Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. RNA Isolation and Analysis of the GH17 Gene Expression by RT-PCR
2.3. Construction of the Single and Multiple Mutants of the GH17 Family with the β-Rec/Six System
2.4. Growth, Sporulation, Germination, Viability, and Morphology of the Mutant Strains
2.5. Carbohydrate Analysis of the Cell Wall Fractions
2.6. Phylogenetic Analysis
2.7. Statistical Analyses
3. Results
3.1. Molecular Characterization of the GH17 Family of A. fumigatus
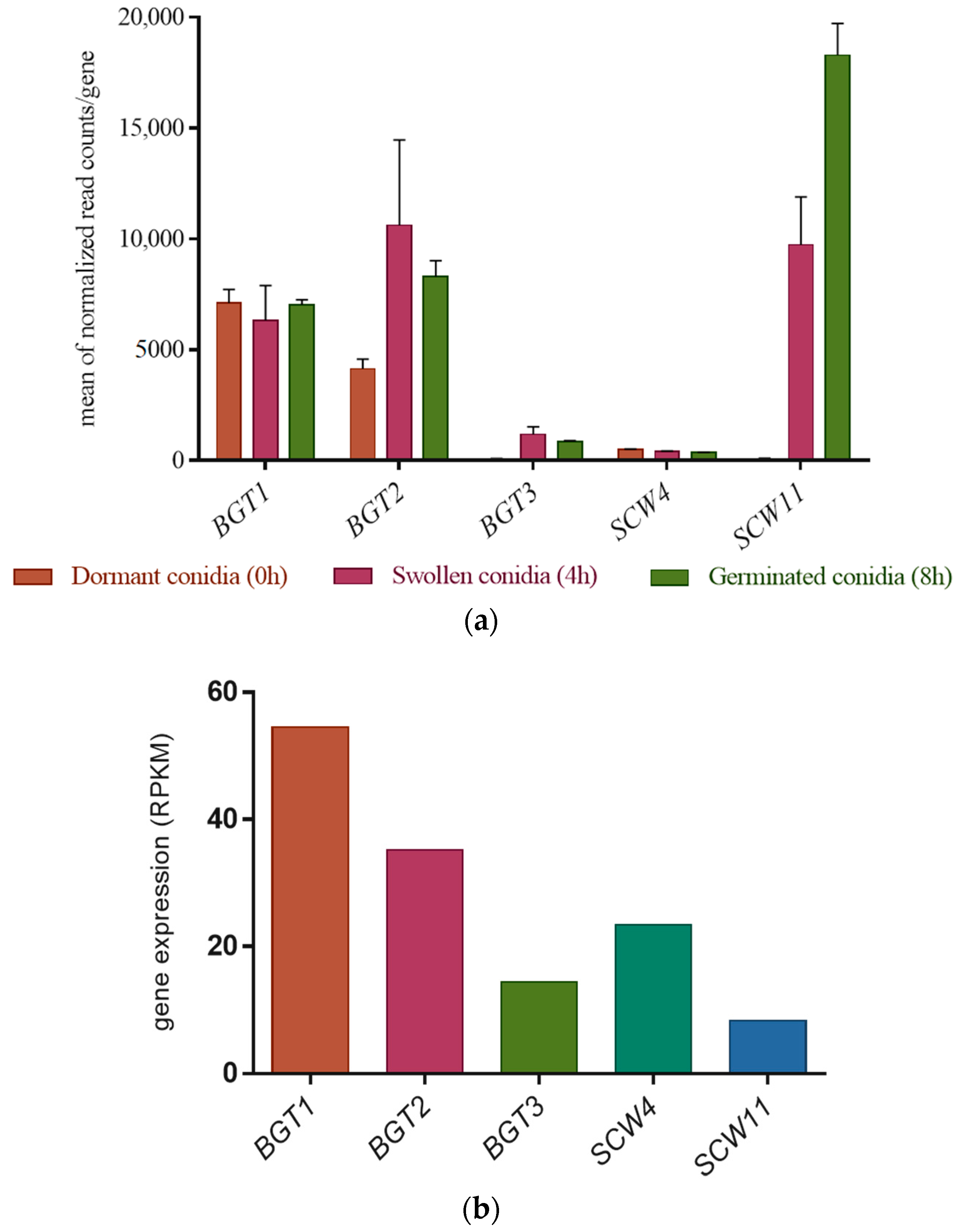
3.2. Gene Expression Level of the GH17 Family during Growth
3.3. Phenotypic Analyses of the Single and Multiple Mutants
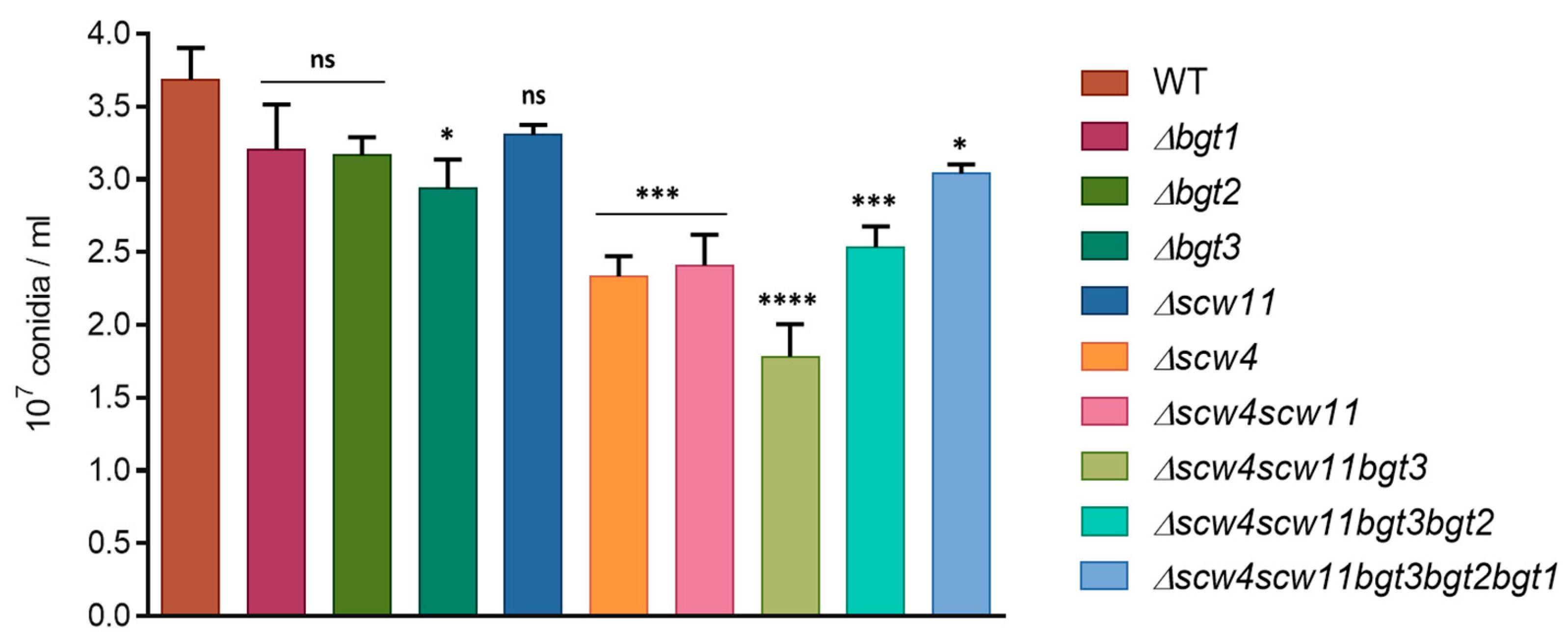
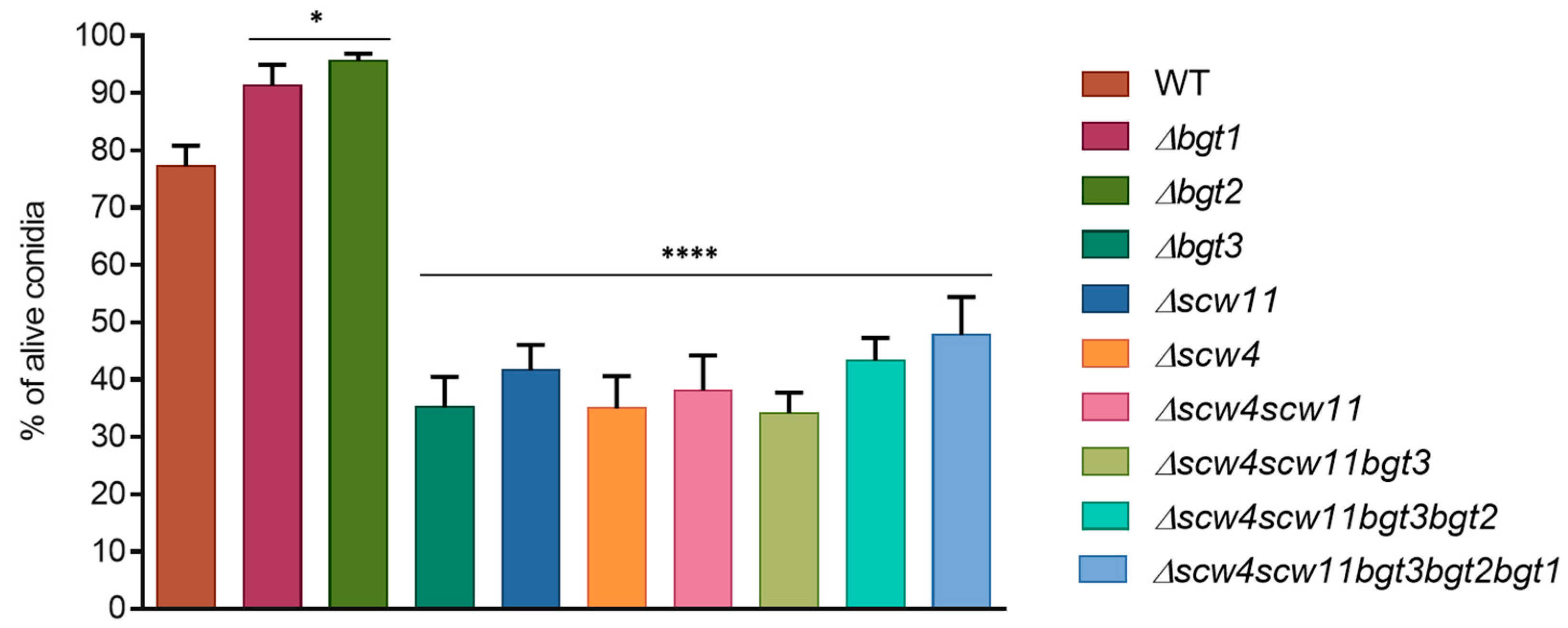
3.3.1. Conidiation, Viability, and Morphology of the Mutants
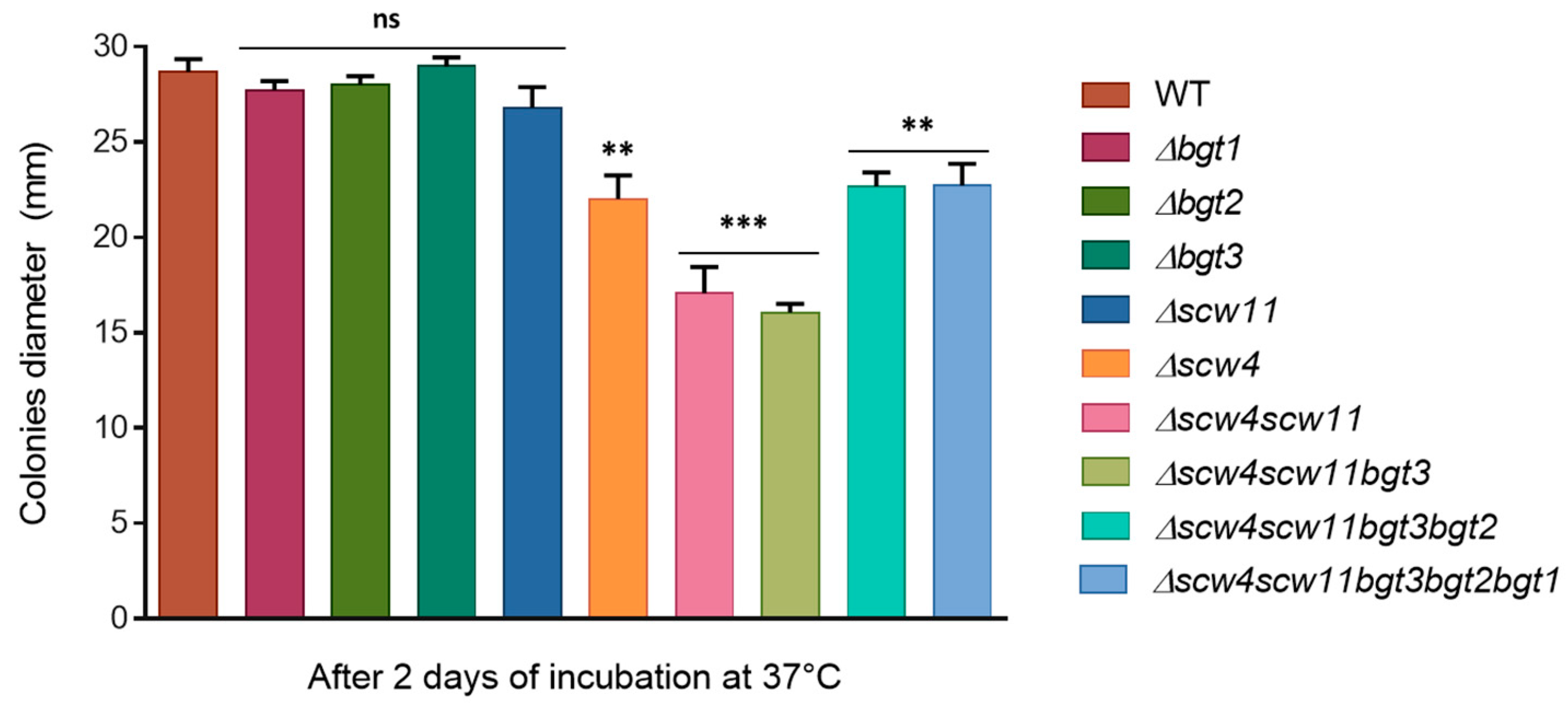
3.3.2. Mycelia Defects
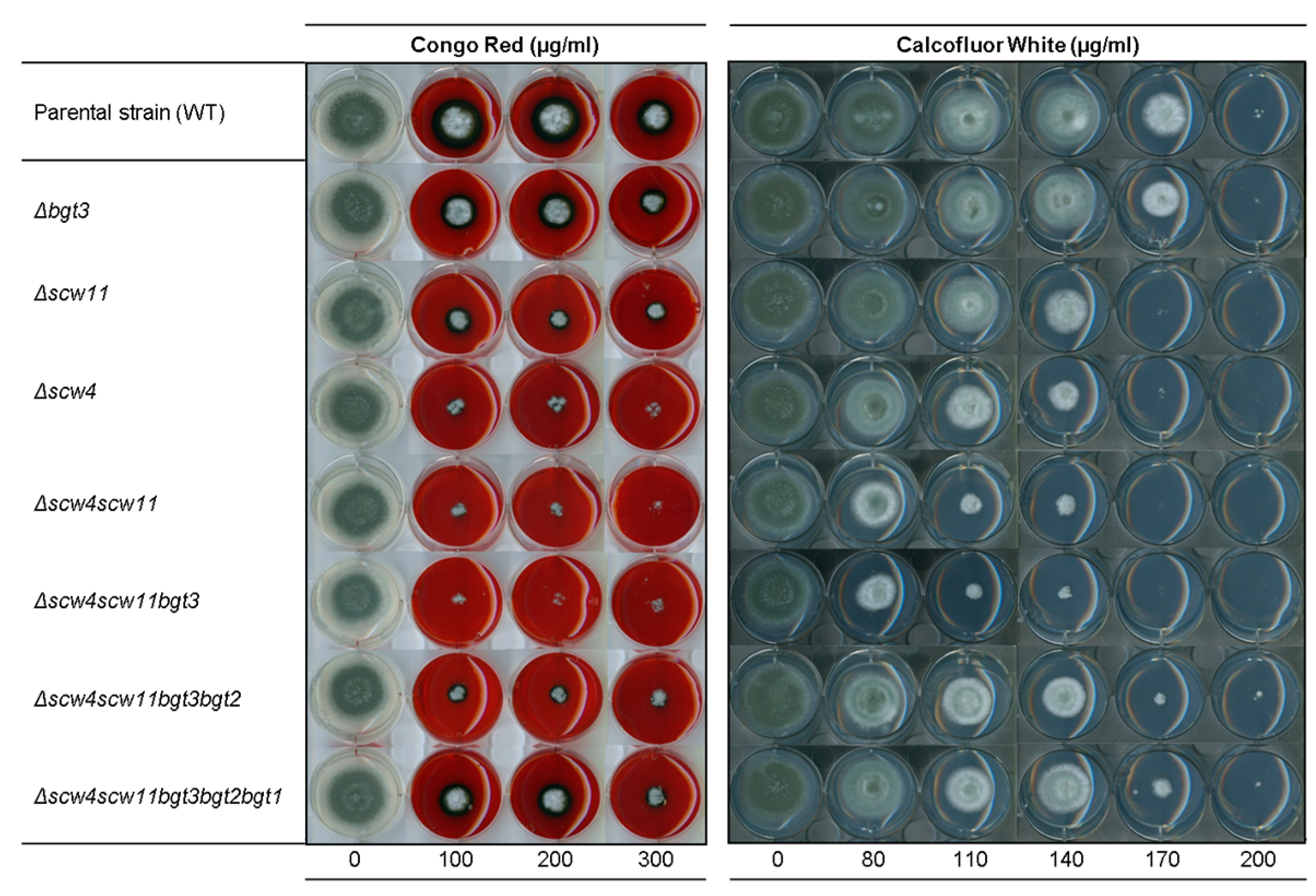
3.3.3. Sensitivity to Cell Wall Disturbing Agents
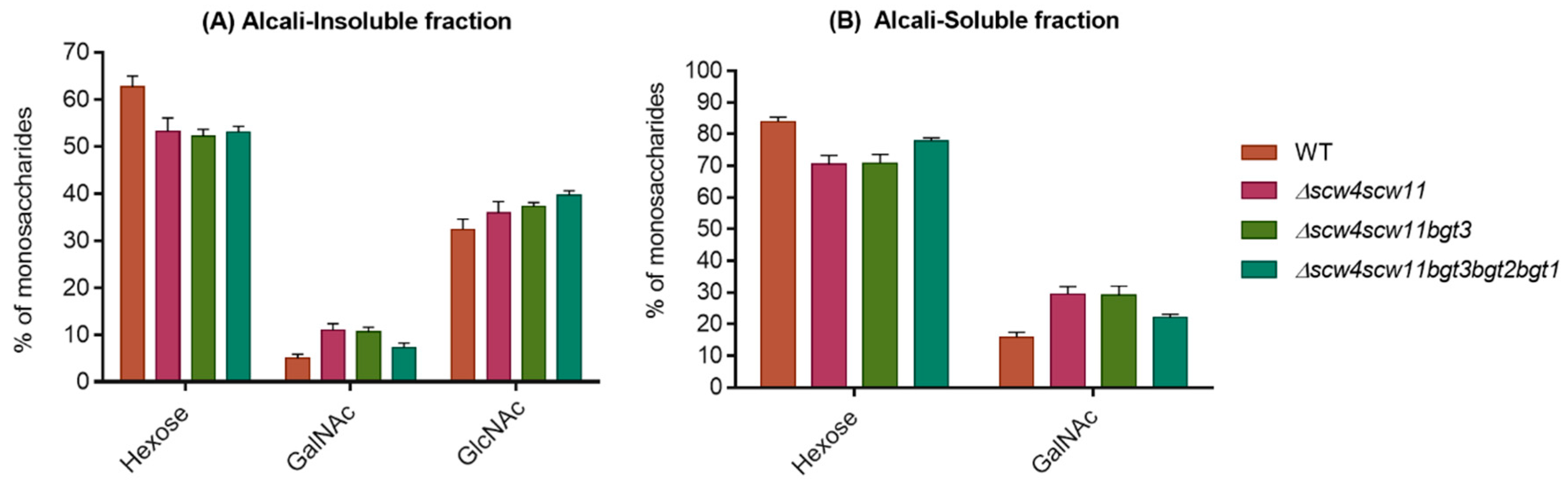
3.4. Analysis of the Cell Wall Polysaccharides
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Latgé, J.P. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 2001, 9, 382–389. [Google Scholar] [CrossRef]
- Beauvais, A.; Fontaine, T.; Aimanianda, V.; Latgé, J.-P. Aspergillus Cell Wall and Biofilm. Mycopathologia 2014, 178, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Free, S.J. Fungal Cell Wall Organization and Biosynthesis. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2013; Volume 81, pp. 33–82. ISBN 978-0-12-407677-8. [Google Scholar]
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latgé, J.-P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Fontaine, T.; Simenel, C.; Dubreucq, G.; Adam, O.; Delepierre, M.; Lemoine, J.; Vorgias, C.E.; Diaquin, M.; Latgé, J.P. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000, 275, 27594–27607. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Drake, R.; Ng, K.; Diaquin, M.; Latgé, J.P. Characterization of the 1,3-β-glucan synthase of Aspergillus fumigatus. Microbiology 1993, 139, 3071–3078. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Bruneau, J.M.; Mol, P.C.; Buitrago, M.J.; Legrand, R.; Latgé, J.P. Glucan Synthase Complex of Aspergillus fumigatus. J. Bacteriol. 2001, 183, 2273–2279. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P.; Steinbach, W.J. Aspergillus fumigatus and Aspergillosis, 1st ed.; ASM Press: Washington, DC, USA, 2008; ISBN 978-1-55581-438-0. [Google Scholar]
- Carbohydrate-Active enZYmes. Available online: http://www.cazy.org (accessed on 19 January 2018).
- Henrissat, B.; Bairoch, A. Updating the sequence-based classification of glycosyl-hydrolases. Biochem. J. 1996, 316, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Hartl, L.; Latgé, J.-P. β-1,3-glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 2013, 4, 81. [Google Scholar] [CrossRef] [PubMed]
- Hartland, R.P.; Fontaine, T.; Debeaupuis, J.P.; Simenel, C.; Delepierre, M.; Latgé, J.P. A novel β-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 1996, 271, 26843–26849. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Monod, M.; Fontaine, T.; Henrissat, B.; Léchenne, B.; Latgé, J.P. Identification of the catalytic residues of the first family of β(1,3)glucanosyltransferases identified in fungi. Biochem. J. 2000, 347, 741–747. [Google Scholar] [PubMed]
- Nakazawa, T.; Horiuchi, H.; Ohta, A.; Takagi, M. Isolation and Characterization of EPD1, an Essential Gene for Pseudohyphal Growth of a Dimorphic Yeast, Candida maltosa. J. Bacteriol. 1998, 180, 2079–2086. [Google Scholar] [PubMed]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latgé, J.-P. Glycosylphosphatidylinositol-anchored Glucanosyltransferases Play an Active Role in the Biosynthesis of the Fungal Cell Wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Morelle, W.; Vai, M.; Monod, M.; Léchenne, B.; Fontaine, T.; Beauvais, A.; Sarfati, J.; Prévost, M.-C.; Henry, C.; et al. Deletion of GEL2 encoding for a β(1–3) glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 2005, 56, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Gastebois, A.; Fontaine, T.; Latgé, J.-P.; Mouyna, I. β(1,3)Glucanosyltransferase Gel4p Is Essential for Aspergillus fumigatus. Eukaryot. Cell 2010, 9, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Hartland, R.P.; Fontaine, T.; Diaquin, M.; Simenel, C.; Delepierre, M.; Henrissat, B.; Latge, J.-P. A 1,3-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bgl2p. Microbiology 1998, 144, 3171–3180. [Google Scholar] [CrossRef] [PubMed]
- Gastebois, A.; Mouyna, I.; Simenel, C.; Clavaud, C.; Coddeville, B.; Delepierre, M.; Latgé, J.-P.; Fontaine, T. Characterization of a New β(1–3)-Glucan Branching Activity of Aspergillus fumigatus. J. Biol. Chem. 2010, 285, 2386–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sestak, S.; Hagen, I.; Tanner, W.; Strahl, S. Scw10p, a cell-wall glucanase/transglucosidase important for cell-wall stability in Saccharomyces cerevisiae. Microbiology 2004, 150, 3197–3208. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Ferreira, M.E.; Kress, M.R.V.Z.; Savoldi, M.; Goldman, M.H.S.; Hartl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuBKU80 Mutant Deficient for Nonhomologous End Joining Is a Powerful Tool for Analyzing Pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T.; Dümig, M.; Jaber, B.M.; Szewczyk, E.; Olbermann, P.; Morschhäuser, J.; Krappmann, S. Validation of a Self-Excising Marker in the Human Pathogen Aspergillus fumigatus by Employing the β-Rec/six Site-Specific Recombination System. Appl. Environ. Microbiol. 2010, 76, 6313. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, O.; Aguirre, J. Efficient Transformation of Aspergillus nidulans by Electroporation of Germinated Conidia. Fungal Genet. Rep. 1996, 43, 48–51. [Google Scholar] [CrossRef]
- Gastebois, A.; Aimanianda, V.; Bachellier-Bassi, S.; Nesseir, A.; Firon, A.; Beauvais, A.; Schmitt, C.; England, P.; Beau, R.; Prévost, M.-C.; et al. SUN Proteins Belong to a Novel Family of β-(1,3)-Glucan-modifying Enzymes Involved in Fungal Morphogenesis. J. Biol. Chem. 2013, 288, 13387–13396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Johnson, A.R. Improved method of hexosamine determination. Anal. Biochem. 1971, 44, 628–635. [Google Scholar] [CrossRef]
- EnsemblFungi. Available online: http://fungi.ensembl.org/Aspergillus_fumigatusa1163/Info/Index.
- SignalP 4.1 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 14 August 2017).
- Big-PI Fungal Predictor GPI Modification Site Prediction in Fungi. Available online: http://mendel.imp.ac.at/gpi/fungi_server.html (accessed on 12 January 2004).
- PredGPI Predictor. Available online: http://gpcr.biocomp.unibo.it/predgpi/pred.htm (accessed on 23 September 2008).
- Mouyna, I.; Aimanianda, V.; Hartl, L.; Prevost, M.C.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Legendre, R.; Coppee, J.Y.; Latgé, J.-P. GH16 and GH81 family β-(1,3)-glucanases in Aspergillus fumigatus are essential for conidial cell wall morphogenesis. Cell. Microbiol. 2016, 18, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, I.; Sarikaya-Bayram, Ö.; Wong Sak Hoi, J.; Muszkieta, L.; Gibbons, J.; Prevost, M.-C.; Mallet, A.; Krijnse-Locker, J.; Ibrahim-Granet, O.; Mouyna, I.; et al. MybA, a transcription factor involved in conidiation and conidial viability of the human pathogen Aspergillus fumigatus. Mol. Microbiol. 2017, 105, 880–900. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Mrsa, V.; Tanner, W. New Potential Cell Wall Glucanases of Saccharomyces cerevisiae and Their Involvement in Mating. J. Bacteriol. 1998, 180, 5030–5037. [Google Scholar] [PubMed]
- Teparic, R. Increased mortality of Saccharomyces cerevisiae cell wall protein mutants. Microbiology 2004, 150, 3145–3150. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Samantaray, S.; Aimanianda, V.; Zhu, Z.; Prévost, M.-C.; Latgé, J.-P.; Ebel, F.; Wagener, J. Aspergillus fumigatus devoid of cell wall β-1,3-glucan is viable, massively sheds galactomannan and is killed by septum formation inhibitors. Mol. Microbiol. 2015, 95, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Latgé, J.-P.; Beauvais, A. α1,3 Glucans Are Dispensable in Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Mrsa, V.; Klebl, F.; Tanner, W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-1,3-glucanase. J. Bacteriol. 1993, 175, 2102–2106. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Simenel, C.; Garnaud, C.; Clavaud, C.; Tada, R.; Barbin, L.; Mouyna, I.; Heddergott, C.; Popolo, L.; Ohya, Y.; et al. The Dual Activity Responsible for the Elongation and Branching of β-(1,3)-Glucan in the Fungal Cell Wall. mBio 2017, 8, e00619-17. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, M.J.; Robbins, P.W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 19758–19767. [Google Scholar] [PubMed]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 Activate Daughter-Specific Genetic Programs to Induce Asymmetric Cell Fates. Cell 2001, 107, 739–750. [Google Scholar] [CrossRef]

| Proteins | Size (aa) | Signal Peptide | GPI Anchor | AFUA Gene Number |
|---|---|---|---|---|
| Bgt1p | 305 | 1-18 | NO | AFUA_1G11460 |
| Bgt2p | 446 | 1-18 | Yes (Ω site: 423) | AFUA_3G00270 |
| Bgt3p | 688 | No | No | AFUA_5G08780 |
| Scw4p | 369 | 1-18 | No | AFUA_6G12380 |
| Scw11p | 565 | 1-18 | No | AFUA_8G05610 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millet, N.; Latgé, J.-P.; Mouyna, I. Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis. J. Fungi 2018, 4, 18. https://doi.org/10.3390/jof4010018
Millet N, Latgé J-P, Mouyna I. Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis. Journal of Fungi. 2018; 4(1):18. https://doi.org/10.3390/jof4010018
Chicago/Turabian StyleMillet, Nicolas, Jean-Paul Latgé, and Isabelle Mouyna. 2018. "Members of Glycosyl-Hydrolase Family 17 of A. fumigatus Differentially Affect Morphogenesis" Journal of Fungi 4, no. 1: 18. https://doi.org/10.3390/jof4010018





