Determination of Ploidy Levels and Nuclear DNA Content in Cryptococcus neoformans by Flow Cytometry: Drawbacks with Variability
Abstract
:1. Introduction
2. Methods
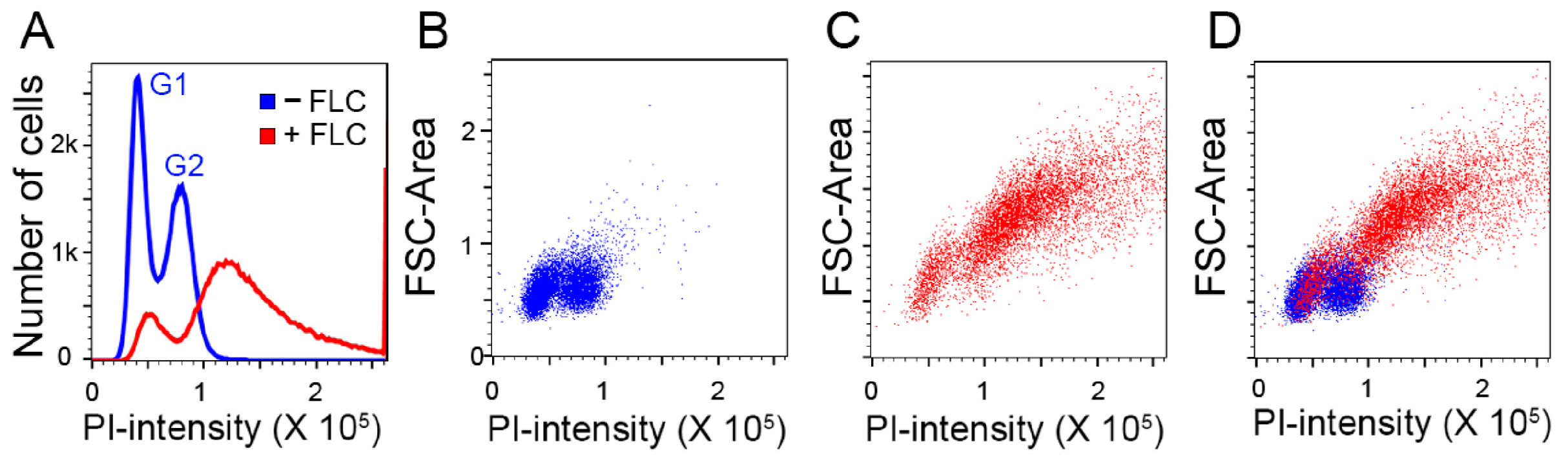
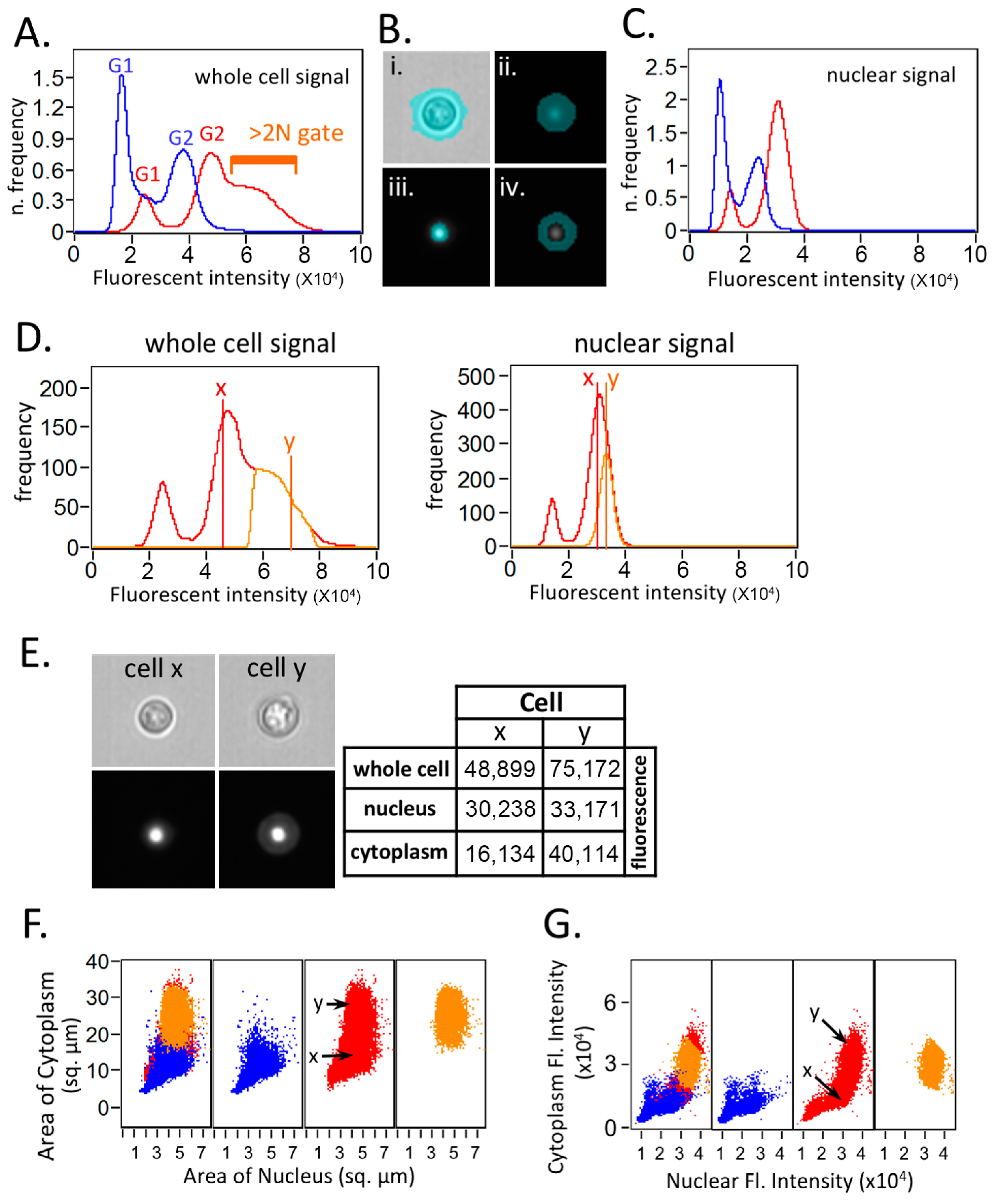
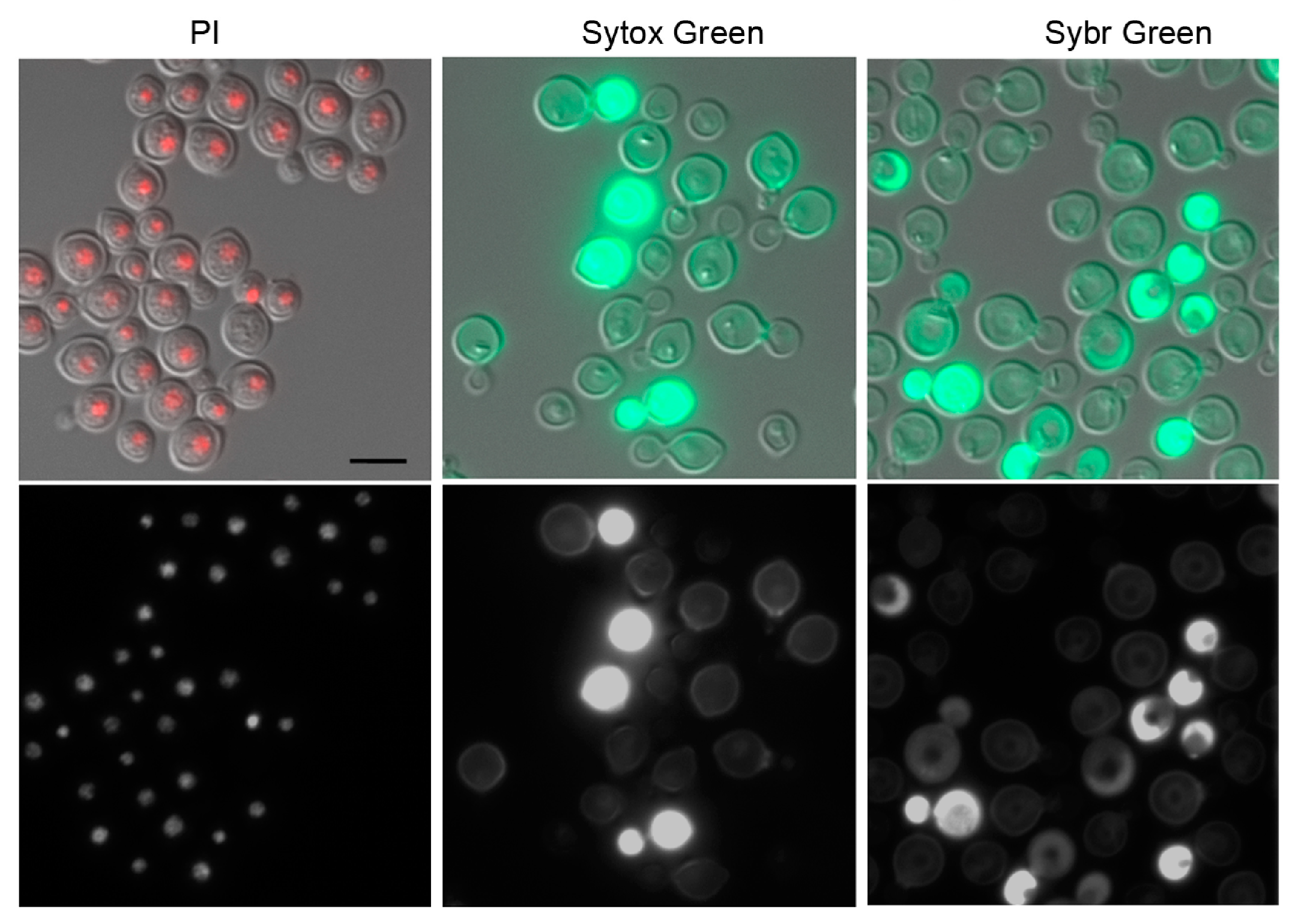
3. Results and Discussions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ormerod, K.L.; Fraser, J.A. Balancing stability and flexibility within the genome of the pathogen Cryptococcus neoformans. PLoS Pathog. 2013, 9, e1003764. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Liu, I.; Sham, A.; Stajich, J.E.; Dietrich, F.S.; Kronstad, J.W. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biol. 2008, 9, R41. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Lee, H.; Chang, Y.C.; Kwon-Chung, K.J. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010, 6, e1000848. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Cox, G.M.; Heitman, J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 2001, 69, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, J.; Choi, J.; Jung, W.H.; Liu, I.; Litvintseva, A.P.; Bicanic, T.; Aurora, R.; Mitchell, T.G.; Perfect, J.R.; et al. Variation in chromosome copy number influences the virulence of Cryptococcus neoformans and occurs in isolates from AIDS patients. BMC Genom. 2011, 12, 526. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Chang, Y.C.; Kwon-Chung, K.J. Azole heteroresistance in Cryptococcus neoformans: Emergence of resistant clones with chromosomal disomy in the mouse brain during fluconazole treatment. Antimicrob. Agents Chemother. 2013, 57, 5127–5130. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Varma, A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006, 6, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Saidykhan, L.; Correia, J.; Romanyuk, A.; Peacock, A.F.A.; Desanti, G.E.; Taylor-Smith, L.; Makarova, M.; Ballou, E.R.; May, R.C. An in vitro method for inducing titan cells reveals novel features of yeast-to-titan switching in the human fungal pathogen Cryptococcus gattii. PLoS Pathog. 2022, 18, e1010321. [Google Scholar] [CrossRef] [PubMed]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chretien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Zaragoza, O.; Nielsen, K. Titan cells in Cryptococcus neoformans: Cells with a giant impact. Curr. Opin. Microbiol. 2013, 16, 409–413. [Google Scholar] [CrossRef]
- Francis, V.I.; Liddle, C.; Camacho, E.; Kulkarni, M.; Junior, S.R.S.; Harvey, J.A.; Ballou, E.R.; Thomson, D.D.; Hardwick, J.M.; Casadevall, A.; et al. Cryptococcus neoformans rapidly invades the murine brain by sequential breaching of airway and endothelial tissues barriers, followed by engulfment by microglia. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Darzynkiewicz, Z.; Huang, X.; Zhao, H. Analysis of Cellular DNA Content by Flow Cytometry. Curr. Protoc. Cytom. 2017, 82, 7.5.1–7.5.20. [Google Scholar] [CrossRef]
- Poot, M. Nucleic acid probes. Curr. Protoc. Cytom. 2004, 4, 4.3. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.P. Handbook of Fluorescent Probes and Research Chemicals, 6th ed.; Molecular Probes: Eugene, OR, USA, 1996. [Google Scholar]
- Tanaka, R.; Taguchi, H.; Takeo, K.; Miyaji, M.; Nishimura, K. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J. Med. Vet. Mycol. 1996, 34, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, S.; Fang, D.; Simmons, C.; Sridhar, S.; Wu, P.; Sanyal, K.; Kozubowski, L. Fluconazole-Induced Ploidy Change in Cryptococcus neoformans Results from the Uncoupling of Cell Growth and Nuclear Division. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Morris, I.R.; Wickes, B.L. The production of monokaryotic hyphae by Cryptococcus neoformans can be induced by high temperature arrest of the cell cycle and is independent of same-sex mating. PLoS Pathog. 2013, 9, e1003335. [Google Scholar] [CrossRef] [PubMed]
- Hedley, D.W.; Friedlander, M.L.; Taylor, I.W.; Rugg, C.A.; Musgrove, E.A. Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J. Histochem. Cytochem. 1983, 31, 1333–1335. [Google Scholar] [CrossRef]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analyses. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef] [PubMed]
- Otto, F. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods Cell Biol. 1990, 33, 105–110. [Google Scholar]
- Haase, S.B.; Reed, S.I. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 2002, 1, 132–136. [Google Scholar] [CrossRef]
- Chang, Y.C.; Khanal Lamichhane, A.; Kwon-Chung, K.J. Cryptococcus neoformans, Unlike Candida albicans, Forms Aneuploid Clones Directly from Uninucleated Cells under Fluconazole Stress. mBio 2018, 9. [Google Scholar] [CrossRef]
- Fortuna, M.; Sousa, M.J.; Corte-Real, M.; Leao, C.; Salvador, A.; Sansonetty, F. Cell cycle analysis of yeasts. Curr. Protoc. Cytom. 2001, 11, 11–13. [Google Scholar] [CrossRef]
- Suzuki, T.; Nishibayashi, S.; Kuroiwa, T.; Kanbe, T.; Tanaka, K. Variance of ploidy in Candida albicans. J. Bacteriol. 1982, 152, 893–896. [Google Scholar] [CrossRef]
- Chang, Y.C.; Khanal Lamichhane, A.; Garraffo, H.M.; Walter, P.J.; Leerkes, M.; Kwon-Chung, K.J. Molecular mechanisms of hypoxic responses via unique roles of Ras1, Cdc24 and Ptp3 in a human fungal pathogen Cryptococcus neoformans. PLoS Genet. 2014, 10, e1004292. [Google Scholar] [CrossRef]
- Ballou, E.R.; Kozubowski, L.; Nichols, C.B.; Alspaugh, J.A. Ras1 acts through duplicated Cdc42 and Rac proteins to regulate morphogenesis and pathogenesis in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 2013, 9, e1003687. [Google Scholar] [CrossRef] [PubMed]
- Lebaron, P.; Catala, P.; Parthuisot, N. Effectiveness of SYTOX Green stain for bacterial viability assessment. Appl. Environ. Microbiol. 1998, 64, 2697–2700. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.C.; Davis, M.J.; Kwon-Chung, K.J. Determination of Ploidy Levels and Nuclear DNA Content in Cryptococcus neoformans by Flow Cytometry: Drawbacks with Variability. J. Fungi 2024, 10, 296. https://doi.org/10.3390/jof10040296
Chang YC, Davis MJ, Kwon-Chung KJ. Determination of Ploidy Levels and Nuclear DNA Content in Cryptococcus neoformans by Flow Cytometry: Drawbacks with Variability. Journal of Fungi. 2024; 10(4):296. https://doi.org/10.3390/jof10040296
Chicago/Turabian StyleChang, Yun C., Michael J. Davis, and Kyung J. Kwon-Chung. 2024. "Determination of Ploidy Levels and Nuclear DNA Content in Cryptococcus neoformans by Flow Cytometry: Drawbacks with Variability" Journal of Fungi 10, no. 4: 296. https://doi.org/10.3390/jof10040296




