Toxins Institute, University of Minnesota: Minneapolis, MN, USA, 2003; pp. 1–20. [Google Scholar]
- Scott, M.; Jones, M. The biodegradation of surfactants in the environment. Biochim. Biophys. Acta. 2000, 1508, 235–251. [Google Scholar] [CrossRef]
- Mousavi, S.; Khodadoost, F. Effects of detergents on natural ecosystems and wastewater treatment processes: A review. Environ. Sci. Pollut. Res. 2019, 26, 26439–26448. [Google Scholar] [CrossRef]
- Woodard & Curran, Inc. Synthetic Detergent. In Industrial Waste Treatment Handbook, 2nd ed.; Fundamental; Elsivier: Amsterdam, The Netherlands, 2006; pp. 29–49. [Google Scholar] [CrossRef]
- Manga, M.; Willis, D.; Ali, N.; York, D. The impact of raw material properties and process conditions on the color of a powdered formulated detergent product. Particuology 2019, 45, 35–41. [Google Scholar] [CrossRef]
- Ganz, C.; Schulze, J.; Stensby, P.; Lyman, F.; Macek, K. Accumulation and elimination studies of four detergent fluorescent whitening agents in bluegill (Lepomismacrochirus). Environ. Sci. Technol. 1975, 9, 738–744. [Google Scholar] [CrossRef]
- Chen, H.; Han, X.; Zhu, C.; Du, B.; Tan, L.; He, R.; Shen, M.; Liu, L.-Y.; Zeng, L. Identification of Fluorescent Brighteners as Another Emerging Class of Abundant, Ubiquitous Pollutants in the Indoor Environment. Environ. Sci. Technol. 2022, 56, 10131–10140. [Google Scholar] [CrossRef] [PubMed]
- Kover, F. Enviromentals Contaminants—Optical Brighteners, Methyl Chloroform, Tricholorethylene, Tetrachloroethylene, Ions Exchange Resins. Doctoral Thesis, Franklin Institute, Washington, DC, USA, July 1975. EPA-560/2-75-002. [Google Scholar]
- Tiki, A. Chemistry of optical brighteners and uses in textile industries. Pakistan Text. J. 2010, 59, 42–43. [Google Scholar]
- Sadek, S.; Lee, J.; Lund, R.; McConnell, W. Waste Reduction by Process Improvement in the Brightener Intermediates Production. Water Sci. Technol. 1992, 26, 309–317. [Google Scholar] [CrossRef]
- Mondragón, G.; Pineda, S.; Martínez, A.; Martínez, A.M. Optical brightener Tinopal C1101 as an ultraviolet protectant for a nucleopolyhedrovirus. Commun. Agric. Appl. Biol. Sci. 2007, 72, 543–547. [Google Scholar] [PubMed]
- Rusell, G.; Delgado, V. Determinación de la concentración letal media (CL50) de cuatro detergentes domésticos biodegradables en Laeonereis culveri (Webster 1879) (polychaeta: Annelida). Rev. Int. Contam. Ambient. 2012, 28, 137–144. [Google Scholar]
- Espina, S.; Días, F.; Rosas, C. Detergent influence on the energy balance of Ctenopharyngodon idella through a chronicc bioassay. Rev. Int. Contam. Ambient. 1986, 2, 25–37. [Google Scholar]
- De la Parra-Guerra, A.; Stürzenbaum, S.; Olivero-Verbel, J. Intergenerational toxicity of nonylphenol ethoxylate (NP-9) in Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 2020, 197, 110588. [Google Scholar] [CrossRef]
- Leung, M.C.; Williams, P.L.; Benedetto, A.; Au, C.; Helmcke, K.J.; Aschner, M.; Meyer, J.N. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008, 106, 5–28. [Google Scholar] [CrossRef]
- Hunt, P.R. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017, 37, 50–59. [Google Scholar] [CrossRef]
- Williams, P.L.; Dusenbery, D.B. Aquatic toxicity testing using the nematode, Caenorhabditis elegans. Environ. Toxicol. Chem. 1990, 9, 1285–1290. [Google Scholar] [CrossRef]
- Dennington, R.D.; Keith, T.A.; Millam, J.M. GaussView 5.0; Gaussian, Inc.: Wallingford, CT, USA, 2008; p. 340. [Google Scholar]
- Becke, A.D. Density-Functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- McGrath, M.P.; Radom, L. Extension of Gaussian-1 (G1) theory to bromine-containing molecules. J. Chem. Phys. 1991, 94, 511. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, S.; Yu, B.; Lui, Y.; Huang, K.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic. Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef] [PubMed]
- Österberg, F.; Morris, G.; Sanner, M.; Olson, A.; Goodsell, D. Automated docking to multiple target structures: Incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins 2002, 46, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Ghersi, D.; Sanche, R. Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins 2009, 20, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for ligand-receptor docking. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; Chapter 8. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Durdagi, S.; Şentürk, M.; Ekinci, D.; Balaydın, H.; Göksu, S.; Küfrevioğlu, Ö.İ.; Innocenti, A.; Scozzafava, A.; Supuran, C.T. Kinetic and docking studies of phenol-based inhibitors of carbonic anhydrase isoforms I, II, IX and XII evidence a new binding mode within the enzyme active site. Bioorganic Med. Chem. 2011, 19, 1381–1389. [Google Scholar] [CrossRef]
- Coronado-Posada, N.; Mercado-Camargo, J.; Olivero-Verbel, J. In Silico Analysis to Identify Molecular Targets for Chemicals of Concern: The Case Study of Flocoumafen, an Anticoagulant Pesticide. Environ. Toxicol. Chem. 2021, 40, 2034–2043. [Google Scholar] [CrossRef]
- Montes-Grajales, D.; Olivero-Verbel, J. Computer-aided identification of novel protein targets of Bisphenol A. Toxicol. Lett. 2013, 222, 312–320. [Google Scholar] [CrossRef]
- Mark, J.A.; Teemu, M.; Roland, S.; Szilárd, P.; Jeremy, C.S.; Berk, H.; Erik, L. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Poger, D.; Mark, A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: A comparison with experiment. J. Chem. Theory Comput. 2010, 6, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef]
- Stiernagle, T. Maintenance of C. elegans; WormBook: Pasadena, CA, USA, 2006; pp. 1–11. [Google Scholar] [CrossRef]
- Tejeda-Benitez, L.; Olivero-Verbel, J. Caenorhabditis elegans, a biological model for research in toxicology. Rev. Environ. Contam. Toxicol. 2016, 237, 1–35. [Google Scholar] [CrossRef]
- Olivero-Verbel, J.; De la Parra-Guerra, A.; Caballero-Gallardo, K.; Sierra-Marquez, L.; Fuentes-Lopez, K.; Franco-Marmolejo, J.; Jannasch, A.; Sepulveda, M.; Stashenko, E. The aqueous extract of Fridericia chica grown in northern Colombia ameliorates toxicity induced by Tergitol on Caenorhabditis elegans. Comp. Biochem. Physiol. C 2021, 244, 109026. [Google Scholar] [CrossRef]
- Roh, J.; Park, Y.; Park, K.; Choi, J. Ecotoxicological investigation of CeO2 and TiO2 nanoparticles on the soil nematode Caenorhabditis elegans using gene expression, growth, fertility, and survival as endpoints. Environ. Toxicol. Pharmacol. 2010, 29, 167–172. [Google Scholar] [CrossRef]
- Duran-Izquierdo, M.; Taboada-Alquerque, M.; Sierra-Marquez, L.; Alvarez-Ortega, N.; Stashenko, E.; Olivero-Verbel, J. Hydroalcoholic extract of Haematoxylum brasiletto protects Caenorhabditis elegans from cadmium-induced toxicity. BMC Complement. Med. Ther. 2022, 22, 184. [Google Scholar] [CrossRef]
- Sharma, J.; Bhardwaj, V.K.; Singh, R.; Rajendran, V.; Purohit, R.; Kumar, S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non-structural protein-15 of SARS-CoV-2. Food Chem. 2021, 346, 128933. [Google Scholar] [CrossRef] [PubMed]
- Furman, M.; Krueger, L.; Linden, M. Release of soluble CD40L from platelets is regulated by glycoprotein IIb/IIIa and actin polymerization. J. Am. Coll. Cardiol. 2004, 43, 2319–2325. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, T.; Matsushima, M.; Hashimoto, N.; Imaizumi, K.; Hasegawa, Y. CD40/CD40 ligand interactions in immune responses and pulmonary immunity. Nagoya J. Med. Sci. 2011, 73, 69–78. [Google Scholar] [PubMed]
- BASF—Optical brighteners. Focus Pigment. 2022, 2022, 4. [CrossRef]
- Zhu, Y.; Lu, H.; He, D.; Yang, Z. Synthesis, fluorescence properties and applications of two novel oxadiazole-based stilbene optical brighteners as UV protectants for insect baculovirus. J. Photochem. Photobiol. B Biol. 2013, 125, 8–12. [Google Scholar] [CrossRef]
- Liu, M.; Lin, H.; Yang, M.; Lai, M.; Chang Ch Liu, H.; Shiao, P.; Chen, I.; Chen, J. Thermal and fluorescent properties of optical brighteners and their whitening effect for pelletization of cycloolefin copolymers. Mater. Lett. 2006, 60, 2132–2137. [Google Scholar] [CrossRef]
- Salas, H.; Gutiérrez-Bouzán, C.; López-Grimau, V.; Vilaseca, M. Respirometric Study of Optical Brighteners in Textile Wastewater. Materials 2019, 12, 785. [Google Scholar] [CrossRef]
- Blanco, M.; Jiménez, L.; Valverde, I. Degradation of a stilbene-type fluorescent whitening agent with hypochlorite: Identification of the degradation products by capillary electrophoresis. Environ. Toxicol. Chem. 2001, 20, 2193–2197. [Google Scholar]
- Wisniewska, M.; Karlberg, T.; Lehtiö, L.; Johansson, I.; Kotenyova, T.; Moche, M.; Schüler, H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS ONE 2010, 5, e8625. [Google Scholar] [CrossRef]
- Fang, C.; Kuo, H.; Pan, T.; Yu, F.; Yih, L. HSP70 regulates the function of mitotic centrosomes, Cell. Mol. Life Sci. 2016, 73, 3949–3960. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, C.; Steinbeck, C.; Wohlfahrt, G. Classification and comparison of ligand-binding sites derived from grid-mapped knowledge-based potentials. J. Mol. Graph. Model. 2006, 24, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Mejía, M. Estudio in silico e in vivo de la Actividad Farmacológica de Nuevos Compuestos de Tipo Pirazolopiridinas. Master’s Thesis, Universidad de Valle, Cali, Colombia, 27 January 2023. [Google Scholar]
- Gul, A.; Pastuszak, A.; Kabasakal, L.; Bates, J.; Altinay, S.; Celik, D.; Semercioz, A.; Serefoglu, E. Tadalafil Preserves Penile Nitric Oxide Synthase from Detrimental Effect of Paroxetine in Rats. Eurasian J. Emerg. Med. 2019, 51, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Sengupta, P. The Role of Nitric Oxide on Male and Female Reproduction. Malays. J. Med. Sci. 2022, 29, 18–30. [Google Scholar] [CrossRef]
- Croce, R.; Cinà, F.; Lombardo, A.; Crispeyn, G.; Cappelli, C.I.; Vian, M.; Maiorana, S.; Benfenati, E.; Baderna, D. Aquatic toxicity of several textile dye formulations: Acute and chronic assays with Daphnia magna and Raphidocelis subcapitata. Ecotoxicol. Environ. Saf. 2017, 144, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Matschke, G.H.; Fagerstone, K.A. Effects of a new rodenticide, benzenesulfonic acid hydrazide, on prenatal mice. J. Toxicol. Environ. Health 1977, 3, 407–411. [Google Scholar] [CrossRef]
- Schafer, E. The acute oral toxicity of 369 pesticidal, pharmaceutical, and other chemicals to wild birds. Toxicol. Appl. Pharmacol. 1972, 21, 315–330. [Google Scholar] [CrossRef]
- Inokuchi, A.; Yamamoto, R.; Morita, F.; Takumi, S.; Matsusaki, H.; Ishibashi, H.; Arizono, K. Effects of lithium on growth, maturation, reproduction and gene expression in the nematode Caenorhabditis elegans. J. Appl. Toxicol. 2015, 35, 999–1006. [Google Scholar] [CrossRef]
- Kapulkin, W.J.; Hiester, B.G.; Link, C.D. Compensatory regulation among ER chaperones in C. elegans. FEBS Lett. 2007, 581, 5952. [Google Scholar] [CrossRef]
- Avila, D.; Benedetto, A.; Au, C.; Bornhorst, J.; Aschner, M. Involvement of heat shock proteins on Mn-induced toxicity in Caenorhabditis elegans. BMC Pharmacol. Toxicol. 2016, 17, 54. [Google Scholar] [CrossRef]
- Morrison, J.; Friday, A.; Henderson, M.; Hao, E.; Keiper, B. Induction of cap-independent BiP (hsp-3) and Bcl-2 (ced-9) translation in response to eIF4G (IFG-1) depletion in C. elegans. Translation 2014, 2, e28935. [Google Scholar] [CrossRef]
- Zha, J.; Ying, M.; Alexander-Floyd, J.; Gidalevitz, T. HSP-4/BiP expression in secretory cells is regulated by a developmental program and not by the unfolded protein response. PLoS Biol. 2019, 17, e3000196. [Google Scholar] [CrossRef]
- Deng, L. The Roles of Inhibition in C. elegans Locomotion. Doctoral Dissertations, New Jersey Institute of Technology, Newark, NJ, USA, 2020; p. 1558. Available online: https://digitalcommons.njit.edu/dissertations/1558 (accessed on 10 November 2022).
- Zhen, M.; Aravinthan, S. C. elegans locomotion: Small circuits, complex functions. Curr. Opin. Neurobiol. 2015, 33, 117–126. [Google Scholar] [CrossRef]
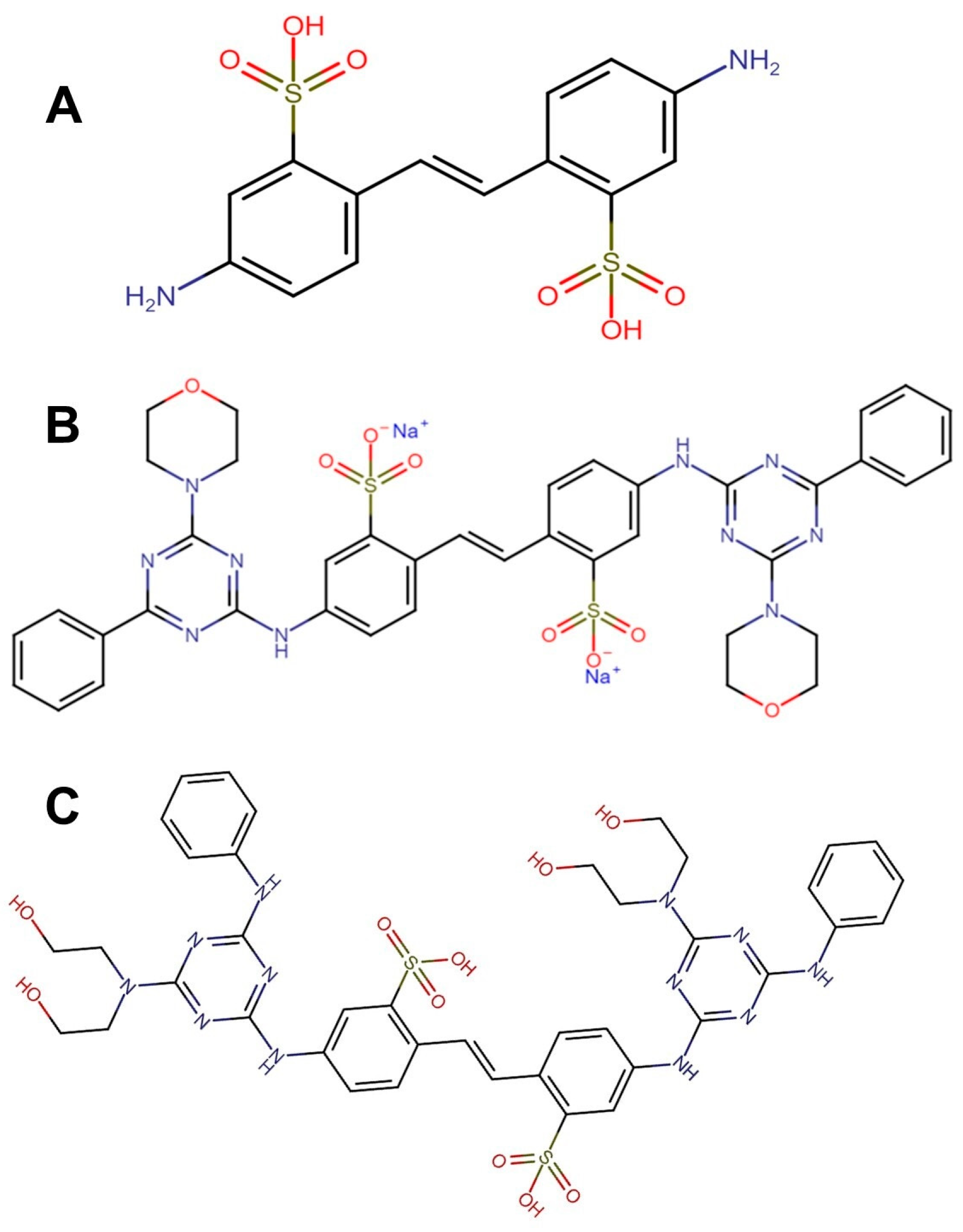



| DAST | FB-71 | FB-28 | ||||||
|---|---|---|---|---|---|---|---|---|
| PDB ID | Description | Average Affinity Values * (kcal/mol) | PDB ID | Description | Average Affinity Values (kcal/mol) | PDB ID | Description | Average Affinity Values (kcal/mol) |
| 3JXU | Heat shock 70 kDa protein 1A | −11.2 | 3LKJ | TNF Family Cytokine CD40 ligand | −15.3 | 3LKJ | TNF Family Cytokine CD40 ligand | −13.3 |
| 1P8D | Oxysterols receptor LXR-beta | −10.3 | 1NSI | Nitric oxide synthase, inducible | −14.5 | 2VQQ | Histone deacetylase 4 | −12.6 |
| 4MJH | Heat shock protein beta-1 (Hsp27) | −10.3 | 1F9Q | Platelet factor 4 | −14.5 | 3O96 | RAC-alpha serine/threonine-protein kinase | −11.9 |
| 3LKJ | TNF Family Cyokine CD40 ligand | −10.2 | 1PL1 | glutathione transferase (GST) A1-1 | −14.4 | 1LYW | Cathepsin D | −11.9 |
| 4EY7 | Acetylcholinesterase | −10.2 | 2OC2 | Angiotensin-converting enzyme | −14.4 | 1O8A | Angiotensin-converting enzyme | −11.8 |
| 1S3X | Heat shock 70 kDa protein 1A ATPase domain | −10.2 | 2VQQ | Histone deacetylase 4 | −14.4 | 3EAH | Nitric oxide synthase, endothelial | −11.7 |
| 4XRY | Human Cytochrome P450 2D6 | −10.2 | 1O8A | Angiotensin-converting enzyme | −14.3 | 3CO6 | Forkhead box protein O1 | −11.7 |
| 4EY6 | Acetylcholinesterase | −10.1 | 3E7G | Nitric oxide synthase, inducible | −14.2 | 4QTD | Mitogen-activated protein kinase 8 | −11.7 |
| 2XSI | Human serum albumin | −10.1 | 5GPG | Peptidyl-prolyl cis-trans isomerase FKBP3 | −14.0 | 4D75 | Cytochrome P450 3A4 | −11.6 |
| 1RY8 | Prostaglandin F synthase | −10.0 | 1LYW | Cathepsin D | −14.0 | 1NSI | Nitric oxide synthase, inducible | −11.6 |
| 4IDT | Mitogen-activated protein kinase kinase kinase 14 | −10.0 | 3CO6 | Forkhead box protein O1 | −13.8 | 1Z57 | Dual specificity protein kinase CLK1 | −11.5 |
| 4NRE | UGGGGU quadruplexes | −9.8 | 3LN1 | Prostaglandin G/H synthase 2 | −13.7 | 3UA1 | Cytochrome P450 3A4 | −11.4 |
| 1NO6 | Protein Tyrosine Phosphatase 1B | −9.8 | 3UA1 | Cytochrome P450 3A4 | −13.7 | 4EY7 | Acetylcholinesterase | −11.4 |
| 3MDY | Bone morphogenetic protein receptor type-1B | −9.8 | 3O96 | RAC-alpha serine/threonine-protein kinase | −13.7 | 5KDI | Pleckstrin homology domain-containing family A | −11.4 |
| 3EAH | Nitric oxide synthase, endothelial | −9.7 | 2PVR | Casein kinase II subunit alpha | −13.7 | 4D6Z | Cytochrome P450 3A4 | −11.3 |
| 5E6H | Lysine-specific demethylase 5A | −9.7 | 3PAG | Beta-lactamase | −13.7 | 4IDT | Mitogen-activated protein kinase kinase kinase 14 | −11.3 |
| 4R0A | Toll-like receptor 8 | −9.7 | 4QBZ | Toll-like receptor 8 | −13.7 | 3HOK | Heme oxygenase 1 | −11.3 |
| 1NSI | Nitric oxide synthase, inducible | −9.7 | 3W3J | Toll-like receptor 8 | −13.6 | 3NXQ | Angiotensin-converting enzyme | −11.2 |
| 1UHL | Retinoic acid receptor RXR-beta | −9.7 | 3QX3 | DNA topoisomerase 2-beta | −13.5 | 2OC2 | Angiotensin-converting enzyme | −11.1 |
| 2PQ5 | Dual specificity protein phosphatase 13 isoform B | −9.7 | 4IDT | Mitogen-activated protein kinase kinase kinase 14 | −13.5 | 4XII | Cholinesterase | −11.1 |
| 3O3U | Advanced glycosylation end product-specific receptor | −9.7 | 1JQE | Histamine Methyltransferase | −13.5 | 1ROS | Macrophage metalloelastase | −11.1 |
| 3UA1 | Cytochrome P450 3A4 | −9.7 | 4C8B | Receptor-interacting serine/threonine-protein | −13.5 | 2D0T | Indoleamine 2,3-dioxygenase 1 | −11.1 |
| 1P0I | Butyryl cholinesterase | −9.6 | 4FM9 | DNA topoisomerase 2-alpha | −13.5 | 4FYR | Aminopeptidase N | −11.1 |
| 2C66 | Amine oxidase [flavin-containing] B | −9.6 | 4TUH | Bcl-2-like protein 1 | −13.5 | 4R6A | Toll-like receptor 8 | −11.0 |
| 3O96 | RAC-alpha serine/threonine-protein kinase | −9.6 | 3EAH | Nitric oxide synthase, endothelial | −13.4 | 3W3J | Toll-like receptor 8 | −11.0 |
| Complexes | |||||
|---|---|---|---|---|---|
| Energy Components | Hsp70-1-ADP | Hsp70-1-DAST | CD40 Ligand-LKJ | CD40 Ligand-FB-71 | CD40 Ligand-FB-28 |
| Total Binding Free Energy (kcal/mol) | −25.96 ± 1.02 | −10.23 ± 0.55 | −17.56 ± 0.63 | −10.53 ±0.50 | −18.03 ± 0.67 |
| Van der Waals Energy (kcal/mol) | −27.17 ± 0.94 | −38.76 ± 0.46 | −39.98 ± 0.58 | −28.71 ± 0.72 | −44.29 ± 1.30 |
| Electrostatic Energy (kcal/mol) | −72.35 ± 2.54 | −53.78 ± 1.09 | −116.92 ± 1.98 | 111.31 ± 1.76 | −34.88 ± 1.37 |
| Polar solvation energy (kcal/mol) | 77.92 ± 2.24 | 88.19 ± 0.83 | 144.13 ± 0.07 | −89.34 ± 1.78 | 67.51 ± 1.34 |
| SASA energy (kcal/mol) | −4.36 ± 0.092 | −5.88 ± 0.035 | −4.78 ± 0.50 | −3.78 ± 0.13 | −6.36 ± 0.14 |
| ΔG gas | −99.5 ± 2.66 | −92.55 ± 1.01 | −156.90 ± 2.28 | 82.59 ± 1.82 | −79.17 ± 1.63 |
| ΔG solv. | 73.56 ± 2.18 | 82.32 ± 0.83 | 139.34 ± 1.90 | −93.13 ± 1.71 | 61.14 ± 1.25 |
| OB Concentration (µM) | Relative Expression for GFP Transgenic Strains | |||||
|---|---|---|---|---|---|---|
| Cellular Stress | Oxidative Stress | Metabolism | ||||
| hsp-3 | hsp-4 | gpx-4 | sod-4 | gst-1 | ||
| DAST | 0 | 1.00 ± 0.04 | 1.00 ± 0.06 | 1.00 ± 0.05 | 1.00 ± 0.04 | 1.00 ± 0.09 |
| 50 | 1.04 ± 0,16 | 1.06 ± 0.13 | 1.19 ± 0.10 | 0.84 ± 0.26 | 0.86 ± 0.24 | |
| 100 | 1.00 ± 0.17 | 1.08 ± 0.19 | 1.60 ± 0.60 * | 0.98 ± 0.38 | 1.62 ± 0.24 * | |
| 250 | 1.21 ± 0.13 | 1.24 ± 0.19 * | 2.09 ± 0.43 * | 1.30 ± 0.29 | 1.04 ± 0.20 | |
| 500 | 1.55 ± 0.32 * | 1.41 ± 0.20 * | 3.20 ± 0.61 * | 3.06 ± 0.90 * | 2.11 ± 0.27 * | |
| FB-71 | 0 | 1.00 ± 0.05 | 1.00 ± 0.00 | 1.00 ± 0.06 | 1.00 ± 0.11 | 1.00 ± 0.04 |
| 50 | 1.11 ± 0.17 | 1.21 ± 0.19 | 1.03 ± 0.25 | 1.32 ± 0.36 | 0.97 ± 0.09 | |
| 100 | 1.14 ± 0.15 | 1.20 ± 0.14 | 1.17 ± 0.37 | 1.45 ± 0.48 * | 0.96 ± 0.13 | |
| 250 | 1.14 ± 0.16 | 1.23 ± 0.31 * | 1.35 ± 0.43 | 1.40 ± 0.23 * | 1.03 ± 0.07 | |
| 500 | 1.46 ± 0.19 * | 1.39 ± 0.18 * | 1.70 ± 0.28 * | 1.70 ± 0.29 * | 1.08 ± 0.08 | |
| FB-28 | 0 | 1.00 ± 0.04 | 1.00 ± 0.02 | 1.00 ± 0.06 | 1.00 ± 0.05 | 1.00 ± 0.09 |
| 50 | 1.05 ± 0.19 | 1.27 ± 0.23 | 1.04 ± 0.24 | 1.15 ± 0.24 | 1.23 ± 0.20 | |
| 100 | 1.06 ± 0.19 | 1.26 ± 0.34 | 0.93 ± 0.17 | 1.08 ± 0.15 | 1.27± 0.20 | |
| 250 | 1.10 ± 0.17 | 1.20 ± 0.28 | 0.98 ± 0.10 | 1.16 ± 0.13 | 1.28 ± 0.41 | |
| 500 | 1.12 ± 0.13 | 1.19 ± 0.23 | 0.96 ± 0.16 | 1.11 ± 0.14 | 1.20 ± 0.26 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro-Sierra, I.; Duran-Izquierdo, M.; Sierra-Marquez, L.; Ahumedo-Monterrosa, M.; Olivero-Verbel, J. Toxicity of Three Optical Brighteners: Potential Pharmacological Targets and Effects on Caenorhabditis elegans. Toxics 2024, 12, 51. https://doi.org/10.3390/toxics12010051
Castro-Sierra I, Duran-Izquierdo M, Sierra-Marquez L, Ahumedo-Monterrosa M, Olivero-Verbel J. Toxicity of Three Optical Brighteners: Potential Pharmacological Targets and Effects on Caenorhabditis elegans. Toxics. 2024; 12(1):51. https://doi.org/10.3390/toxics12010051
Chicago/Turabian StyleCastro-Sierra, Isel, Margareth Duran-Izquierdo, Lucellys Sierra-Marquez, Maicol Ahumedo-Monterrosa, and Jesus Olivero-Verbel. 2024. "Toxicity of Three Optical Brighteners: Potential Pharmacological Targets and Effects on Caenorhabditis elegans" Toxics 12, no. 1: 51. https://doi.org/10.3390/toxics12010051





