The Diversity and Community Composition of Three Plants’ Rhizosphere Fungi in Kaolin Mining Areas
Abstract
:1. Introduction
2. Methods and Materials
2.1. Rhizosphere Soil Sample Collection
2.2. Genomic DNA Extraction
2.3. PCR Amplification
2.4. Library Preparation, Sequencing, and Raw Data Processing
2.5. Species Composition Analysis
2.6. Alpha Diversity and Beta Diversity Analysis
2.7. Community Difference Analysis and Functional Prediction
2.8. Statistical Analysis
3. Results
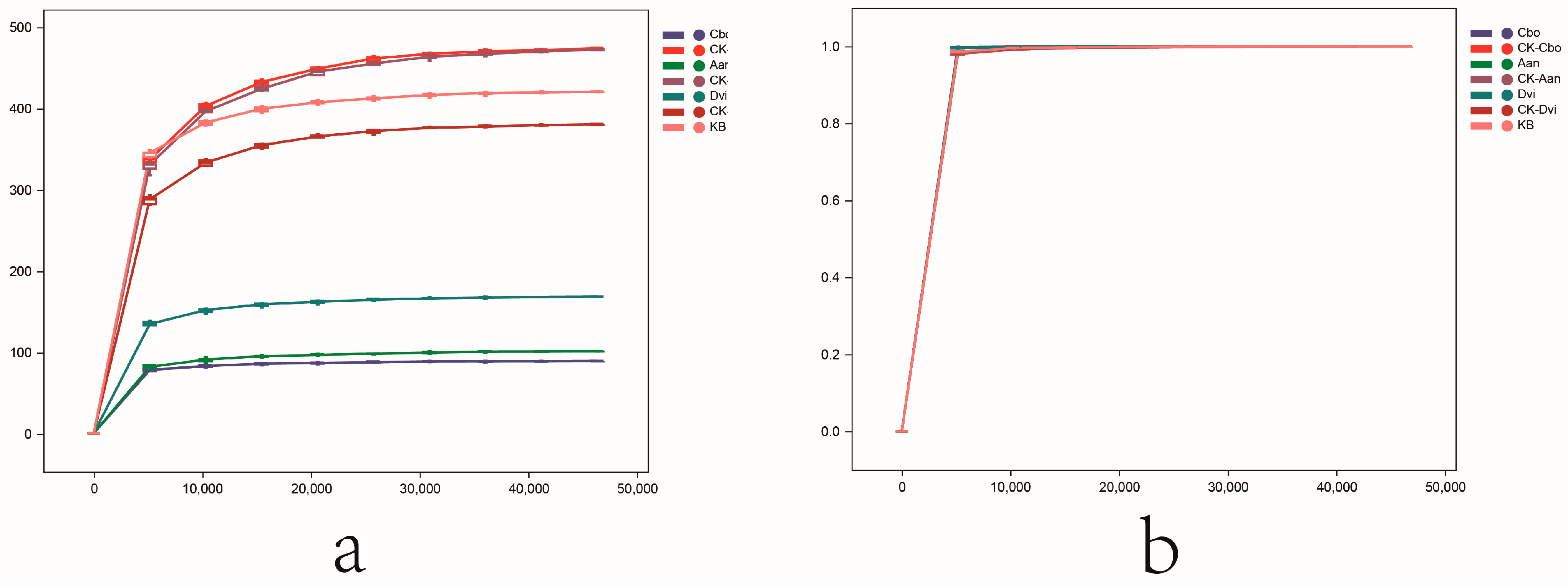
3.1. Quality Control and Processing of Sequencing Data
3.2. Species Composition Analysis
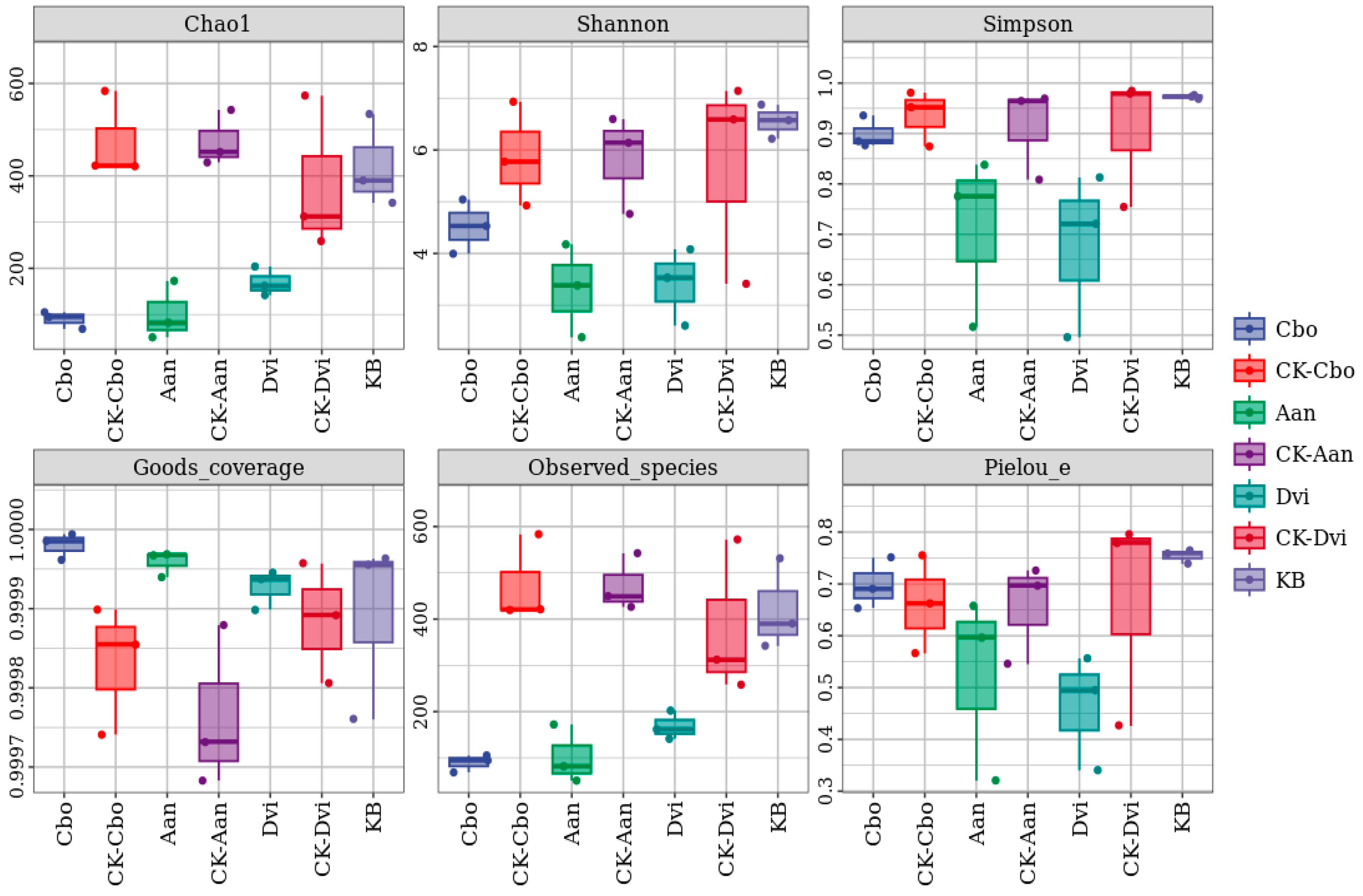
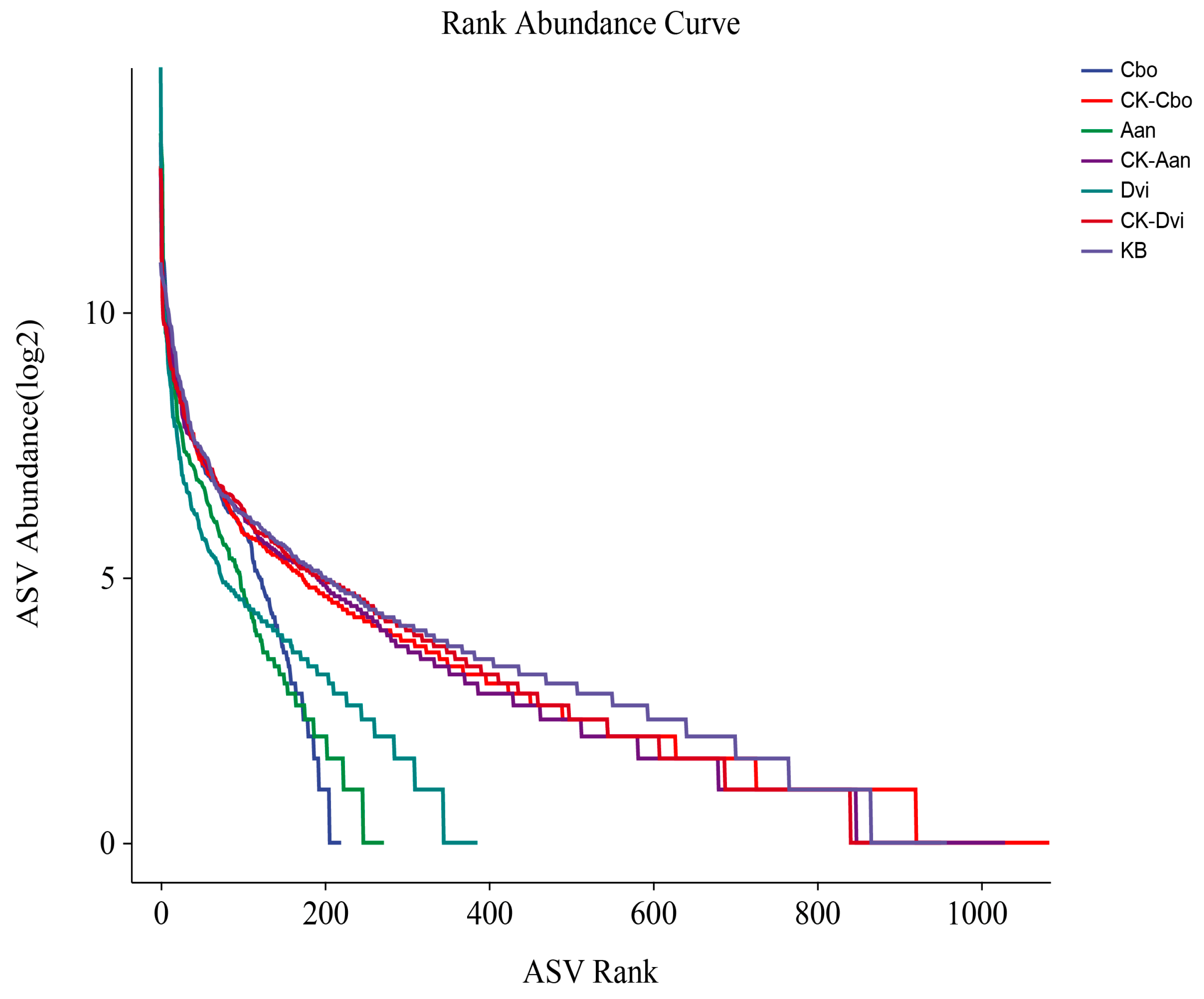
3.3. Alpha Diversity
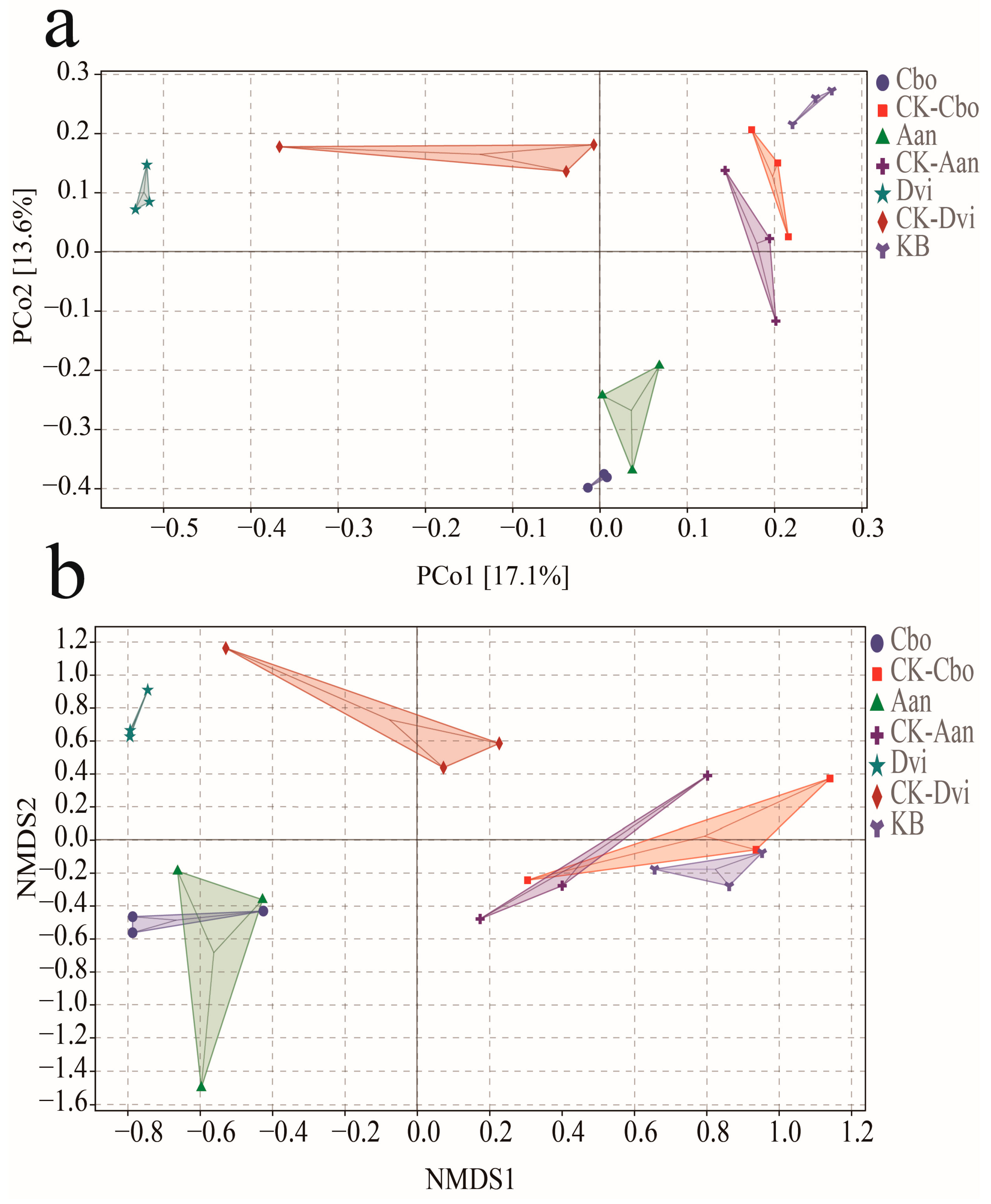
3.4. Beta Diversity
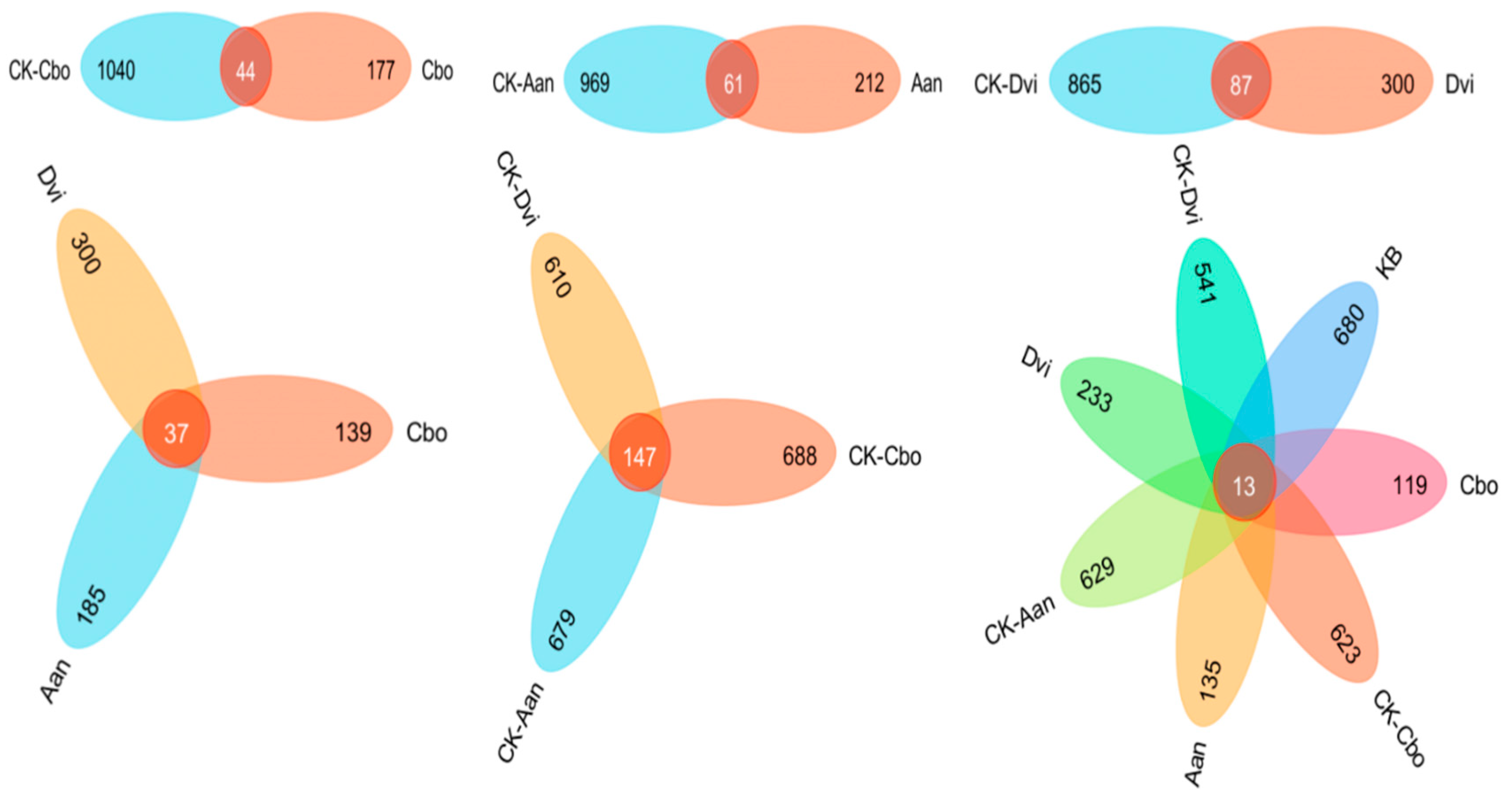
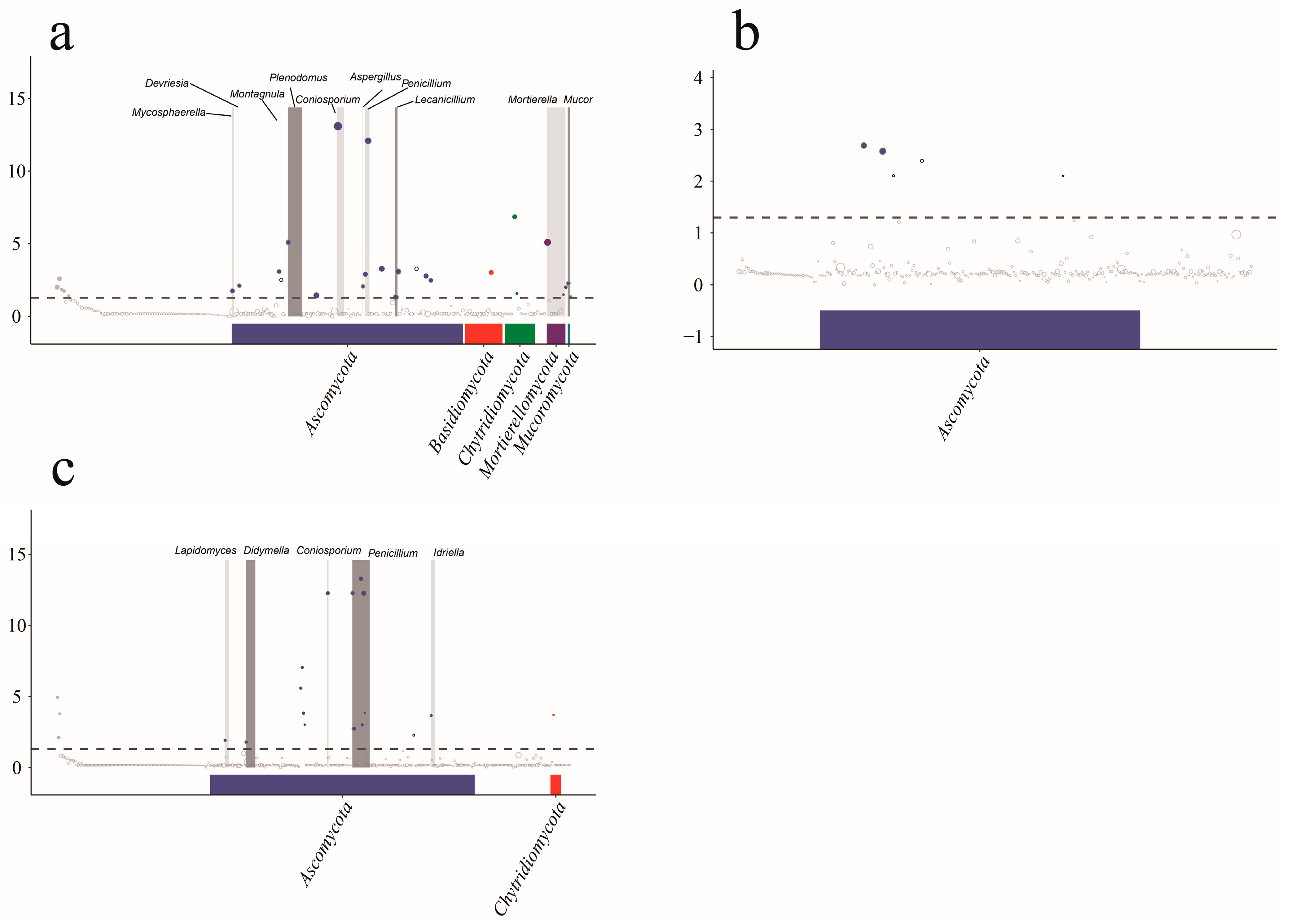
3.5. Community Species Composition and Difference Analysis
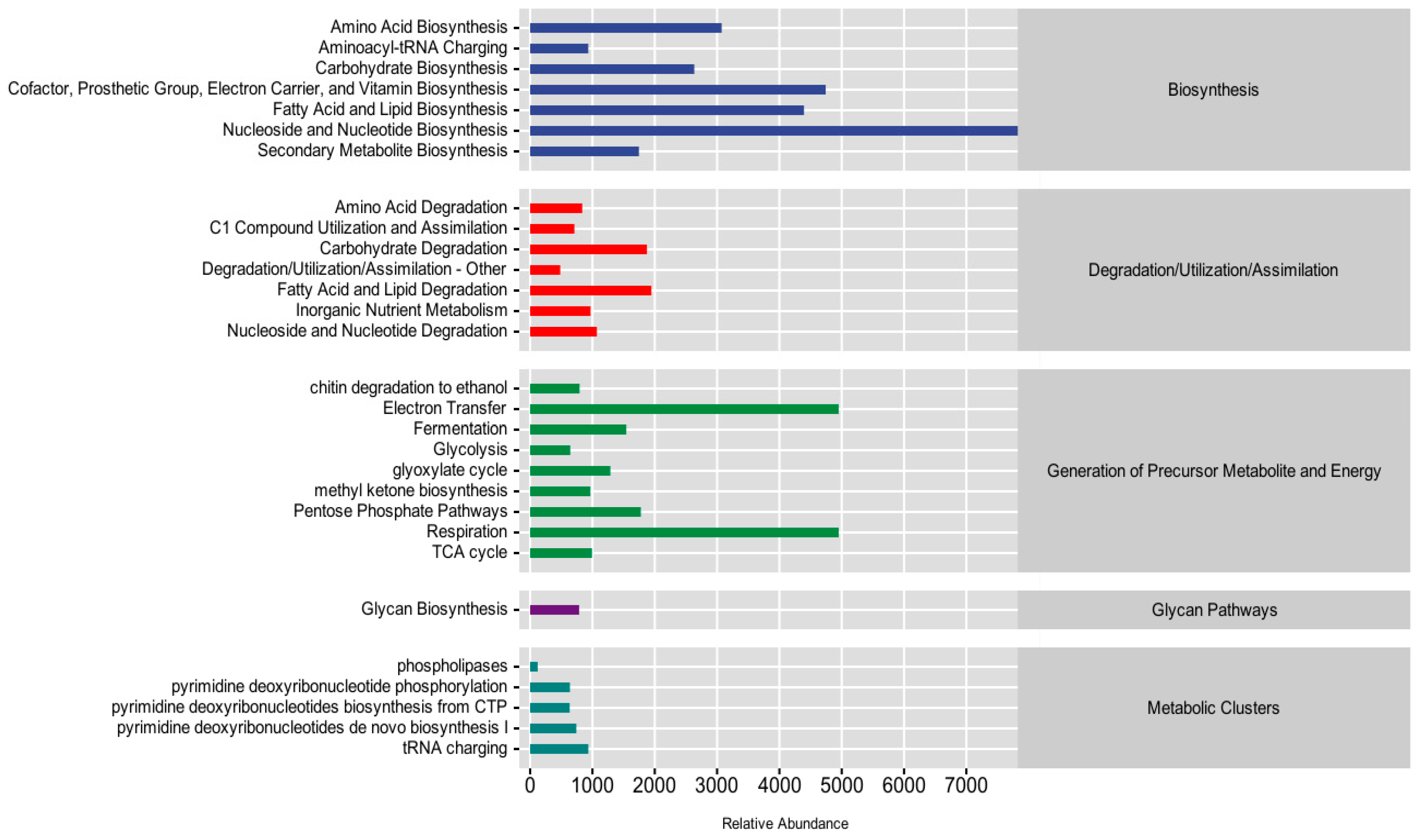
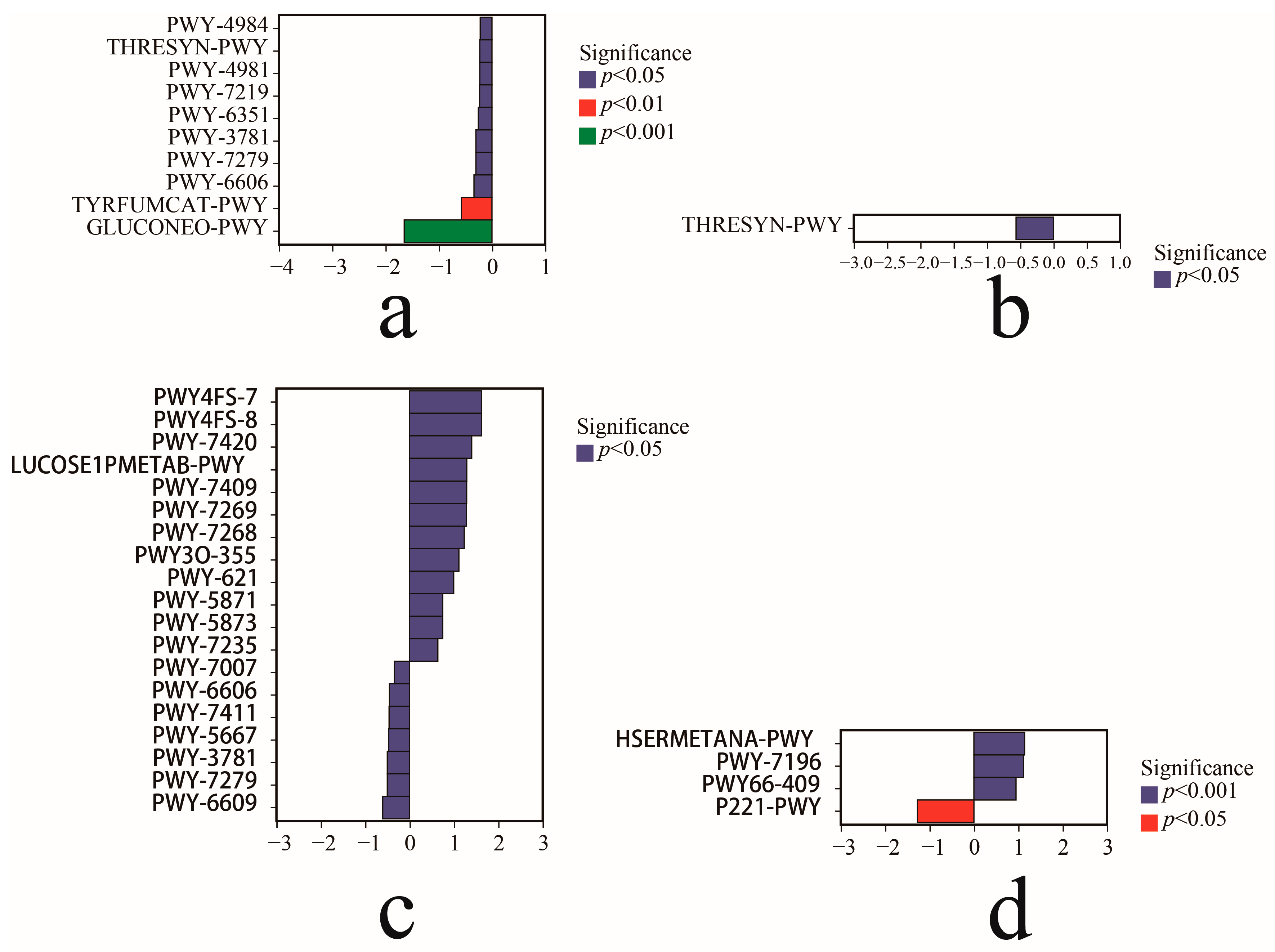
3.6. Community Function Prediction and Difference Analysis
4. Discussion
4.1. Rhizosphere Fungal Diversity and Community Composition
4.2. Functions of the Rhizosphere Fungal Community
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Loganathan, N.; Wilson, A.K. Adsorption, structure, and dynamics of short-and long-chain PFAS molecules in kaolinite: Molecular-level insights. Environ. Sci. Technol. 2022, 56, 8043–8052. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, A.; Geatches, D.L.; Clark, S.J.; Greenwell, H.C. Understanding cationic polymer adsorption on mineral surfaces: Kaolinite in cement aggregates. Minerals 2018, 8, 130. [Google Scholar] [CrossRef]
- Pak, V.I.; Kirov, S.S.; Nalivaiko, A.Y.; Ozherelkov, D.Y.; Gromov, A.A. Obtaining alumina from kaolin clay via aluminum chloride. Materials 2019, 12, 3938. [Google Scholar] [CrossRef] [PubMed]
- Awwad, A.; Amer, M.; Al-aqarbeh, M. TiO2-kaolinite nanocomposite prepared from the Jordanian Kaolin clay: Adsorption and thermodynamics of Pb (II) and Cd (II) ions in aqueous solution. Chem. Int. 2020, 6, 168–178. [Google Scholar]
- Banu, H.A.T.; Karthikeyan, P.; Vigneshwaran, S.; Meenakshi, S. Adsorptive performance of lanthanum encapsulated biopolymer chitosan-kaolin clay hybrid composite for the recovery of nitrate and phosphate from water. Int. J. Biol. Macromol. 2020, 154, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Abbas, R.; Khereby, M.A.; Ghorab, H.Y.; Elkhoshkhany, N. Preparation of geopolymer concrete using Egyptian kaolin clay and the study of its environmental effects and economic cost. Clean Technol. Environ. Policy 2020, 22, 669–687. [Google Scholar] [CrossRef]
- Du, H.; Dai Pang, S.J.C.; Materials, B. High-performance concrete incorporating calcined kaolin clay and limestone as cement substitute. Constr. Build. Mater. 2020, 264, 120152. [Google Scholar] [CrossRef]
- Qi, Y.; Ju, Y.; Huang, C.; Zhu, H.; Bao, Y.; Wu, J.; Meng, S.; Chen, W. Influences of organic matter and kaolinite on pore structures of transitional organic-rich mudstone with an emphasis on S2 controlling specific surface area. Fuel 2019, 237, 860–873. [Google Scholar] [CrossRef]
- Yang, F.; Xu, Z.; Yu, L.; Gao, B.; Xu, X.; Zhao, L.; Cao, X. Kaolinite enhances the stability of the dissolvable and undissolvable fractions of biochar via different mechanisms. Environ. Sci. Technol. 2018, 52, 8321–8329. [Google Scholar] [CrossRef]
- Kallenbach, C.M.; Frey, S.D.; Grandy, A.S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 2016, 7, 13630. [Google Scholar] [CrossRef]
- Andrade, G.R.P.; Cuadros, J.; Partiti, C.S.M.; Cohen, R.; Vidal-Torrado, P. Sequential mineral transformation from kaolinite to Fe-illite in two Brazilian mangrove soils. Geoderma 2018, 309, 84–99. [Google Scholar] [CrossRef]
- Mustapha, S.; Ndamitso, M.; Abdulkareem, A.; Tijani, J.; Mohammed, A.; Shuaib, D.J.H. Potential of using kaolin as a natural adsorbent for the removal of pollutants from tannery wastewater. Heliyon 2019, 5, e02923. [Google Scholar] [CrossRef] [PubMed]
- Li, G.L.; Zhou, C.H.; Fiore, S.; Yu, W.H. Interactions between microorganisms and clay minerals: New insights and broader applications. Appl. Clay Sci. 2019, 177, 91–113. [Google Scholar] [CrossRef]
- Kyaschenko, J.; Clemmensen, K.E.; Karltun, E.; Lindahl, B.D. Below-ground organic matter accumulation along a boreal forest fertility gradient relates to guild interaction within fungal communities. Ecol. Lett. 2017, 20, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, Z.; van der Kuijp, T.J.; Yuan, Z.; Huang, L. A review of soil heavy metal pollution from mines in China: Pollution and health risk assessment. Sci. Total Environ. 2014, 468, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, P.; Sobczyk, Ł. Effects of heavy metal pollution from mining and smelting on enchytraeid communities under different land management and soil conditions. Sci. Total Environ. 2015, 536, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Narendrula, R.; Nkongolo, K.K. Biodegredation. Fatty acids profile of microbial populations in a mining reclaimed region contaminated with metals: Relation with ecological characteristics and soil respiration. J. Bioremediation Biodegrad. 2015, 6, 1. [Google Scholar] [CrossRef]
- Shrestha, R.K.; Lal, R. Changes in physical and chemical properties of soil after surface mining and reclamation. Geoderma 2011, 161, 168–176. [Google Scholar] [CrossRef]
- Warhate, S.R.; Yenkie, M.K.N.; Chaudhari, M.D.; Pokale, W.K. Impacts of mining activities on water and soil. J. Environ. Sci. Eng. 2006, 48, 81–90. [Google Scholar]
- Feng, Y.; Wang, J.; Bai, Z.; Reading, L. Effects of surface coal mining and land reclamation on soil properties: A review. Earth-Sci. Rev. 2019, 191, 12–25. [Google Scholar] [CrossRef]
- de Quadros, P.D.; Zhalnina, K.; Davis-Richardson, A.G.; Drew, J.C.; Menezes, F.B.; Camargo, F.A.D.O.; Triplett, E.W. Coal mining practices reduce the microbial biomass, richness and diversity of soil. Appl. Soil Ecol. 2016, 98, 195–203. [Google Scholar] [CrossRef]
- Walter, J.; Hradská, I.; Kout, J.; Bureš, J.; Konvička, M. The impact of abandoned kaolin quarries on macromycetes (Fungi: Basidiomycota, Ascomycota), carabid beetle (Coleoptera: Carabidae), and spider (Araneae) assemblages. Biodivers. Conserv. 2023, 32, 1437–1449. [Google Scholar] [CrossRef]
- Wang, K.; Bi, Y.; Cao, Y.; Peng, S.; Christie, P.; Ma, S.; Zhang, J.; Xie, L. Shifts in composition and function of soil fungal communities and edaphic properties during the reclamation chronosequence of an open-cast coal mining dump. Sci. Total Environ. 2021, 767, 144465. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nan, J.; Xu, D.; Mo, L.; Zheng, Y.; Chao, L.; Qu, H.; Guo, Y.; Li, F.; Bao, Y.J.C. Response differences between soil fungal and bacterial communities under opencast coal mining disturbance conditions. Catena 2020, 194, 104779. [Google Scholar] [CrossRef]
- Rosenfeld, C.E.; James, B.R.; Santelli, C.M. Persistent bacterial and fungal community shifts exhibited in selenium-contaminated reclaimed mine soils. Appl. Environ. Microbiol. 2018, 84, e01394-18. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Li, M.; Shao, D.; Zhou, J.; Gu, J.; Qin, J.; Chen, W.; Wei, W. Signatures within esophageal microbiota with progression of esophageal squamous cell carcinoma. Chin. J. Cancer Res. 2020, 32, 755. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K.J.N.a.r. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.; Kopylova, E.; Morton, J.; Xu, Z.J.P.P.C. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, X.; Guo, F.; Wu, W.; Zhang, T. Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 2013, 47, 4207–4216. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xiong, Z.; Xiang, P.; Zhou, L.; Zhang, T.; Wu, Q.; Zhao, C. Effects of uranium mining on soil bacterial communities and functions in the Qinghai-Tibet plateau. Chemosphere 2024, 347, 140715. [Google Scholar] [CrossRef]
- Xiang, P.; Liao, W.; Xiong, Z.; Xiao, W.; Luo, Y.; Peng, L.; Zou, L.; Zhao, C.; Li, Q. Effects of polystyrene microplastics on the agronomic traits and rhizosphere soil microbial community of highland barley. Sci. Total Environ. 2023, 907, 167986. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Wang, N.; Wang, A.; Xie, J.; He, M. Responses of soil fungal and archaeal communities to environmental factors in an ongoing antimony mine area. Sci. Total Environ. 2019, 652, 1030–1039. [Google Scholar] [CrossRef]
- Maron, J.L.; Marler, M.; Klironomos, J.N.; Cleveland, C.C. Soil fungal pathogens and the relationship between plant diversity and productivity. Ecol. Lett. 2011, 14, 36–41. [Google Scholar] [CrossRef]
- Shi, L.-L.; Mortimer, P.E.; Ferry Slik, J.; Zou, X.-M.; Xu, J.; Feng, W.-T.; Qiao, L.J. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Divers. 2014, 64, 305–315. [Google Scholar] [CrossRef]
- Choi, I.Y.; Choi, J.N.; Choi, D.C.; Sharma, P.K.; Lee, W.H. Identification and characterization of the causal organism of gummy stem blight in the muskmelon (Cucumis melo L.). Mycobiology 2010, 38, 166–170. [Google Scholar] [CrossRef]
- Doohan, F.; Zhou, B. Fungal pathogens of plants. Fungi Biol. Appl. 2017, 355–387. [Google Scholar] [CrossRef]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.; Castillejo, M.A.; Singh, K.B.; Rispail, N. Achievements and challenges in legume breeding for pest and disease resistance. Crit. Rev. Plant Sci. 2015, 34, 195–236. [Google Scholar] [CrossRef]
- Lamb, E.M.; Rosskopf, E.N.; Koblegard, C. Use of kaolin clay for disease control in greenhouse cucumbers. Proc. Fla. State Hortic. Soc. 2002, 185, 180–182. [Google Scholar]
- Crocker, F.H.; Jung, C.M.; Indest, K.J.; Everman, S.J.; Carr, M.R. Effects of chitin and temperature on sub-Arctic soil microbial and fungal communities and biodegradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) and 2, 4-dinitrotoluene (DNT). Biodegradation 2019, 30, 415–431. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, X.; Yang, H.; Sun, L. Bioremediation of heavy metal pollution utilizing composite microbial agent of Mucor circinelloides, Actinomucor sp. and Mortierella sp. J. Environ. Chem. Eng. 2017, 5, 3616–3621. [Google Scholar] [CrossRef]
- Tao, J.; Cao, P.; Xiao, Y.; Wang, Z.; Huang, Z.; Jin, J.; Liu, Y.; Yin, H.; Liu, T.; Zhou, Z. Distribution of the potential pathogenic Alternaria on plant leaves determines foliar fungal communities around the disease spot. Environ. Res. 2021, 200, 111715. [Google Scholar] [CrossRef] [PubMed]
- Rocha, P.d.S.d.; Paula, V.M.B.; Olinto, S.C.F.; Dos Santos, E.L.; de Picoli Souza, K.; Estevinho, L.M. Diversity, chemical constituents and biological activities of endophytic fungi isolated from Schinus terebinthifolius Raddi. Microorganisms 2020, 8, 859. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Susca, A. Penicillium species and their associated mycotoxins. Mycotoxigenic Fungi Methods Protoc. 2017, 1542, 107–119. [Google Scholar]
- Houbraken, J.; Frisvad, J.C.; Seifert, K.; Overy, D.P.; Tuthill, D.; Valdez, J.; Samson, R. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia Mol. Phylogeny Evol. Fungi 2012, 29, 78–100. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Verma, P.; Kumar, V.; Sangwan, P.; Mishra, S.; Panjiar, N.; Gupta, V.K.; Saxena, A.K. Chapter 1—Biodiversity of the Genus Penicillium in Different Habitats. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Rodriguez-Couto, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 3–18. [Google Scholar] [CrossRef]
- Das, B.K.; Roy, A.; Koschorreck, M.; Mandal, S.M.; Wendt-Potthoff, K.; Bhattacharya, J. Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Water Res. 2009, 43, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Marescotti, P.; Roccotiello, E.; Zotti, M.; De Capitani, L.; Carbone, C.; Azzali, E.; Mariotti, M.G.; Lucchetti, G. Influence of soil mineralogy and chemistry on fungi and plants in a waste-rock dump from the Libiola mine (eastern Liguria, Italy). Period. Mineral. 2013, 82, 141–162. [Google Scholar]
- Wang, F. Occurrence of arbuscular mycorrhizal fungi in mining-impacted sites and their contribution to ecological restoration: Mechanisms and applications. Crit. Rev. Environ. Sci. Technol. 2017, 47, 1901–1957. [Google Scholar] [CrossRef]
- Leung, H.M.; Ye, Z.H.; Wong, M.H. Survival strategies of plants associated with arbuscular mycorrhizal fungi on toxic mine tailings. Chemosphere 2007, 66, 905–915. [Google Scholar] [CrossRef]
- de Moura, M.A.; Oki, Y.; Arantes-Garcia, L.; Cornelissen, T.; Nunes, Y.R.F.; Fernandes, G.W. Mycorrhiza fungi application as a successful tool for worldwide mine land restoration: Current state of knowledge and the way forward. Ecol. Eng. 2022, 178, 106580. [Google Scholar] [CrossRef]
- Ortega-Larrocea, M.d.P.; Xoconostle-Cázares, B.; Maldonado-Mendoza, I.E.; Carrillo-González, R.; Hernández-Hernández, J.; Garduño, M.D.; López-Meyer, M.; Gómez-Flores, L.; González-Chávez, M.d.C.A. Plant and fungal biodiversity from metal mine wastes under remediation at Zimapan, Hidalgo, Mexico. Environ. Pollut. 2010, 158, 1922–1931. [Google Scholar] [CrossRef]
- Zhang, T.; Jiang, R.; Fang, L.; Liu, X.; Jiang, L. Chlorination of L-tyrosine and metal complex: Degradation kinetics and disinfection by-products generation. Environ. Technol. 2023, 44, 3532–3543. [Google Scholar] [CrossRef] [PubMed]
- Berto, S.; De Laurentiis, E.; Tota, T.; Chiavazza, E.; Daniele, P.G.; Minella, M.; Isaia, M.; Brigante, M.; Vione, D.J. Properties of the humic-like material arising from the photo-transformation of l-tyrosine. Sci. Total. Environ. 2016, 545, 434–444. [Google Scholar] [CrossRef]
- Refat, M.S.; El-Korashy, S.A.; Ahmed, A.S. Preparation, structural characterization and biological evaluation of L-tyrosinate metal ion complexes. J. Mol. Struct. 2008, 881, 28–45. [Google Scholar] [CrossRef]
- Irshad, K.; Rehman, K.; Fiayyaz, F.; Sharif, H.; Murtaza, G.; Kamal, S.; Akash, M.S.H. Role of heavy metals in metabolic disorders. Endocr. Disrupting Chem.-Induc. Metab. Disord. Treat. Strateg. 2021, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Wang, B.; Cui, Y.; Wang, Y.; Zhang, K.; Chen, C.; Xia, F.; Ye, L.; Wang, L.; Wang, N. Low-level lead exposure promotes hepatic gluconeogenesis and contributes to the elevation of fasting glucose level. Chemosphere 2021, 276, 130111. [Google Scholar] [CrossRef]
- Mostafalou, S.; Baeeri, M.; Bahadar, H.; Soltany-Rezaee-Rad, M.; Gholami, M.; Abdollahi, M. Molecular mechanisms involved in lead induced disruption of hepatic and pancreatic glucose metabolism. Environ. Toxicol. Pharmacol. 2015, 39, 16–26. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Bashir, S.; Javed, S.; Al-Anazi, K.M.; Farah, M.A.; Ali, S.J. Bioremediation of Cadmium Toxicity in Wheat (Triticum aestivum L.) Plants Primed with L-Proline, Bacillus subtilis and Aspergillus niger. Int. J. Environ. Res. Public Health 2022, 19, 12683. [Google Scholar] [CrossRef]
- Huang, L.; Fang, Z.; Gao, J.; Wang, J.; Li, Y.; Sun, L.; Wang, Y.; Liao, J.; Gooneratne, R. Protective role of l-threonine against cadmium toxicity in Saccharomyces cerevisiae. J. Basic Microbiol. 2021, 61, 339–350. [Google Scholar] [CrossRef]
- Ayangbenro, A.S.; Babalola, O.O. A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef]
- Nanda, M.; Kumar, V.; Sharma, D. Multimetal tolerance mechanisms in bacteria: The resistance strategies acquired by bacteria that can be exploited to ‘clean-up’heavy metal contaminants from water. Aquat. Toxicol. 2019, 212, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Słaba, M.; Bernat, P.; RÓżalska, S.; Nykiel, J.; Długoński, J. Comparative study of metal induced phospholipid modifications in the heavy metal tolerant filamentous fungus Paecilomyces marquandii and implications for the fungal membrane integrity. Acta Biochim. Pol. 2013, 60, 695–700. [Google Scholar] [CrossRef]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal Biodiversity and Their Role in Soil Health. Front. Microbiol. 2018, 9, 316246. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.M.; Irzykowski, W.; Jedryczka, M.; Han, F.X. Analysis of the Genetic Structure of Sclerotinia sclerotiorum (Lib.) de Bary Populations from Different Regions and Host Plants by Random Amplified Polymorphic DNA Markers. J. Integr. Plant Biol. 2005, 47, 385–395. [Google Scholar] [CrossRef]
- Fomina, M.; Skorochod, I. Microbial interaction with clay minerals and its environmental and biotechnological implications. Minerals 2020, 10, 861. [Google Scholar] [CrossRef]
- Held, B.W.; Salomon, C.E.; Blanchette, R.A. Diverse subterranean fungi of an underground iron ore mine. PLoS ONE 2020, 15, e0234208. [Google Scholar] [CrossRef]
- Ye, F.; Gong, D.; Pang, C.; Luo, J.; Zeng, X.; Shang, C. Analysis of fungal composition in mine-contaminated soils in Hechi City. Curr. Microbiol. 2020, 77, 2685–2693. [Google Scholar] [CrossRef]
- Narendrula-Kotha, R.; Nkongolo, K.K. Bacterial and fungal community structure and diversity in a mining region under long-term metal exposure revealed by metagenomics sequencing. Ecol. Genet. Genom. 2017, 2, 13–24. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Liang, J.-L.; Liu, J.; Yang, T.-T.; Wang, P.-D.; Zhang, S.-C.; Jia, P.; Liao, B.; Shu, W.-S.; Li, J.-T. Contrasting soil fungal communities at different habitats in a revegetated copper mine wasteland. Soil Ecol. Lett. 2020, 2, 8–19. [Google Scholar] [CrossRef]
- Ozimek, E.; Hanaka, A. Mortierella Species as the Plant Growth-Promoting Fungi Present in the Agricultural Soils. Agriculture 2021, 11, 7. [Google Scholar] [CrossRef]
- Babu, A.G.; Shim, J.; Shea, P.J.; Oh, B.-T. Penicillium aculeatum PDR-4 and Trichoderma sp. PDR-16 promote phytoremediation of mine tailing soil and bioenergy production with sorghum-sudangrass. Ecol. Eng. 2014, 69, 186–191. [Google Scholar] [CrossRef]
- Zhou, L.S.; Tang, K.; Guo, S.X. The Plant Growth-Promoting Fungus (PGPF) Alternaria sp. A13 Markedly Enhances Salvia miltiorrhiza Root Growth and Active Ingredient Accumulation under Greenhouse and Field Conditions. Int. J. Mol. Sci. 2018, 19, 270. [Google Scholar] [CrossRef] [PubMed]
- Osorio, N.W.; Habte, M. Phosphate desorption from the surface of soil mineral particles by a phosphate-solubilizing fungus. Biol. Fertil. Soils 2013, 49, 481–486. [Google Scholar] [CrossRef]
- Onweremadu, E. Predicting phosphorus sorption characteristics in highly weathered soils of South-Eastern Nigeria. Res. J. Environ. Sci. 2007, 1, 47–55. [Google Scholar]
- Zhang, K.; Bonito, G.; Hsu, C.-M.; Hameed, K.; Vilgalys, R.; Liao, H.-L. Mortierella elongata Increases Plant Biomass among Non-Leguminous Crop Species. Agronomy 2020, 10, 754. [Google Scholar] [CrossRef]
- Ozimek, E.; Jaroszuk-Ściseł, J.; Bohacz, J.; Korniłłowicz-Kowalska, T.; Tyśkiewicz, R.; Słomka, A.; Nowak, A.; Hanaka, A. Synthesis of Indoleacetic Acid, Gibberellic Acid and ACC-Deaminase by Mortierella Strains Promote Winter Wheat Seedlings Growth under Different Conditions. Int. J. Mol. Sci. 2018, 19, 3218. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Chai, L.; Yang, Z.; Tang, C.; Tong, H.; Yuan, P. Bioleaching of heavy metals from a contaminated soil using indigenous Penicillium chrysogenum strain F1. J. Hazard. Mater. 2012, 233, 25–32. [Google Scholar] [CrossRef]
- Carrasco, L.; Azcón, R.; Kohler, J.; Roldán, A.; Caravaca, F. Comparative effects of native filamentous and arbuscular mycorrhizal fungi in the establishment of an autochthonous, leguminous shrub growing in a metal-contaminated soil. Sci. Total Environ. 2011, 409, 1205–1209. [Google Scholar] [CrossRef]
- Renpeng, T.; Qiao, Y.; Guoling, Z.; Jingquan, T.; Luozhen, Z.; Chengxiang, F. Taxonomic study on a taxol producing fungus isolated from bark of Taxus chinensis var. mairei. Wuhan Zhi Wu Xue Yan Jiu Wuhan Bot. Res. 2006, 24, 541–545. [Google Scholar]
- Zhao, J.; Shan, T.; Mou, Y.; Zhou, L. Plant-derived bioactive compounds produced by endophytic fungi. Mini-Rev. Med. Chem. 2011, 11, 159–168. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, W.; Zhang, Y.; Chen, X.; Sha, A.; Xiong, Z.; Luo, Y.; Peng, L.; Zou, L.; Zhao, C.; Li, Q. The Diversity and Community Composition of Three Plants’ Rhizosphere Fungi in Kaolin Mining Areas. J. Fungi 2024, 10, 306. https://doi.org/10.3390/jof10050306
Xiao W, Zhang Y, Chen X, Sha A, Xiong Z, Luo Y, Peng L, Zou L, Zhao C, Li Q. The Diversity and Community Composition of Three Plants’ Rhizosphere Fungi in Kaolin Mining Areas. Journal of Fungi. 2024; 10(5):306. https://doi.org/10.3390/jof10050306
Chicago/Turabian StyleXiao, Wenqi, Yunfeng Zhang, Xiaodie Chen, Ajia Sha, Zhuang Xiong, Yingyong Luo, Lianxin Peng, Liang Zou, Changsong Zhao, and Qiang Li. 2024. "The Diversity and Community Composition of Three Plants’ Rhizosphere Fungi in Kaolin Mining Areas" Journal of Fungi 10, no. 5: 306. https://doi.org/10.3390/jof10050306





