Cysteine Dioxygenase Enzyme Activity and Gene Expression in the Dimorphic Pathogenic Fungus Histoplasma capsulatum Is in both the Mold and Yeast Morphotypes and Exhibits Substantial Strain Variation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth
2.2. DNA and RNA Extraction and Blotting
2.3. Quantitative PCR
2.4. Standard and Fusion PCR
2.5. CDO Assay
2.6. CDO Flag-Tag Analysis
2.7. Intracellular Cysteine Analysis
3. Results
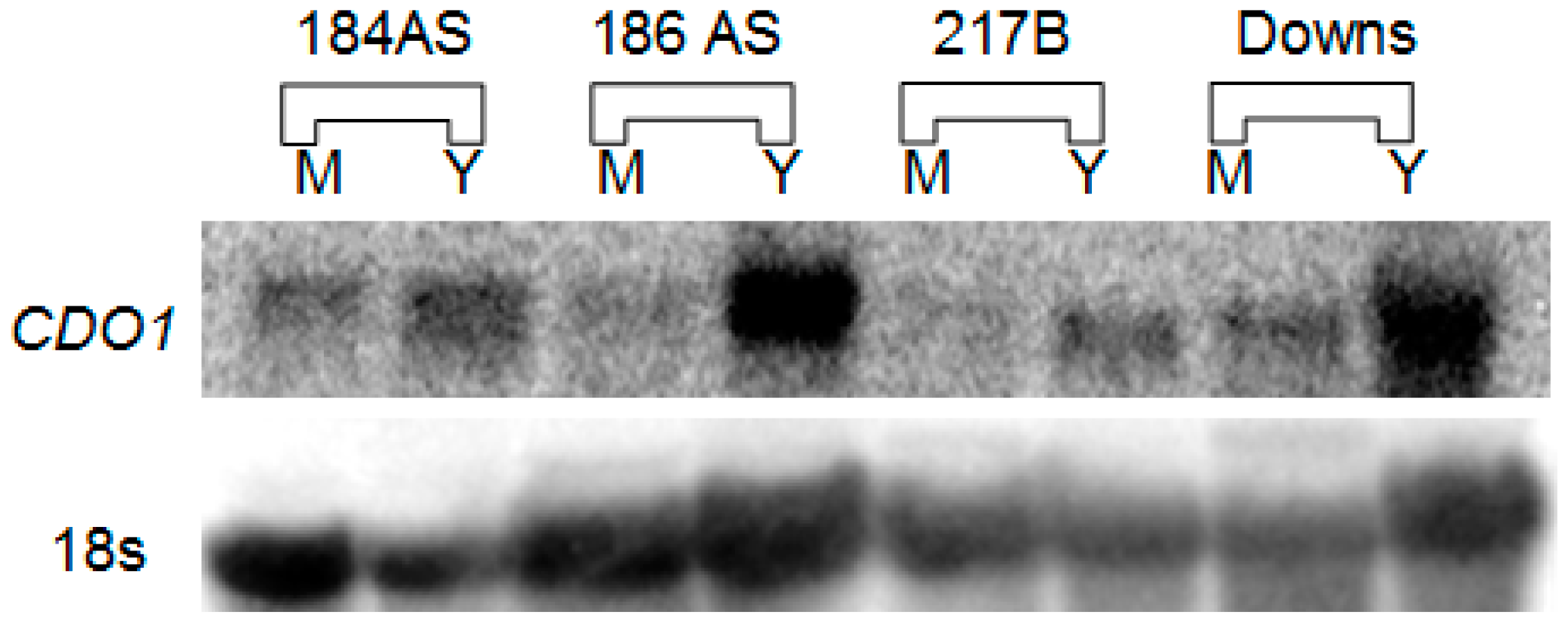
3.1. CDO1 Expression
3.2. CDO Enzyme Activity
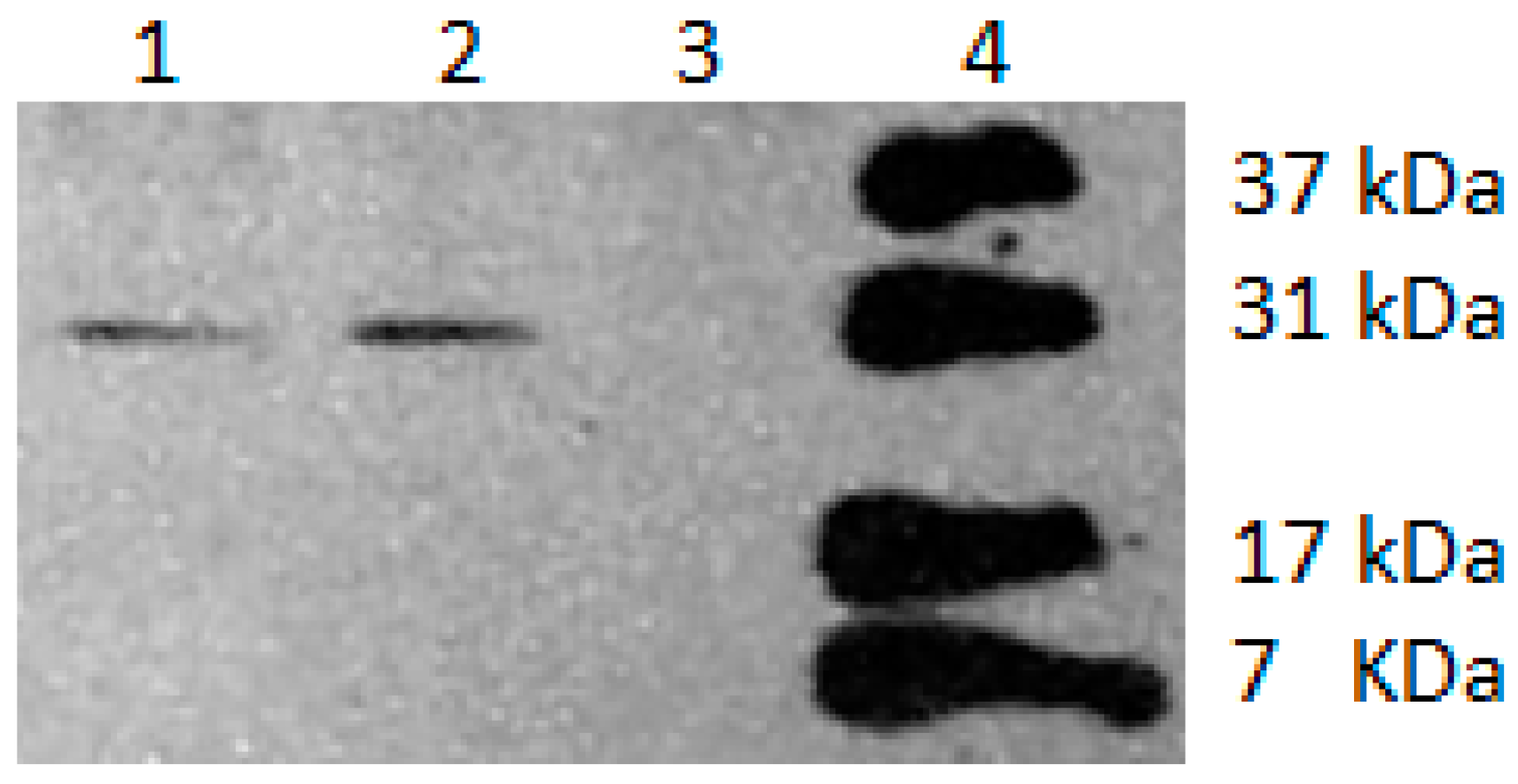
3.3. CDO Molecular Weight
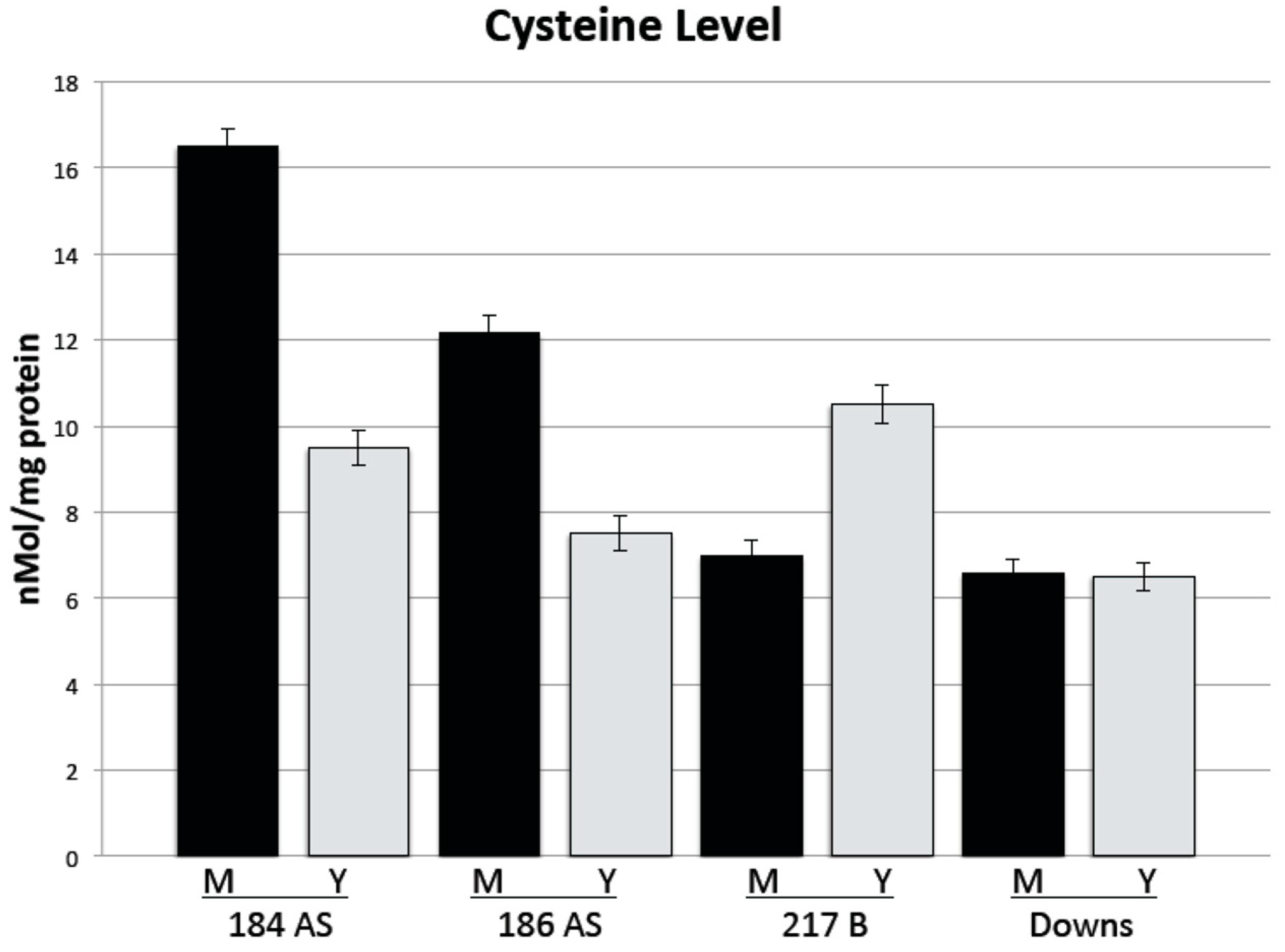
3.4. Intracellular Cysteine Levels
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hammerman, K.J.; Powell, K.E.; Tosh, F.E. The incidence of hospitalized cases of systemic mycotic infections. Sabouraudia 1974, 12, 33–45. [Google Scholar] [CrossRef]
- Bahr, N.C.; Antinori, S.; Wheat, L.J.; Sarosi, G.A. Histoplasmosis infections worldwide: Thinking outside of the Ohio River valley. Curr. Trop. Med. Rep. 2015, 2, 70–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medoff, G.; Sacco, M.; Maresca, B.; Schlessinger, D.; Painter, A.; Kobayashi, G.S.; Carratu, L. Irreversible bl ock of the mycelial-to-yeast phase transition of Histoplasma capsulatum. Science 1986, 231, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Maresca, B.; Lambowitz, A.M.; Kumar, V.B.; Grant, G.A.; Kobayashi, G.S.; Medoff, G. Role of cysteine in regulating morphogenesis and mit Chondrial activity in the dimorphic fungus Histoplasma capsulatum. Proc. Natl. Acad. Sci. USA 1981, 78, 4596–4600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, M.; Medoff, G.; Lambowitz, A.M.; Kumar, B.V.; Kobayashi, G.S.; Painter, A. Sulfhydryl induced respiratory “shunt” pathways and their role in morphogenesis in the fungus, Histoplasma capsulatum. J. Biol. Chem. 1983, 258, 8223–8230. [Google Scholar]
- Maresca, B.; Kobayashi, G.S. Dimorphism in Histoplasma capsulatum: A model for the study of cell differentiation in pathogenic fungi. Microbiol. Rev. 1989, 53, 186–209. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Maresca, B.; Sacco, M.; Goewert, R.; Kobayashi, G.S.; Medoff, G. Purification and characterization of a cysteine dioxygenase from the yeast phase of Histoplasma capsulatum. Biochemistry 1983, 22, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Keath, E.J.; Kobayashi, G.S.; Medoff, G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J. Clin. Microbiol. 1992, 30, 2104–2107. [Google Scholar] [CrossRef] [Green Version]
- Kasuga, T.; White, T.J.; Koenig, G.; McEwen, J.; Restrepo, A.; Castaneda, E.; Da Silva Lacaz, C.; Heins-Vaccari, E.M.; De Freitas, R.S.; Zancope-Oliveira, R.M.; et al. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 2003, 12, 3383–3401. [Google Scholar] [CrossRef] [Green Version]
- Vincent, R.D.; Goewert, R.; Goldman, W.E.; Kobayashi, G.S.; Lambowitz, A.M.; Medoff, G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J. Bacteriol. 1986, 165, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Sepulveda, V.E.; Williams, C.L.; Goldman, W.E. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. MBio 2014, 5, e01376-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, M.; Patane, J.S.; Taylor, M.L.; Gomez, B.L.; Theodoro, R.C.; de Hoog, S.; Engelthaler, D.M.; Zancope-Oliveira, R.M.; Felipe, M.S.; Barker, B.M. Worldwide phylogenetic distributions and population dynamics of the genus Histoplasma. PLoS Negl. Trop. Dis. 2016, 10, e0004732. [Google Scholar] [CrossRef] [Green Version]
- Worsham, P.L.; Goldman, W.E. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 1988, 26, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Shearer, G. The mold-specific MS8 gene is required for normal hypha formation in the dimorphic pathogenic fungus Histoplasma capsulatum. Eukaryot. Cell 2002, 1, 249–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Dominguez, Y.; Scazz Cchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Stipanuk, M.H.; Dominy, J.E.J.; Ueki, I.; Hirschberger, L.L. Measurement of Cysteine Dioxygenase Activity and Protein Abundance. Curr Prot. C Toxicol. 2008, 38, 6.15.1–6.15.25. [Google Scholar] [CrossRef]
- Hopp, T.P.; Prickett, K.S.; Price, V.L.; Libby, R.T.; March, C.J.; Pat Douglas, C.; Urdal, D.L.; Conlon, P.J. A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Bio/Technology 1988, 6, 1204–1210. [Google Scholar] [CrossRef]
- Crossley, D.; Naraharisetty, V.; Shearer, G. The Mould-specific M46 gene is not essential for yeast-mould dimorphism in the pathogenic fungus Histoplasma capsulatum. Med. Mycol. 2016, 54, 876–884. [Google Scholar] [CrossRef] [Green Version]
- Dominy, J.E.J.; Hwang, J.; Stipanuk, M.H. Overexpression of cysteine dioxygenase reduces intracellular cysteine and glutathione pools in HepG2/C3A cells. Am. J. Physiol. End. Crinol. Metab. 2007, 293, E62–E69. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.A.; Chen, C.; Kemski, M.M.; Hu, J.; Mitchell, T.K.; Rappleye, C.A. Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles. BMC Genom. 2013, 14, 695. [Google Scholar] [CrossRef] [Green Version]
- Hwang, L.; Hocking-Murray, D.; Bahrami, A.K.; Andersson, M.; Rine, J.; Sil, A. Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol. Biol. Cell 2003, 14, 2314–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rippon, J.W. Monitored environment system to control cell growth, morphology, and metabolic rate in fungi by oxidation-reduction potentials. Appl. Microbiol. 1968, 16, 114–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hennicke, F.; Grumbt, M.; Lermann, U.; Ueberschaar, N.; Palige, K.; Bottcher, B.; Jacobsen, I.D.; Staib, C.; Morschhauser, J.; Monod, M.; et al. Factors supporting cysteine tolerance and sulfite production in Candida albicans. Eukaryot. Cell 2013, 12, 604–613. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adams, M.A.; Shearer, G., Jr. Cysteine Dioxygenase Enzyme Activity and Gene Expression in the Dimorphic Pathogenic Fungus Histoplasma capsulatum Is in both the Mold and Yeast Morphotypes and Exhibits Substantial Strain Variation. J. Fungi 2020, 6, 24. https://doi.org/10.3390/jof6010024
Adams MA, Shearer G Jr. Cysteine Dioxygenase Enzyme Activity and Gene Expression in the Dimorphic Pathogenic Fungus Histoplasma capsulatum Is in both the Mold and Yeast Morphotypes and Exhibits Substantial Strain Variation. Journal of Fungi. 2020; 6(1):24. https://doi.org/10.3390/jof6010024
Chicago/Turabian StyleAdams, Melissa A., and Glenmore Shearer, Jr. 2020. "Cysteine Dioxygenase Enzyme Activity and Gene Expression in the Dimorphic Pathogenic Fungus Histoplasma capsulatum Is in both the Mold and Yeast Morphotypes and Exhibits Substantial Strain Variation" Journal of Fungi 6, no. 1: 24. https://doi.org/10.3390/jof6010024



