Integrating Epigenetics, Proteomics, and Metabolomics to Reveal the Involvement of Wnt/β-Catenin Signaling Pathway in Oridonin-Induced Reproductive Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagent and Cell Culture
2.2. MTT and LDH Release Assay for Determining Cytotoxic Effects
2.3. RNA Extraction and High-Throughput Sequence
2.4. Molecular Docking and Molecular Dynamics (MD) Simulation
2.5. Proteomics Sample Processing and LC-MS/MS Analysis
2.6. Sample Preparation and Analysis for Metabolomics
2.7. ROS Measurement
2.8. DNA Damage Assay by γ-H2AX Immunofluorescence
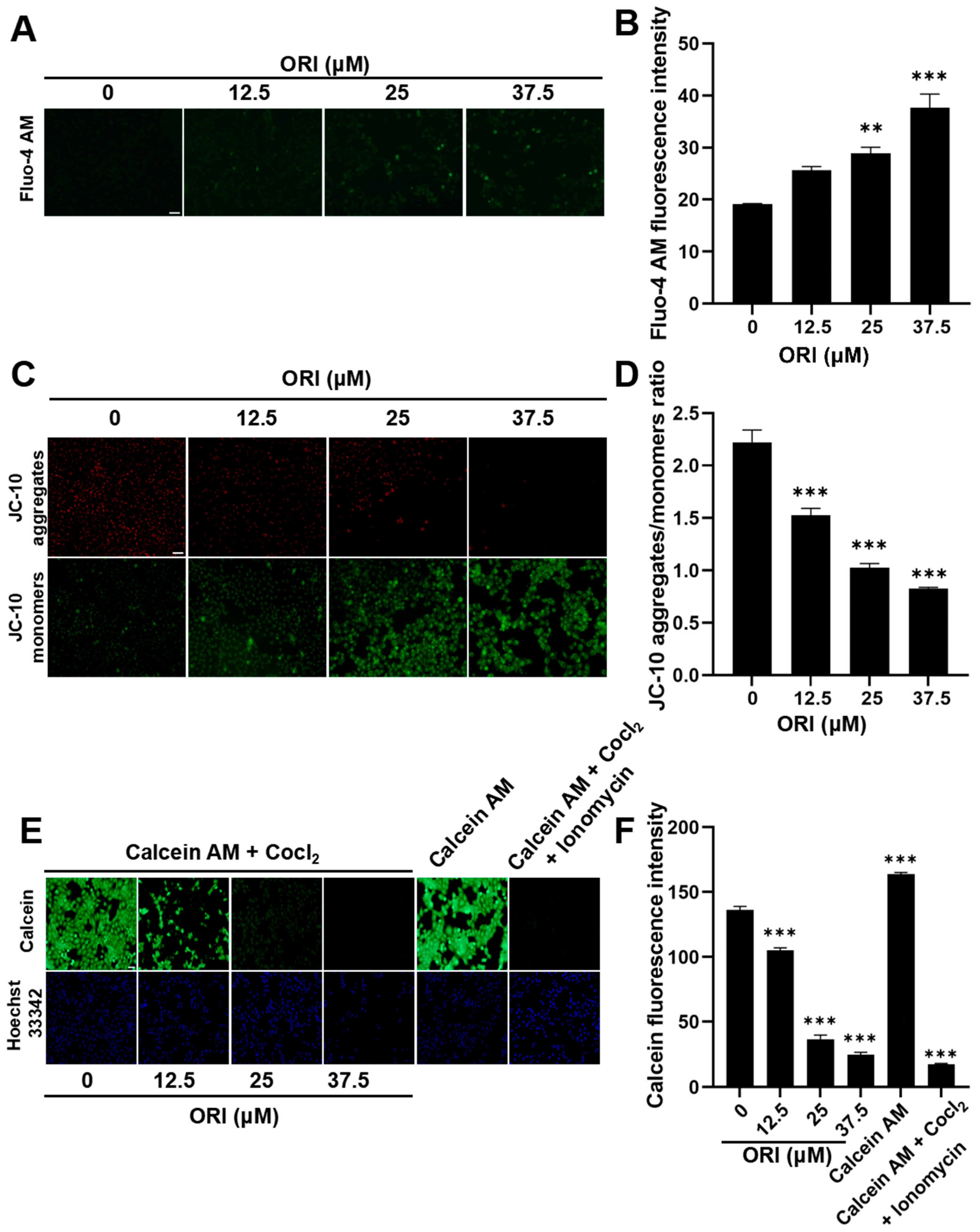
2.9. Detection of Calcium by Fluo-4 AM
2.10. Detection of MMP and mPTP
2.11. RT-qPCR
2.12. Western Blotting Assay
2.13. Statistical Analysis
3. Results
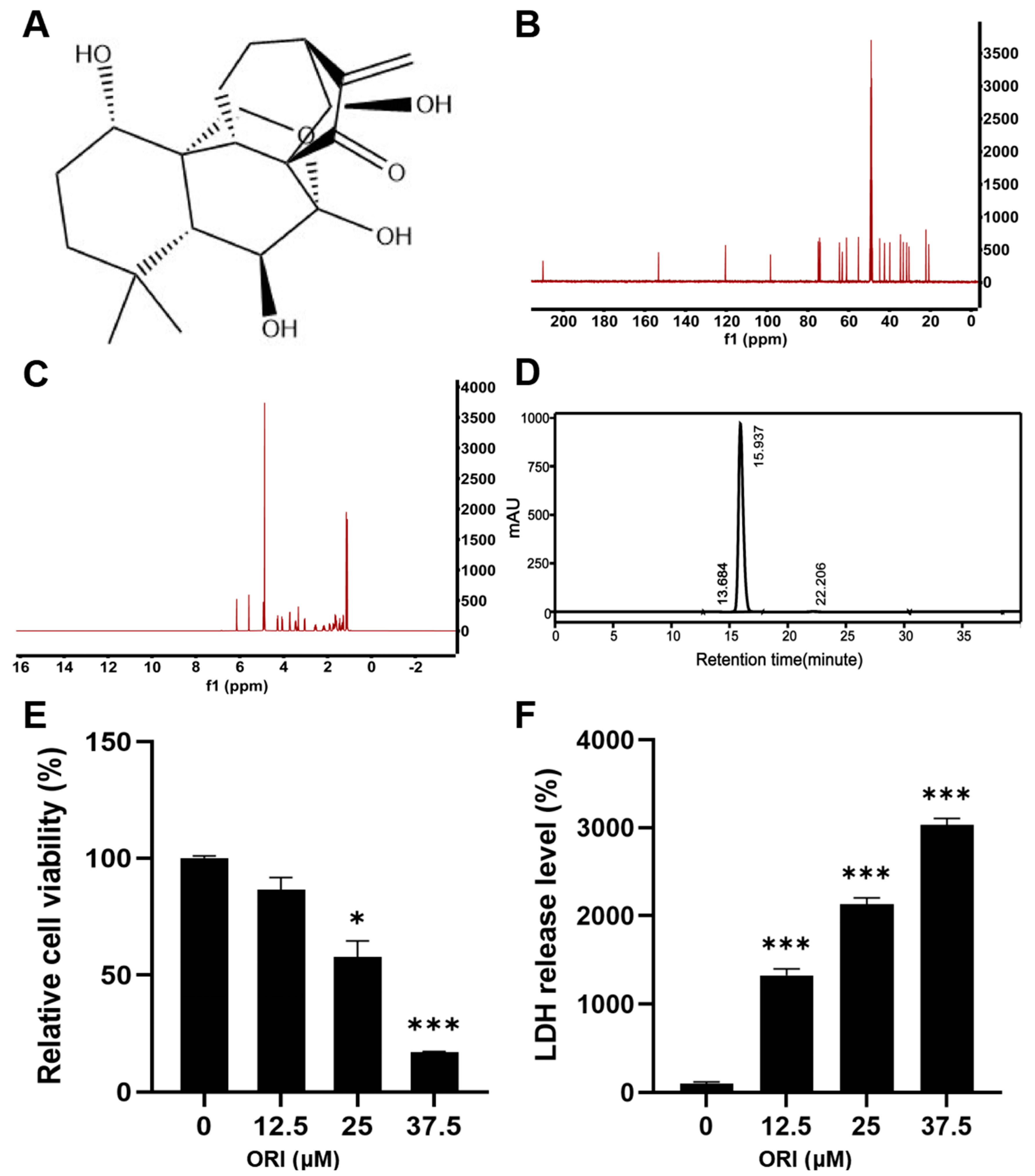
3.1. The Cytotoxic Effect of Oridonin on HTR-8/SVneo Cells
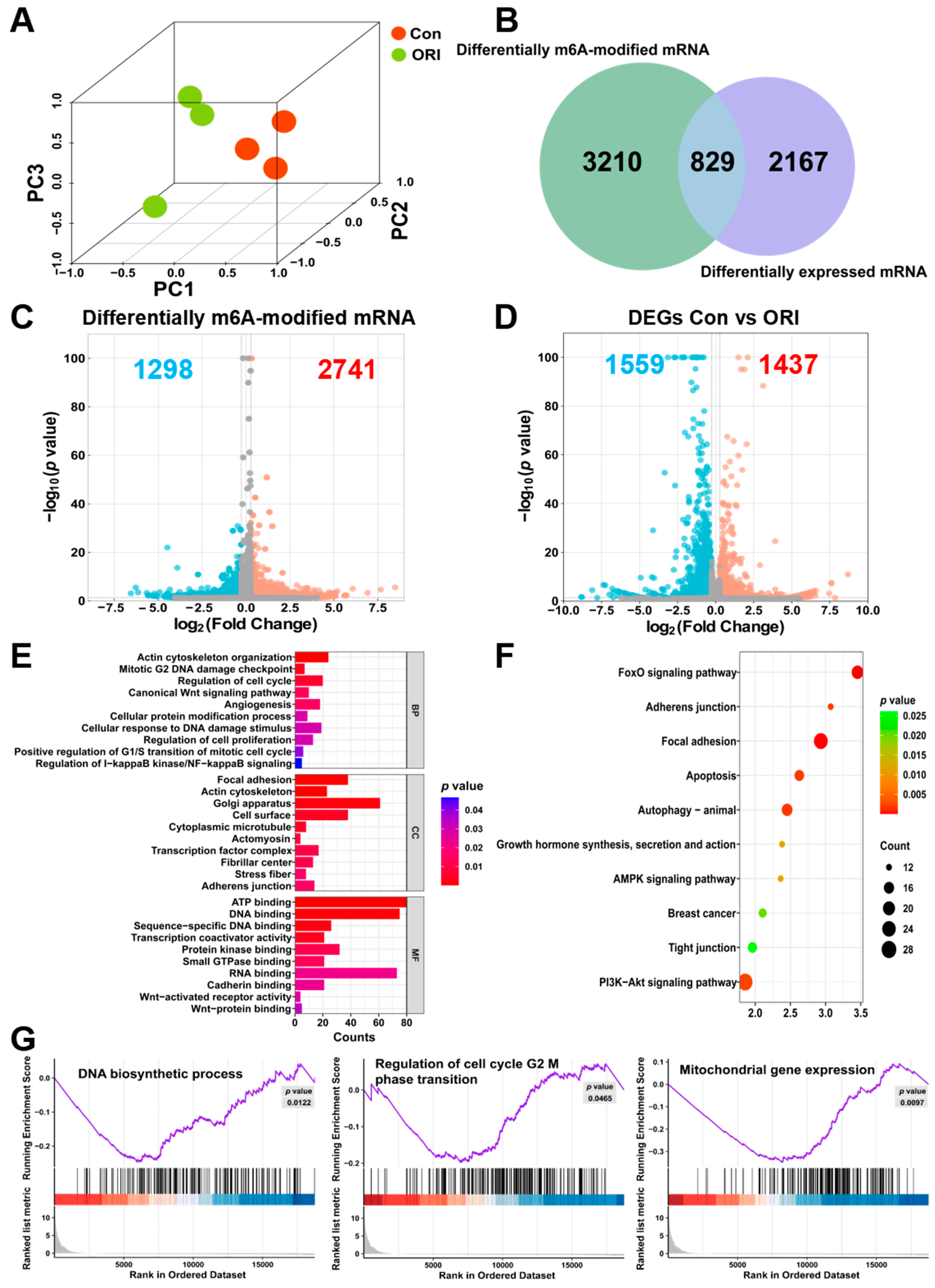
3.2. Transcriptome-Wide Assessment of m6A Modification Post-Oridonin Treatment in HTR-8/SVneo Cells
3.3. Combined Analysis of RNA-seq and MeRIP-seq
3.4. Potential Regulators of RNA m6A Methylation
3.5. Molecular Interactions and Molecular Dynamics between Oridonin and m6A Regulatory Proteins
3.6. Results of the Analysis of Differentially Expressed Proteins (DEPs)
3.7. Analysis Results from GO Annotation and Enrichment of KEGG Pathways
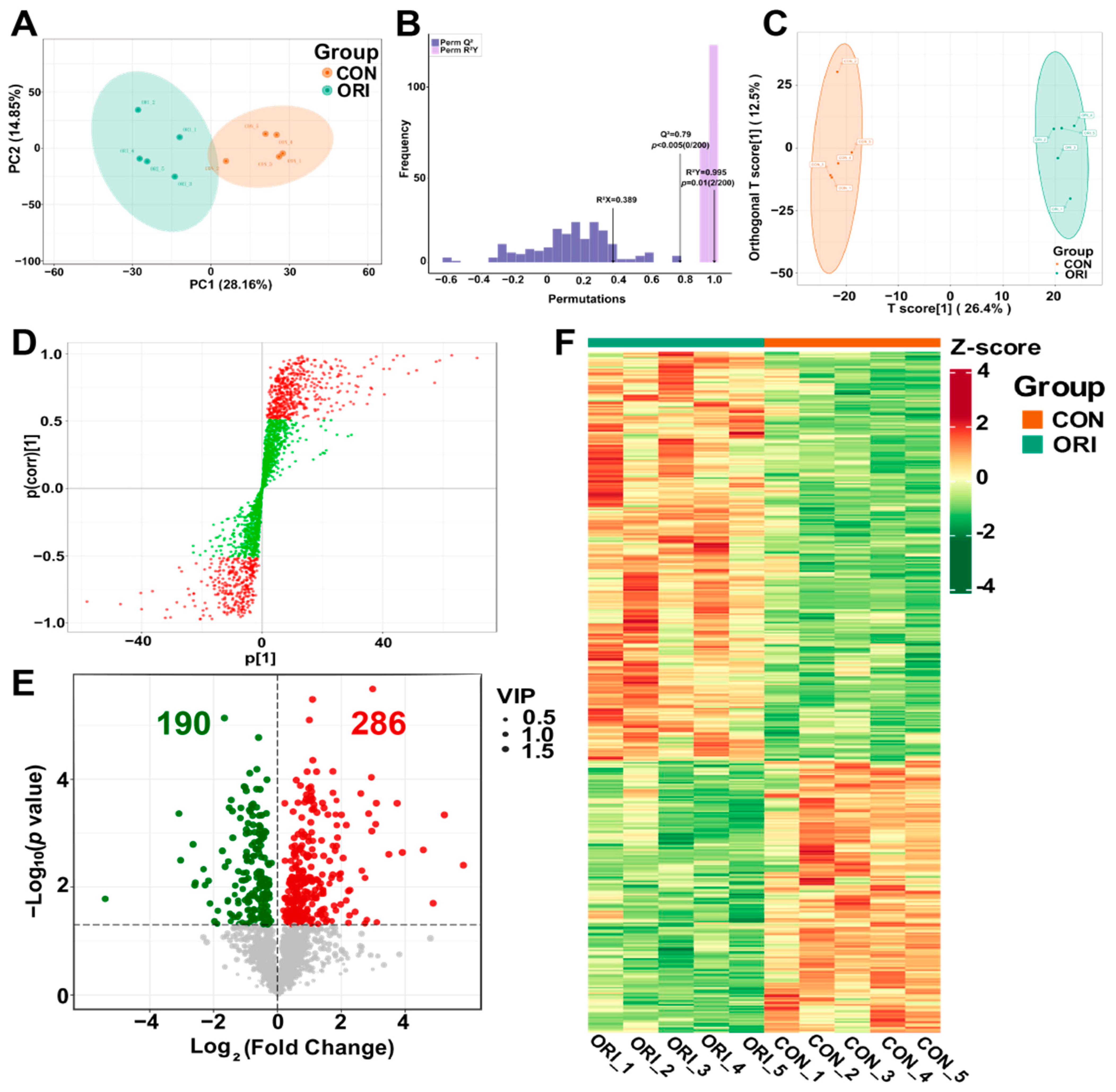
3.8. Analysis of Metabolomics Data between Oridonin-Treated and Control Groups
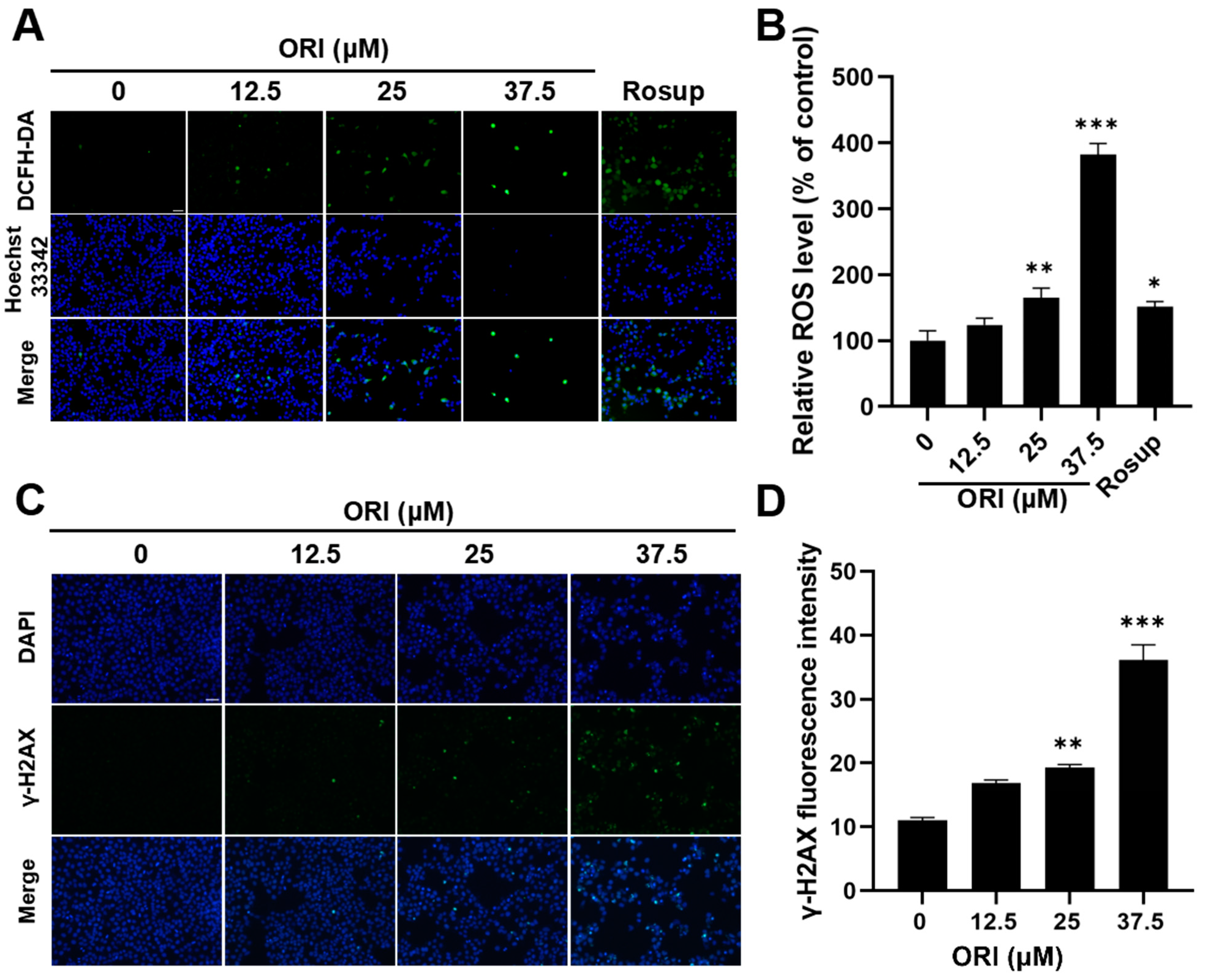
3.9. Oxidative Stress and DNA Damage Induced by Oridonin
3.10. The Adverse Impacts of Oridonin Therapy on Mitochondrial Function
3.11. Expression Levels of mRNA and Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Wang, L.; Zi, Y.; Zhang, L.; Guo, Y.; Huang, Y. Oridonin effectively reverses the drug resistance of cisplatin involving induction of cell apoptosis and inhibition of MMP expression in human acute myeloid leukemia cells. Saudi J. Biol. Sci. 2017, 24, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ding, C.; Ye, N.; Liu, Z.; Wold, E.A.; Chen, H.; Wild, C.; Shen, Q.; Zhou, J. Discovery and development of natural product oridonin-inspired anticancer agents. Eur. J. Med. Chem. 2016, 122, 102–117. [Google Scholar] [CrossRef]
- Li, X.; Zhang, C.T.; Ma, W.; Xie, X.; Huang, Q. Oridonin: A review of its pharmacology, pharmacokinetics and toxicity. Front. Pharmacol. 2021, 12, 645824. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Dai, M.; Nai, J.; Zhu, L.; Sheng, H. Solubility and bioavailability enhancement of oridonin: A review. Molecules 2020, 25, 332. [Google Scholar] [CrossRef]
- Xu, L.; Bi, Y.; Xu, Y.; Zhang, Z.; Xu, W.; Zhang, S.; Chen, J. Oridonin inhibits the migration and epithelial-to-mesenchymal transition of small cell lung cancer cells by suppressing FAK-ERK1/2 signalling pathway. J. Cell. Mol. Med. 2020, 24, 4480–4493. [Google Scholar] [CrossRef]
- Park, H.; Jeong, Y.J.; Han, N.K.; Kim, J.S.; Lee, H.J. Oridonin enhances radiation-induced cell death by promoting DNA damage in non-small cell lung cancer cells. Int. J. Mol. Sci. 2018, 19, 2378. [Google Scholar] [CrossRef]
- Ying, Y. Effects of Oridonin on Estrogen and Antioxidative Function of Ovarian Granulosa Cells. Master’s Thesis, Beijing University of Agriculture, Beijing, China, 2017. [Google Scholar]
- Rosell-Hidalgo, A.; Moore, A.L.; Ghafourian, T. Prediction of drug-induced mitochondrial dysfunction using succinate-cytochrome c reductase activity, QSAR and molecular docking. Toxicology 2023, 485, 153412. [Google Scholar] [CrossRef]
- Kang, N.; Zhang, J.H.; Qiu, F.; Tashiro, S.; Onodera, S.; Ikejima, T. Inhibition of EGFR signaling augments oridonin-induced apoptosis in human laryngeal cancer cells via enhancing oxidative stress coincident with activation of both the intrinsic and extrinsic apoptotic pathways. Cancer Lett. 2010, 294, 147–158. [Google Scholar] [CrossRef]
- Quinlan, C.L.; Orr, A.L.; Perevoshchikova, I.V.; Treberg, J.R.; Ackrell, B.A.; Brand, M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012, 287, 27255–27264. [Google Scholar] [CrossRef]
- Takeyama, N.; Matsuo, N.; Tanaka, T. Oxidative damage to mitochondria is mediated by the Ca2+-dependent inner-membrane permeability transition. Biochem. J. 1993, 294, 719–725. [Google Scholar] [CrossRef]
- Dykens, J.A.; Marroquin, L.D.; Will, Y. Strategies to reduce late-stage drug attrition due to mitochondrial toxicity. Expert Rev. Mol. Diagn. 2007, 7, 161–175. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef]
- Jati, S.; Sarraf, T.R.; Naskar, D.; Sen, M. Wnt signaling: Pathogen incursion and immune defense. Front. Immunol. 2019, 10, 2551. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.; Vincan, E. The Wnt signaling pathways and cell adhesion. Front. Biosci. 2012, 17, 784–804. [Google Scholar] [CrossRef] [PubMed]
- Preda, V.; Larkin, S.J.; Karavitaki, N.; Ansorge, O.; Grossman, A.B. The Wnt signalling cascade and the adherens junction complex in craniopharyngioma tumorigenesis. Endocr. Pathol. 2015, 26, 1–8. [Google Scholar] [CrossRef] [PubMed]
- May-Simera, H.L.; Kelley, M.W. Cilia, Wnt signaling, and the cytoskeleton. Cilia 2012, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Pope, J.L.; Ahmad, R.; Bhat, A.A.; Washington, M.K.; Singh, A.B.; Dhawan, P. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Mol. Cancer 2014, 13, 167. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.K.; Liu, R.H.; Jin, H.Z.; Liu, X.R.; Ye, J.; Shan, L.; Zhang, W.D. “Omics” in pharmaceutical research: Overview, applications, challenges, and future perspectives. Chin. J. Nat. Med. 2015, 13, 3–21. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, J.; Ma, J.; Wei, Z.; Wang, Y.; Song, B.; Meng, J.; Jia, G.; de Magalhães, J.P.; Rigden, D.J.; et al. DirectRMDB: A database of post-transcriptional RNA modifications unveiled from direct RNA sequencing technology. Nucleic Acids Res. 2023, 51, D106–D116. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, X.; Bao, W.; Hong, X.; Li, C.; Lu, J.; Zhang, D.; Zhu, A. Effect of humantenine on mRNA m6A modification and expression in human colon cancer cell line HCT116. Genes 2022, 13, 781. [Google Scholar] [CrossRef]
- Ma, J.; Song, B.; Wei, Z.; Huang, D.; Zhang, Y.; Su, J.; de Magalhães, J.P.; Rigden, D.J.; Meng, J.; Chen, K. m5C-Atlas: A comprehensive database for decoding and annotating the 5-methylcytosine (m5C) epitranscriptome. Nucleic Acids Res. 2022, 50, D196–D203. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Shen, X.; Chen, M.; Zhang, J.; Lin, Y.; Gao, X.; Tu, J.; Chen, K.; Zhu, A.; Xu, S. Unveiling the impact of ApoF deficiency on liver and lipid metabolism: Insights from transcriptome-wide m6A methylome analysis in mice. Genes 2024, 15, 347. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Klungland, A.; Dahl, J.A.; Greggains, G.; Fedorcsak, P.; Filipczyk, A. Reversible RNA modifications in meiosis and pluripotency. Nat. Methods 2016, 14, 18–22. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Wu, Y.; Bao, W.; Ren, J.; Li, C.; Chen, M.; Zhang, D.; Zhu, A. Integrated profiles of transcriptome and mRNA m6A modification reveal the intestinal cytotoxicity of Aflatoxin B1 on HCT116 cells. Genes 2022, 14, 79. [Google Scholar] [CrossRef]
- Reyfman, P.A.; Walter, J.M.; Joshi, N.; Anekalla, K.R.; McQuattie-Pimentel, A.C.; Chiu, S.; Fernandez, R.; Akbarpour, M.; Chen, C.I.; Ren, Z.; et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1517–1536. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ishfaq, M.; Xu, L.; Xia, C.; Chen, C.; Li, J. METTL3/m6A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front. Pharmacol. 2019, 10, 517. [Google Scholar] [CrossRef] [PubMed]
- Budayeva, H.G.; Kirkpatrick, D.S. Monitoring protein communities and their responses to therapeutics. Nat. Rev. Drug Discov. 2020, 19, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Han, Q.B.; Chan, C.Y.; Wang, H.; Liu, Z.; Cheng, C.H.; Yew, D.T.; Lin, M.C.; He, M.L.; et al. Proteomic identification of molecular targets of gambogic acid: Role of stathmin in hepatocellular carcinoma. Proteomics 2009, 9, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Rochfort, S. Metabolomics reviewed: A new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Su, M.; Zhao, L.; Jiang, J.; Liu, P.; Cheng, J.; Lai, Y.; Liu, Y.; Jia, W. Metabonomic study of aristolochic acid-induced nephrotoxicity in rats. J. Proteome Res. 2006, 5, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Shen, X.; Wang, S.; Wang, Q.; Yang, X. Computational and experimental characterization of isomers of escin-induced renal cytotoxicity by inhibiting heat shock proteins. Eur. J. Pharmacol. 2021, 908, 174372. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Sun, Y.; Zhong, Q.; Yang, J.; Zhang, T.; Zhao, J.; Wang, Q. Effect of euphorbia factor L1 on oxidative stress, apoptosis, and autophagy in human gastric epithelial cells. Phytomedicine 2019, 64, 152929. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-P.; Xia, Y.; Yang, L.; Wei, J.; Yang, Y.I.; Gao, Y.Q. SPONGE: A GPU-accelerated molecular dynamics package with enhanced sampling and AI-driven algorithms. Chin. J. Chem. 2022, 40, 160–168. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Debenedetti, P.G.; Stillinger, F.H.; Lynden-Bell, R.M. A computational investigation of thermodynamics, structure, dynamics and solvation behavior in modified water models. J. Chem. Phys. 2008, 128, 124511. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- He, H.; Jiang, H.; Chen, Y.; Ye, J.; Wang, A.; Wang, C.; Liu, Q.; Liang, G.; Deng, X.; Jiang, W.; et al. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat. Commun. 2018, 9, 2550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Gao, H.; Shang, L.; Li, J.; Zhang, M.; Wang, S.; Li, R.; Ye, L.; Yang, S. Oridonin promotes endoplasmic reticulum stress via TP53-repressed TCF4 transactivation in colorectal cancer. J. Exp. Clin. Cancer Res. 2023, 42, 150. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wei, W.; Zhang, N.; Yu, Q.; Xu, W.B.; Yu, W.J.; Chen, G.Q.; Wu, Y.L.; Yan, H. Oridonin stabilizes retinoic acid receptor alpha through ROS-activated NF-κB signaling. BMC Cancer 2015, 15, 248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tan, Y.; Zhao, R.; Liu, Z. DNA damage induced by oridonin involves cell cycle arrest at G2/M phase in human MCF-7 cells. Contemp. Oncol. 2013, 17, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ouyang, L.; Peng, H.; Zhang, W.Z. Oridonin: Targeting programmed cell death pathways as an anti-tumour agent. Cell Prolif. 2012, 45, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lu, X.J.; Zhang, J.; Diao, H.; Li, G.; Xu, L.; Wang, T.; Wei, J.; Meng, W.; Ma, J.L.; et al. Oridonin, a novel lysine acetyltransferases inhibitor, inhibits proliferation and induces apoptosis in gastric cancer cells through p53- and caspase-3-mediated mechanisms. Oncotarget 2016, 7, 22623–22631. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Cao, S.J.; Zhou, Y.; He, H.; Tashiro, S.; Onodera, S.; Qiu, F.; Ikejima, T. Inhibition of caspase-9 by oridonin, a diterpenoid isolated from Rabdosia rubescens, augments apoptosis in human laryngeal cancer cells. Int. J. Oncol. 2015, 47, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, X.; Qu, S.; Yao, B.; Xu, Y.; Wu, J.; Jin, Y.; Ma, C. Oridonin induces G2/M cell cycle arrest and apoptosis via the PI3K/Akt signaling pathway in hormone-independent prostate cancer cells. Oncol. Lett. 2017, 13, 2838–2846. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Zhang, G.; Jiang, S.; Ning, F.; Zhao, Z.; Huang, M.; Jin, J. Integration of metabolomics and transcriptomics to reveal ferroptosis is involved in Tripterygium wilfordii polyglycoside tablet-induced testicular injury. J. Ethnopharmacol. 2023, 304, 116055. [Google Scholar] [CrossRef] [PubMed]
- Rehnitz, J.; Messmer, B.; Bender, U.; Nguyen, X.P.; Germeyer, A.; Hinderhofer, K.; Strowitzki, T.; Capp, E. Activation of AKT/mammalian target of rapamycin signaling in the peripheral blood of women with premature ovarian insufficiency and its correlation with FMR1 expression. Reprod. Biol. Endocrinol. 2022, 20, 44. [Google Scholar] [CrossRef]
- Liu, W.X.; Liu, H.N.; Weng, Z.P.; Geng, Q.; Zhang, Y.; Li, Y.F.; Shen, W.; Zhou, Y.; Zhang, T. Maternal vitamin B1 is a determinant for the fate of primordial follicle formation in offspring. Nat. Commun. 2023, 14, 7403. [Google Scholar] [CrossRef]
- Yang, H.; Lv, H.; Li, H.; Ci, X.; Peng, L. Oridonin protects LPS-induced acute lung injury by modulating Nrf2-mediated oxidative stress and Nrf2-independent NLRP3 and NF-κB pathways. Cell Commun. Signal. 2019, 17, 62. [Google Scholar] [CrossRef]
- Yu, D.; Li, J.; Wang, Y.; Guo, D.; Zhang, X.; Chen, M.; Zhou, Z. Oridonin ameliorates acetaminophen-induced acute liver injury through ATF4/PGC-1α pathway. Drug Dev. Res. 2023, 84, 211–225. [Google Scholar] [CrossRef]
- Pi, J.; Cai, H.; Jin, H.; Yang, F.; Jiang, J.; Wu, A.; Zhu, H.; Liu, J.; Su, X.; Yang, P.; et al. Qualitative and quantitative analysis of ROS-mediated oridonin-induced oesophageal cancer KYSE-150 cell apoptosis by atomic force microscopy. PLoS ONE 2015, 10, e0140935. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, H.; Niu, J.; Luo, M.; Gou, Y.; Miao, L.; Zou, Z.; Cheng, Y. Induction of ROS overload by alantolactone prompts oxidative DNA damage and apoptosis in colorectal cancer cells. Int. J. Mol. Sci. 2016, 17, 558. [Google Scholar] [CrossRef]
- Siemen, D.; Ziemer, M. What is the nature of the mitochondrial permeability transition pore and what is it not? IUBMB Life 2013, 65, 255–262. [Google Scholar] [CrossRef]
- Sidrat, T.; Rehman, Z.U.; Joo, M.D.; Lee, K.L.; Kong, I.K. Wnt/β-catenin pathway-mediated PPARδ expression during embryonic development differentiation and disease. Int. J. Mol. Sci. 2021, 22, 1854. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- He, Y.; Yuan, X.; Zuo, H.; Sun, Y.; Feng, A. Berberine exerts a protective effect on gut-vascular barrier via the modulation of the Wnt/Beta-catenin signaling pathway during depsis. Cell. Physiol. Biochem. 2018, 49, 1342–1351. [Google Scholar] [CrossRef]
- van der Heyden, M.A.; Rook, M.B.; Hermans, M.M.; Rijksen, G.; Boonstra, J.; Defize, L.H.; Destrée, O.H. Identification of connexin43 as a functional target for Wnt signalling. J. Cell Sci. 1998, 111 Pt 12, 1741–1749. [Google Scholar] [CrossRef]






| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| GSK3B | GCACTCTTCAACTTCACCACTCAAG (F) | CTGTCCACGGTCTCCAGTATTAGC (R) |
| β-catenin | ATAGAGGCTCTTGTGCGTACTGTC (F) | TTGGTGTCGGCTGGTCAGATG (R) |
| WNT-6 | TGCCAGTTCCAGTTCCGCTTC (F) | CCGTCTCCCGAATGTCCTGTTG (R) |
| CLDN1 | AGCCAAGGTGTTGACTCAGACTC (F) | AGCCTCCGCATTAGTTCCATAGC (R) |
| TCF7L1 | GGAGCCGAGCAGCGATAGC (F) | CCTCTCCGCCTCCGAGTCC (R) |
| ZO-1 | GCGGATGGTGCTACAAGTGATG (F) | GCCTTCTGTGTCTGTGTCTTCATAG (R) |
| CCND1 | CGCCCTCGGTGTCCTACTTC (F) | GACCTCCTCCTCGCACTTCTG (R) |
| OCLN | ACTTCGCCTGTGGATGACTTCAG (F) | TTCTCTTTGACCTTCCTGCTCTTCC (R) |
| GAPDH | TGACATCAAGAAGGTGGTGAAGCAG (F) | GTGTCGCTGTTGAAGTCAGAGGAG (R) |
| Genes | Regulation | Base Mean | log2Fold Change | p Value |
|---|---|---|---|---|
| IGF2BP2 | reader | 12,299 | −0.42 | 6.2 × 10−16 |
| FMR1 | reader | 5256 | 0.44 | 1.2 × 10−5 |
| HNRNPA2B1 | reader | 190,433 | 0.27 | 1.4 × 10−4 |
| RBM15B | writer | 4641 | −0.15 | 3.2 × 10−3 |
| WTAP | writer | 4354 | −0.19 | 1.2 × 10−2 |
| RBM15 | writer | 810 | −0.68 | 2.3 × 10−2 |
| YTHDF2 | reader | 1870 | 0.17 | 3.2 × 10−2 |
| IGF2BP1 | reader | 8764 | −0.22 | 5.6 × 10−2 |
| HNRNPC | reader | 82,291 | −0.09 | 7.4 × 10−2 |
| YTHDF3 | reader | 883 | 1.15 | 1.1 × 10−1 |
| ZC3H13 | writer | 47,093 | 0.12 | 1.3 × 10−1 |
| IGF2BP3 | reader | 2040 | −0.12 | 1.6 × 10−1 |
| FTO | eraser | 1580 | 0.14 | 1.7 × 10−1 |
| ALKBH5 | eraser | 16,193 | 0.08 | 2.4 × 10−1 |
| YTHDC2 | reader | 1778 | −0.09 | 2.4 × 10−1 |
| METTL14 | writer | 5153 | −0.06 | 3.5 × 10−1 |
| METTL5 | writer | 2978 | −0.06 | 3.7 × 10−1 |
| YTHDF1 | reader | 3137 | 0.04 | 4.6 × 10−1 |
| CBLL1 | writer | 1580 | 0.03 | 7.7 × 10−1 |
| YTHDC1 | reader | 18,686 | −0.08 | 8.0 × 10−1 |
| VIRMA | writer | 10,151 | 0.01 | 8.3 × 10−1 |
| METTL3 | writer | 1885 | 0.01 | 9.4 × 10−1 |
| Protein | PDB ID | Total Score | Crash | Polar | H-Bond Number | Residues Involve in H-Bond Formation | Hydrophobic Contact Number | Residues Involve in Hydrophobic Contacts |
|---|---|---|---|---|---|---|---|---|
| FMR1 | 2QND | 6.5681 | −0.9508 | 6.7019 | 5 | Arg11, Asp13, Glu68 (3 H-bonds) | 2 | Arg100, Glu104 |
| HNRNPA2B1 | 5WWG | 5.9849 | −4.3987 | 3.4371 | 3 | Glu101, Lys104, Ala107 | 8 | Pro105, Gly106, Lys186, Arg185, Val170, Lys173, Leu171, Val197 |
| IGF2BP2 | 6ROL | 4.432 | −0.8333 | 3.5298 | 4 | Lys509 (3 H-bonds), Asn503 | 5 | Phe502, Leu510, Glu511, Ile562, Gol604 |
| RBM15 | 7Z27 | 4.1057 | −0.6563 | 3.4976 | 4 | Lys9, Lys51, Leu12 (2 H-bonds) | 2 | Ala13, Gln15 |
| WTAP | 7YFJ | 3.6498 | −0.9403 | 2.5236 | 3 | Gln61 (3 H-bonds) | 3 | Tyr64, Leu68, Ser65 |
| YTHDF2 | 4WQN | 3.1273 | −0.587 | 2.3001 | 2 | Arg425, Lys428 | 2 | Asp421, His424 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Gao, X.; Lin, Y.; Wu, C.; Zhang, J.; Chen, M.; Wen, J.; Wu, Y.; Tian, K.; Bao, W.; et al. Integrating Epigenetics, Proteomics, and Metabolomics to Reveal the Involvement of Wnt/β-Catenin Signaling Pathway in Oridonin-Induced Reproductive Toxicity. Toxics 2024, 12, 339. https://doi.org/10.3390/toxics12050339
Wu Q, Gao X, Lin Y, Wu C, Zhang J, Chen M, Wen J, Wu Y, Tian K, Bao W, et al. Integrating Epigenetics, Proteomics, and Metabolomics to Reveal the Involvement of Wnt/β-Catenin Signaling Pathway in Oridonin-Induced Reproductive Toxicity. Toxics. 2024; 12(5):339. https://doi.org/10.3390/toxics12050339
Chicago/Turabian StyleWu, Qibin, Xinyue Gao, Yifan Lin, Caijin Wu, Jian Zhang, Mengting Chen, Jiaxin Wen, Yajiao Wu, Kun Tian, Wenqiang Bao, and et al. 2024. "Integrating Epigenetics, Proteomics, and Metabolomics to Reveal the Involvement of Wnt/β-Catenin Signaling Pathway in Oridonin-Induced Reproductive Toxicity" Toxics 12, no. 5: 339. https://doi.org/10.3390/toxics12050339
APA StyleWu, Q., Gao, X., Lin, Y., Wu, C., Zhang, J., Chen, M., Wen, J., Wu, Y., Tian, K., Bao, W., Sun, P., & Zhu, A. (2024). Integrating Epigenetics, Proteomics, and Metabolomics to Reveal the Involvement of Wnt/β-Catenin Signaling Pathway in Oridonin-Induced Reproductive Toxicity. Toxics, 12(5), 339. https://doi.org/10.3390/toxics12050339







