Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus
Abstract
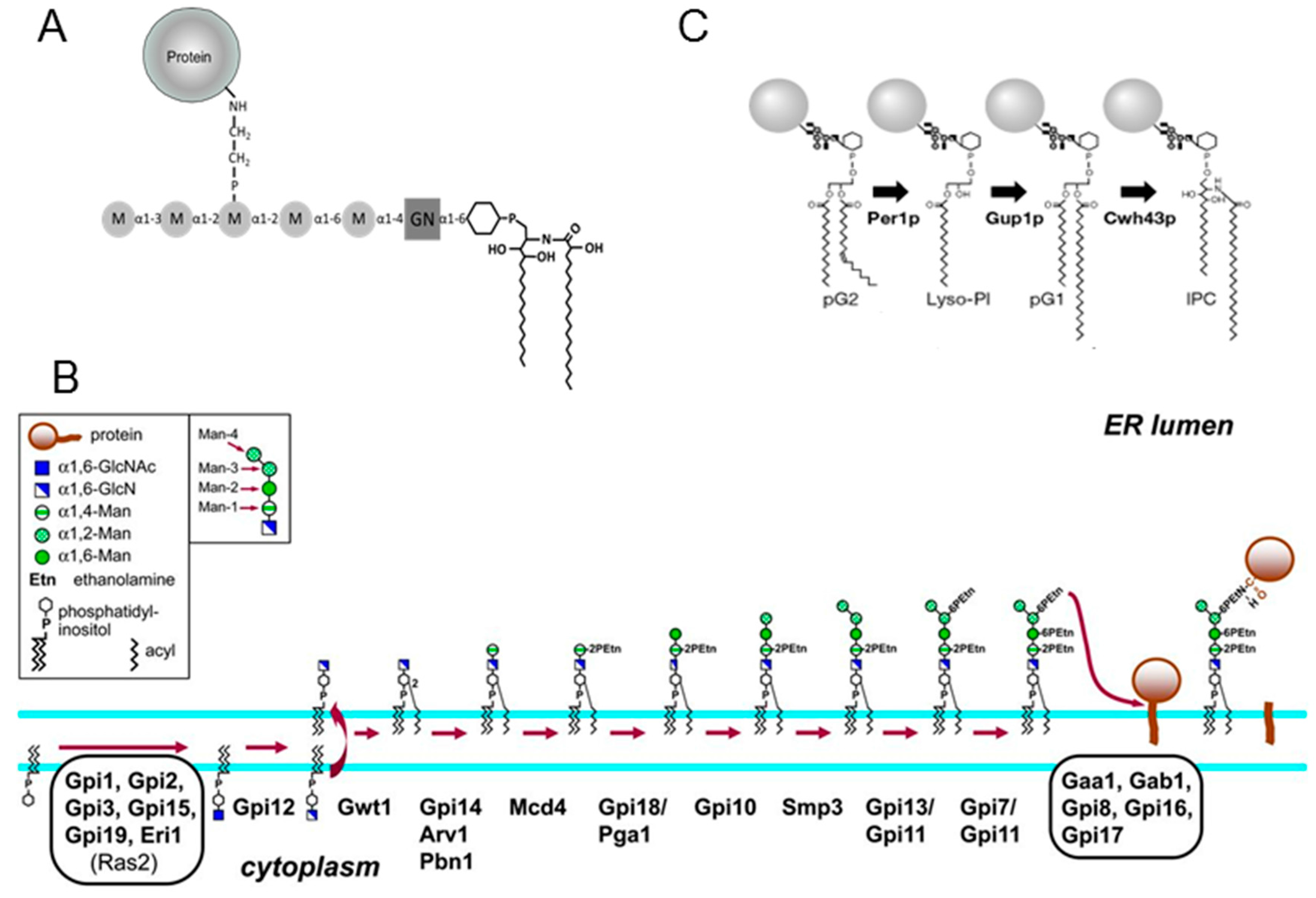
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Construction of the Δper1 Mutant
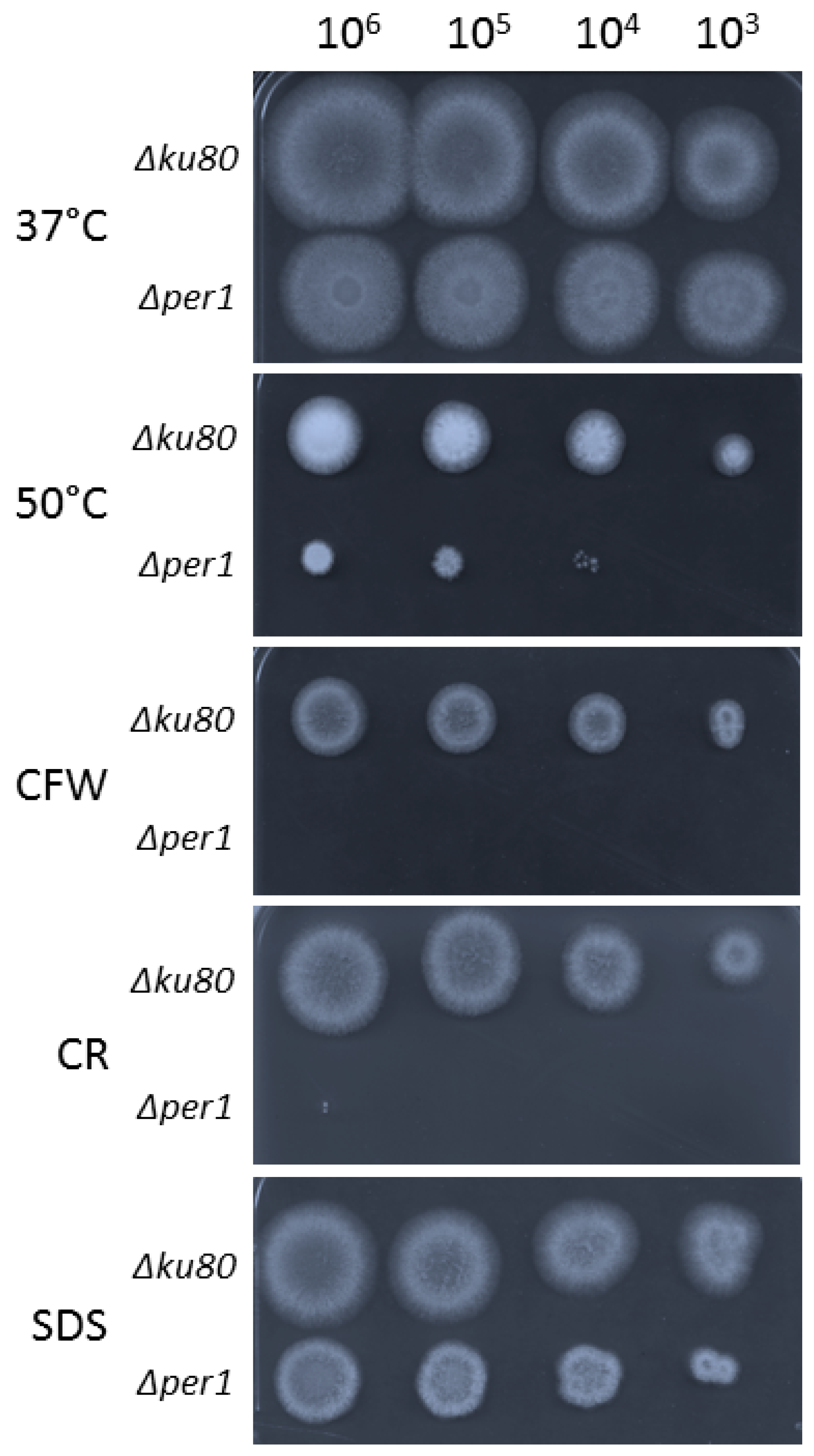
2.3. Fungal Morphotype of the Δper1 Mutant Strain
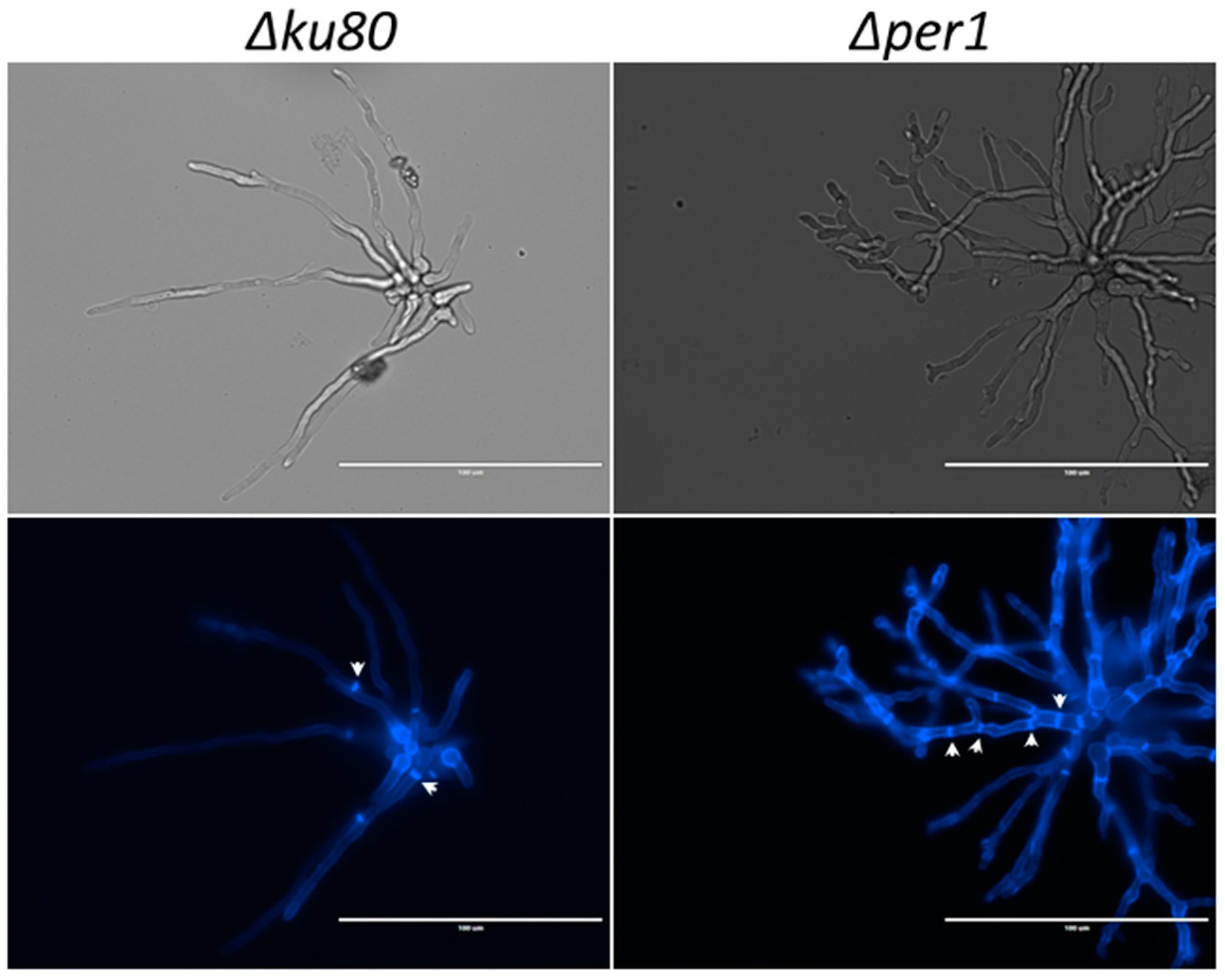
2.4. Microscopy
2.5. Carbohydrate Analysis of the Cell Wall and Culture Supernatant Fractions
2.6. GPI-AP Purification and Detection
2.7. Lipogalactomannan (LGM) and Glycosylinositolphosphoceramide (GIPC) Purification
2.8. MS Analysis of the GPI Lipid Moiety
2.9. Statistical Analysis
3. Results
3.1. Construction of the ∆per1 Mutant, Mycelial Growth and Hyphal Morphology
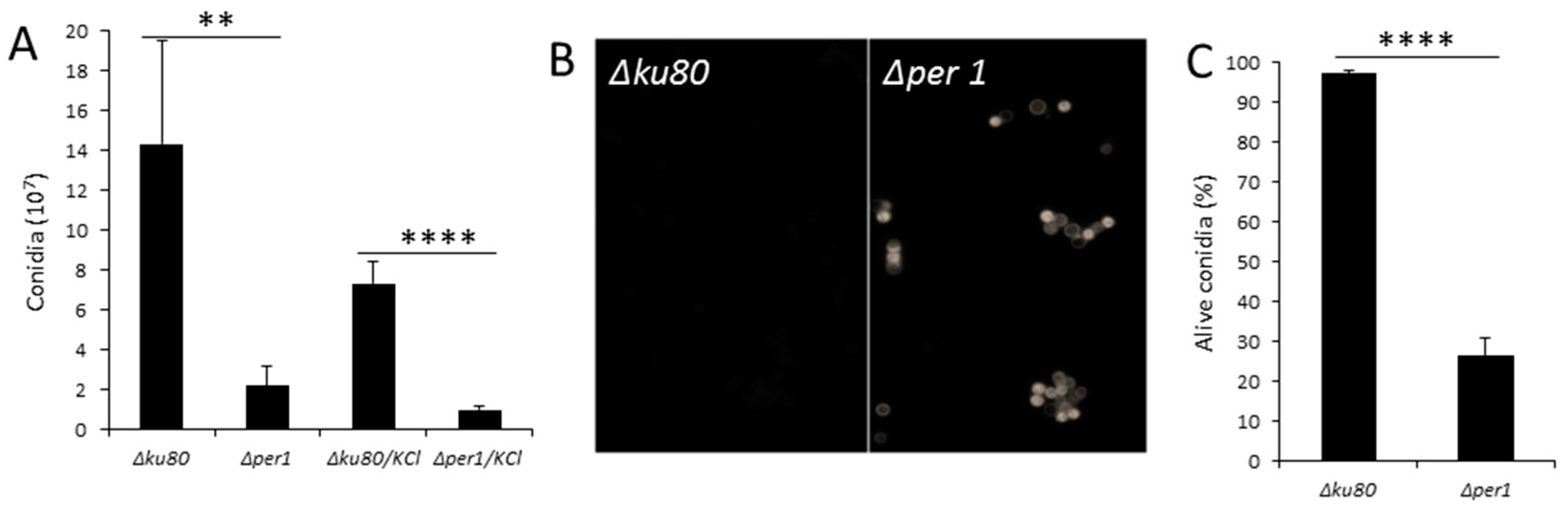
3.2. Conidiation and Conidial Viability
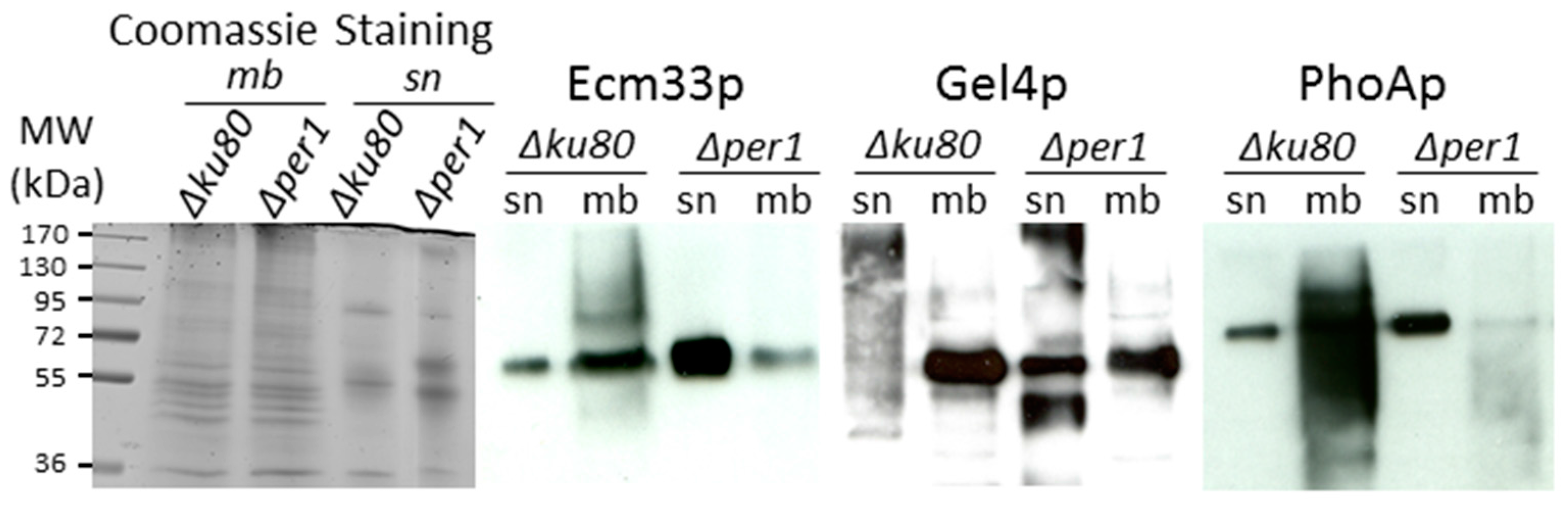
3.3. Localisation of GPI-APs
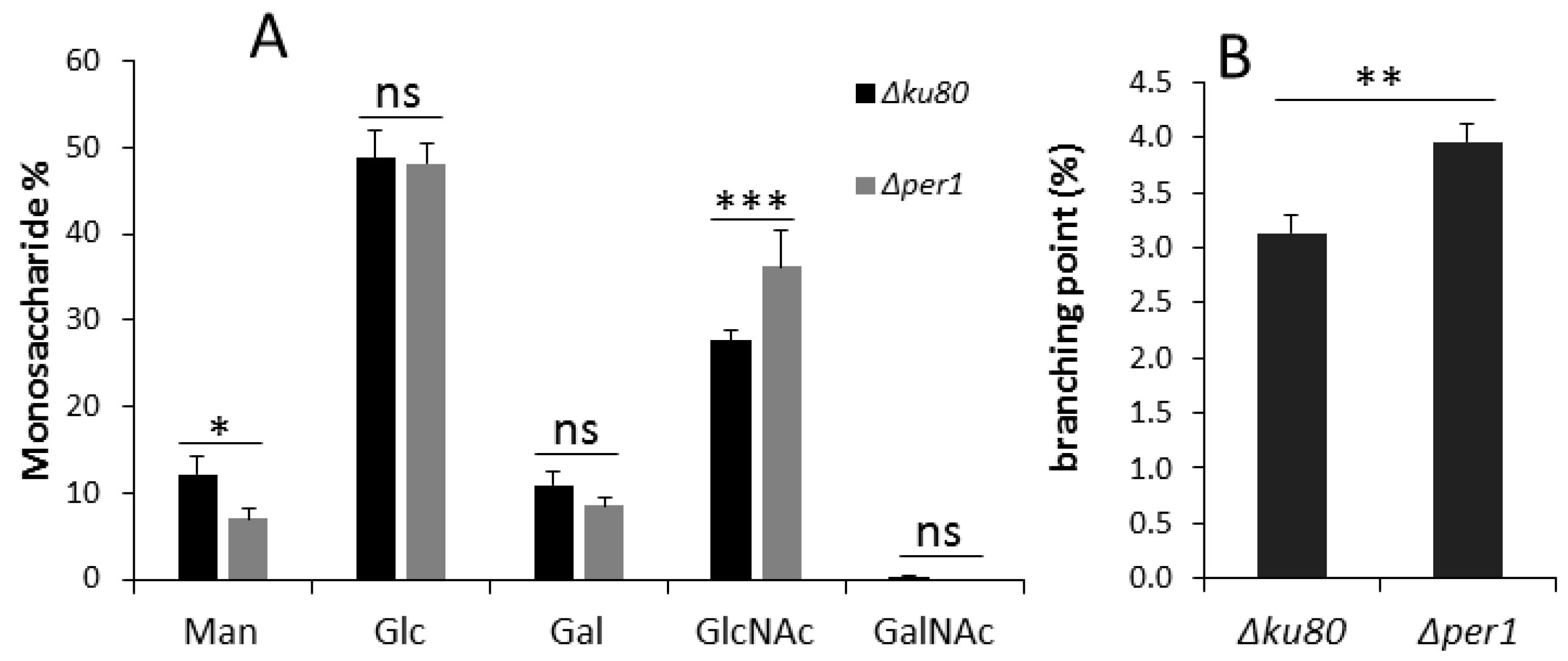
3.4. Cell Wall Composition
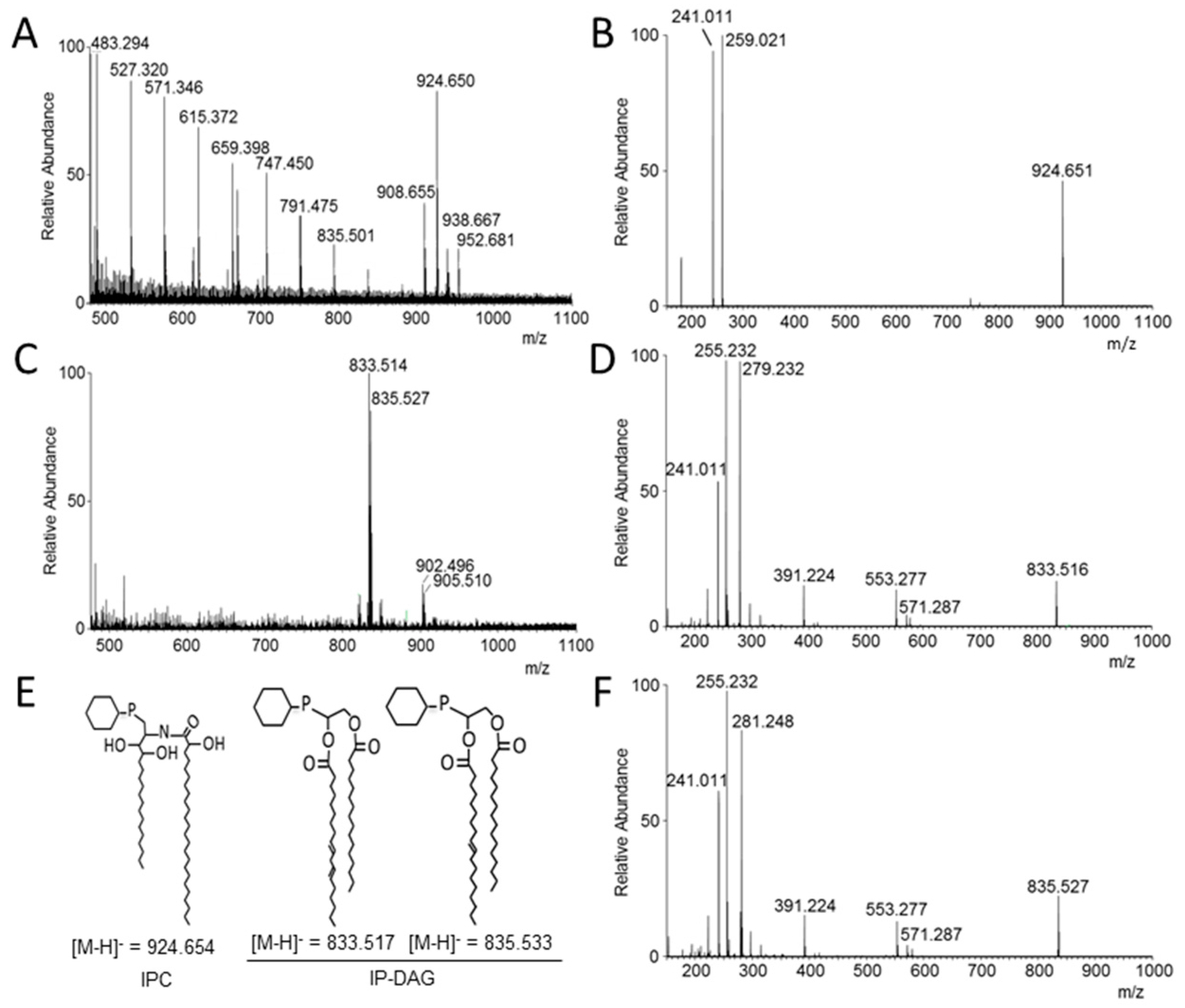
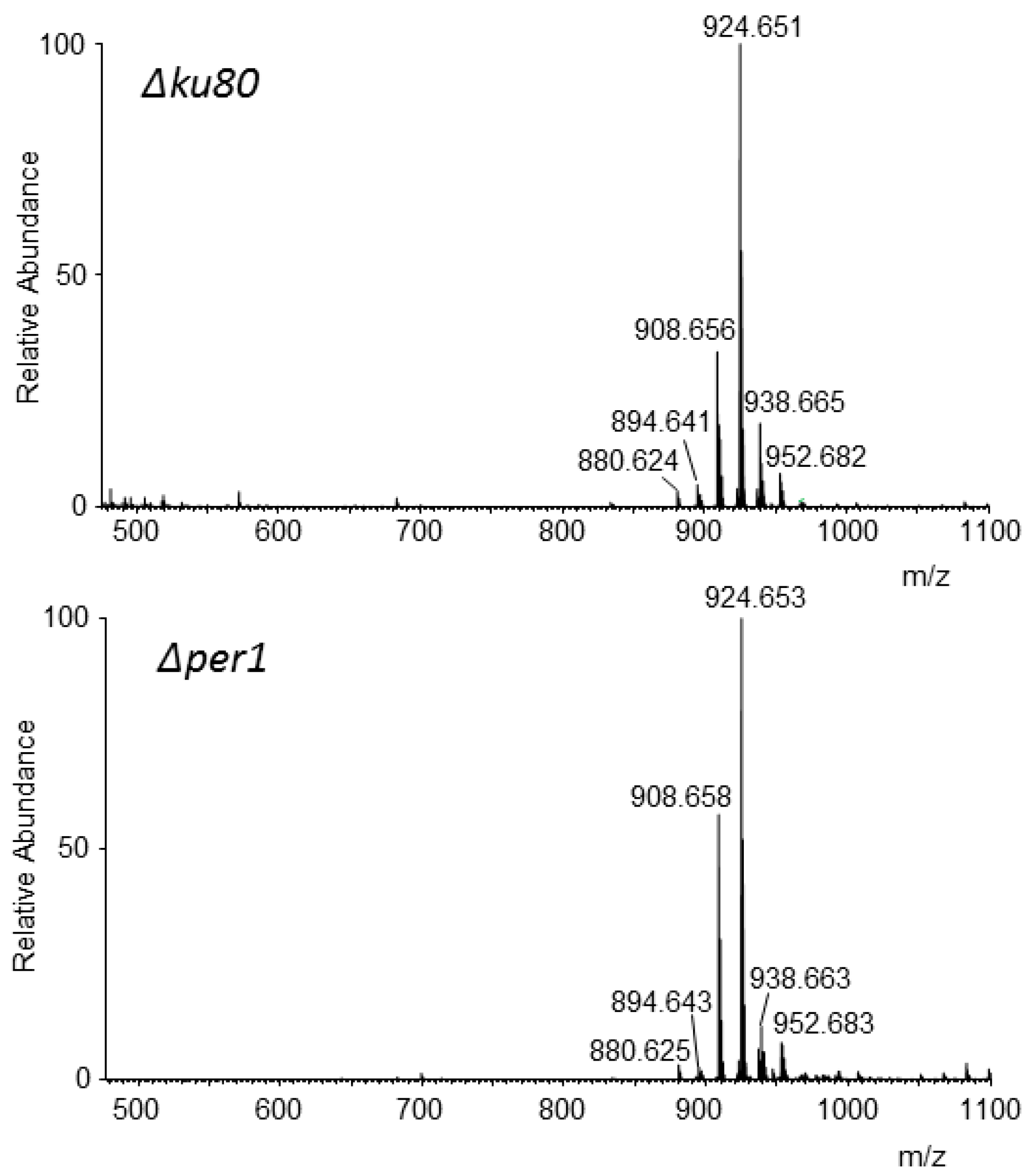
3.5. Analysis of Lipid Moiety of GPI Anchors
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McConville, M.J.; Ferguson, M.A. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 1993, 294, 305–324. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Maruyama, J.; Kitamoto, K.; Sumikoshi, K.; Terada, T.; Nakamura, S.; Shimizu, K. Using a new GPI-anchored-protein identification system to mine the protein databases of Aspergillus fumigatus, Aspergillus nidulans, and Aspergillus oryzae. J. Gen. Appl. Microbiol. 2009, 55, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Gastebois, A.; Mouyna, I.; Simenel, C.; Clavaud, C.; Coddeville, B.; Delepierre, M.; Latgé, J.-P.; Fontaine, T. Characterization of a New β(1–3)-Glucan Branching Activity of Aspergillus fumigatus. J. Biol. Chem. 2010, 285, 2386–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartland, R.P.; Fontaine, T.; Debeaupuis, J.P.; Simenel, C.; Delepierre, M.; Latgé, J.P. A novel beta-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 1996, 271, 26843–26849. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latgé, J.P. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Gastebois, A.; Fontaine, T.; Latgé, J.-P.; Mouyna, I. β(1-3)Glucanosyltransferase Gel4p is Essential for Aspergillus fumigatus. Eukaryot. Cell 2010, 9, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Simenel, C.; Garnaud, C.; Clavaud, C.; Tada, R.; Barbin, L.; Mouyna, I.; Heddergott, C.; Popolo, L.; Ohya, Y.; et al. The Dual Activity Responsible for the Elongation and Branching of β-(1,3)-Glucan in the Fungal Cell Wall. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Farkas, V.; Kosík, O.; Blanco, N.; Arroyo, J.; McPhie, P. Assembly of the Yeast Cell Wall Crh1p and Crh2p Act as Transglycosylases In Vivo and In Vitro. J. Biol. Chem. 2008, 283, 29859–29872. [Google Scholar] [CrossRef] [PubMed]
- Kitagaki, H.; Wu, H.; Shimoi, H.; Ito, K. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 46, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Muszkieta, L.; Fontaine, T.; Beau, R.; Mouyna, I.; Vogt, M.S.; Essen, L.-O.; Jouvion, G.; Latgé, J.-P. The GPI-anchored DFG family is essential for cross-linking galactomannan to cell wall β1–3glucan in Aspergillus fumigatus. J. Biol. Chem. 2018. submitted. [Google Scholar]
- Fankhauser, C.; Homans, S.W.; Thomas-Oates, J.E.; McConville, M.J.; Desponds, C.; Conzelmann, A.; Ferguson, M.A. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J. Biol. Chem. 1993, 268, 26365–26374. [Google Scholar] [PubMed]
- Fontaine, T.; Magnin, T.; Melhert, A.; Lamont, D.; Latgé, J.; Ferguson, M.A.J. Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 2003, 13, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Yoko-o, T.; Ichikawa, D.; Miyagishi, Y.; Kato, A.; Umemura, M.; Takase, K.; Ra, M.; Ikeda, K.; Taguchi, R.; Jigami, Y. Determination and physiological roles of the glycosylphosphatidylinositol lipid remodelling pathway in yeast. Mol. Microbiol. 2013, 88, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Costachel, C.; Coddeville, B.; Latgé, J.-P.; Fontaine, T. Glycosylphosphatidylinositol-anchored Fungal Polysaccharide in Aspergillus fumigatus. J. Biol. Chem. 2005, 280, 39835–39842. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2007, 1771, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Smith, T.K.; Crossman, A.; Brimacombe, J.S.; Latgé, J.-P.; Ferguson, M.A.J. In Vitro Biosynthesis of Glycosylphosphatidylinositol in Aspergillus fumigatus. Biochemistry (Moscow) 2004, 43, 15267–15275. [Google Scholar] [CrossRef] [PubMed]
- Manzano-Lopez, J.; Perez-Linero, A.M.; Aguilera-Romero, A.; Martin, M.E.; Okano, T.; Silva, D.V.; Seeberger, P.H.; Riezman, H.; Funato, K.; Goder, V.; et al. COPII Coat Composition is Actively Regulated by Luminal Cargo Maturation. Curr. Biol. 2015, 25, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, H.M.; Vionnet, C.; Roubaty, C.; Conzelmann, A. Cdc1 removes the ethanolamine phosphate of the first mannose of GPI anchors and thereby facilitates the integration of GPI proteins into the yeast cell wall. Mol. Biol. Cell 2014, 25, 3375–3388. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.T.; Engel, J.; Buettner, F.F.R.; Routier, F.H. Aspergillus fumigatus Cap59-like protein A is involved in α1,3-mannosylation of GPI-anchors. Glycobiology 2016, 26, 30–38. [Google Scholar] [PubMed]
- Fujita, M.; Jigami, Y. Lipid remodeling of GPI-anchored proteins and its function. Biochim. Biophys. Acta BBA Gen. Subj. 2008, 1780, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Ghugtyal, V.; Vionnet, C.; Roubaty, C.; Conzelmann, A. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol. Microbiol. 2007, 65, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Umemura, M.; Yoko-o, T.; Jigami, Y. PER1 is Required for GPI-Phospholipase A2 Activity and Involved in Lipid Remodeling of GPI-anchored Proteins. Mol. Biol. Cell 2006, 17, 5253–5264. [Google Scholar] [CrossRef] [PubMed]
- Bosson, R.; Jaquenoud, M.; Conzelmann, A. GUP1 of Saccharomyces cerevisiae Encodes an O-Acyltransferase Involved in Remodeling of the GPI Anchor. Mol. Biol. Cell 2006, 17, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Umemura, M.; Fujita, M.; Yoko-o, T.; Fukamizu, A.; Jigami, Y. Saccharomyces cerevisiae CWH43 is Involved in the Remodeling of the Lipid Moiety of GPI Anchors to Ceramides. Mol. Biol. Cell 2007, 18, 4304–4316. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Tashima, Y.; Houjou, T.; Fujita, M.; Yoko-o, T.; Jigami, Y.; Taguchi, R.; Kinoshita, T. Fatty Acid Remodeling of GPI-anchored Proteins is Required for Their Raft Association. Mol. Biol. Cell 2007, 18, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Thammahong, A.; Shepardson, K.M.; Blosser, S.J.; Cramer, R.A. Endoplasmic reticulum localized PerA is required for cell wall integrity, azole drug resistance, and virulence in Aspergillus fumigatus. Mol. Microbiol. 2014, 92, 1279–1298. [Google Scholar] [CrossRef] [PubMed]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; van den Hondel, C.A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Da Silva Ferreira, M.E.; Kress, M.R.V.Z.; Savoldi, M.; Goldman, M.H.S.; Härtl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuBKU80 Mutant Deficient for Nonhomologous End Joining is a Powerful Tool for Analyzing Pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Collopy, P.D.; Colot, H.V.; Park, G.; Ringelberg, C.; Crew, C.M.; Borkovich, K.A.; Dunlap, J.C. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol. Biol. Clifton NJ 2010, 638, 33–40. [Google Scholar]
- Muszkieta, L.; Aimanianda, V.; Mellado, E.; Gribaldo, S.; Alcàzar-Fuoli, L.; Szewczyk, E.; Prevost, M.-C.; Latgé, J.-P. Deciphering the role of the chitin synthase families 1 and 2 in the in vivo and in vitro growth of Aspergillus fumigatus by multiple gene targeting deletion. Cell. Microbiol. 2014, 16, 1784–1805. [Google Scholar] [CrossRef] [PubMed]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Stalhberger, T.; Simenel, C.; Clavaud, C.; Eijsink, V.G.H.; Jourdain, R.; Delepierre, M.; Latgé, J.-P.; Breton, L.; Fontaine, T. Chemical Organization of the Cell Wall Polysaccharide Core of Malassezia restricta. J. Biol. Chem. 2014, 289, 12647–12656. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.; Mouyna, I.; Dubreucq, G.; Debeaupuis, J.-P.; Fontaine, T.; Vorgias, C.; Fuglsang, C.; Latgé, J.-P. Characterization of a cell-wall acid phosphatase (PhoAp) in Aspergillus fumigatus. Microbiol. Read. Engl. 2002, 148, 2819–2829. [Google Scholar] [CrossRef] [PubMed]
- Chabane, S.; Sarfati, J.; Ibrahim-Granet, O.; Du, C.; Schmidt, C.; Mouyna, I.; Prevost, M.-C.; Calderone, R.; Latgé, J.-P. Glycosylphosphatidylinositol-Anchored Ecm33p Influences Conidial Cell Wall Biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 2006, 72, 3259–3267. [Google Scholar] [CrossRef] [PubMed]
- Simenel, C.; Coddeville, B.; Delepierre, M.; Latgé, J.-P.; Fontaine, T. Glycosylinositolphosphoceramides in Aspergillus fumigatus. Glycobiology 2008, 18, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, J.M.; Magnin, T.; Tagat, E.; Legrand, R.; Bernard, M.; Diaquin, M.; Fudali, C.; Latgé, J.P. Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 2001, 22, 2812–2823. [Google Scholar] [CrossRef]
- Treumann, A.; Güther, M.L.; Schneider, P.; Ferguson, M.A. Analysis of the carbohydrate and lipid components of glycosylphosphatidylinositol structures. Methods Mol. Biol. Clifton NJ 1998, 76, 213–235. [Google Scholar]
- Zehethofer, N.; Scior, T.; Lindner, B. Elucidation of the fragmentation pathways of different phosphatidylinositol phosphate species (PIPx) using IRMPD implemented on a FT-ICR MS. Anal. Bioanal. Chem. 2010, 398, 2843–2851. [Google Scholar] [CrossRef] [PubMed]
- Toledo, M.S.; Levery, S.B.; Bennion, B.; Guimaraes, L.L.; Castle, S.A.; Lindsey, R.; Momany, M.; Park, C.; Straus, A.H.; Takahashi, H.K. Analysis of glycosylinositol phosphorylceramides expressed by the opportunistic mycopathogen Aspergillus fumigatus. J. Lipid Res. 2007, 48, 1801–1824. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Schmalhorst, P.S.; Krüger, A.T.; Müller, C.T.; Buettner, F.F.R.; Routier, F.H. Characterization of an N-acetylglucosaminyltransferase involved in Aspergillus fumigatus zwitterionic glycoinositolphosphoceramide biosynthesis. Glycobiology 2015, 25, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Kotz, A.; Wagener, J.; Engel, J.; Routier, F.; Echtenacher, B.; Pich, A.; Rohde, M.; Hoffmann, P.; Heesemann, J.; Ebel, F. The mitA gene of Aspergillus fumigatus is required for mannosylation of inositol-phosphorylceramide, but is dispensable for pathogenicity. Fungal Genet. Biol. FG B 2010, 47, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Lamarre, C.; Beau, R.; Balloy, V.; Fontaine, T.; Hoi, J.W.S.; Guadagnini, S.; Berkova, N.; Chignard, M.; Beauvais, A.; Latgé, J.-P. Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 2009, 11, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. GfsA encodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen of O-glycan in Aspergillus nidulans and A. fumigatus. Mol. Microbiol. 2013, 90, 1054–1073. [Google Scholar] [CrossRef] [PubMed]
- Katafuchi, Y.; Li, Q.; Tanaka, Y.; Shinozuka, S.; Kawamitsu, Y.; Izumi, M.; Ekino, K.; Mizuki, K.; Takegawa, K.; Shibata, N.; et al. GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 2017, 27, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Schmalhorst, P.S.; Dörk-Bousset, T.; Ferrières, V.; Routier, F.H. A Single UDP-galactofuranose Transporter is Required for Galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 2009, 284, 33859–33868. [Google Scholar] [CrossRef] [PubMed]
- Jigami, Y. Yeast Glycobiology and Its Application. Biosci. Biotechnol. Biochem. 2008, 72, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Fontaine, T.; Heddergott, C.; Robinet, P.; Aimanianda, V.; Beau, R.; Beauvais, A.; Mouyna, I.; Prevost, M.-C.; Fekkar, A.; et al. Biosynthesis of cell wall mannan in the conidium and the mycelium of Aspergillus fumigatus. Cell. Microbiol. 2016, 18, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Mouyna, I.; Henry, C.; Moyrand, F.; Malosse, C.; Chamot-Rooke, J.; Janbon, G.; Latgé, J.-P.; Fontaine, T. Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus. J. Fungi 2018, 4, 19. https://doi.org/10.3390/jof4010019
Li J, Mouyna I, Henry C, Moyrand F, Malosse C, Chamot-Rooke J, Janbon G, Latgé J-P, Fontaine T. Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus. Journal of Fungi. 2018; 4(1):19. https://doi.org/10.3390/jof4010019
Chicago/Turabian StyleLi, Jizhou, Isabelle Mouyna, Christine Henry, Frédérique Moyrand, Christian Malosse, Julia Chamot-Rooke, Guilhem Janbon, Jean-Paul Latgé, and Thierry Fontaine. 2018. "Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus" Journal of Fungi 4, no. 1: 19. https://doi.org/10.3390/jof4010019





