Abstract
The cedar forests of Lebanon have been threatened by the outbreak caused by climate change of a web-spinning sawfly, Cephalcia tannourinensis (Hymenoptera: Pamphiliidae), which negatively impacted the survival of one of the oldest tree species on earth. In this study, we investigated the occurrence of naturally soil-inhabiting entomopathogenic fungi for their role in containing the massive outbreak of this insect. We used a combination of fungal bioexploration methods, including insect bait and selective media. Morphological features and multilocus phylogeny—based on Sanger sequencing of the transcripts encoding the translation elongation factor 1-alpha (TEF-α), RNA polymerase II second largest subunit (RBP2), and the nuclear intergenic region (Bloc) were used for species identification. The occurrence rate of entomopathogenic fungi (EPF) varied with location, soil structure, forest structure, and isolation method. From 15 soil samples positive for fungal occurrence, a total of 249 isolates was obtained from all locations using different isolation methods. The phylogenetic analysis confirmed the existence of two novel indigenous species: Beauveria tannourinensis sp. nov. and Beauveria ehdenensis sp. nov. In conclusion, the present survey was successful (1) in optimizing the isolation methods for EPF, (2) investigating the natural occurrence of Beauveria spp. in outbreak areas of C. tannourinensis, and (3) in characterizing the presence of new Beauveria species in Lebanese cedar forest soil.
1. Introduction
Soil, the living sphere of microorganisms, provides shelter for a diverse array of bacteria, actinomycetes, fungi, algae, and protozoa [1] including species of the cosmopolitan entomopathogenic fungal genus Beauveria (Hypocreales: Cordycipitaceae) [2]. Beauveria bassiana—the type species of this genus—has been widely studied as a bio-control agent, and currently forms the basis of several commercially available mycoinsecticides [3]. The wide range of hosts susceptible to Beauveria includes over 15 insect [4] and mites [5] orders. Furthermore, Beauveria produces a wide range of mycotoxins, including, but not limited to, beauvericin and bassianolide, that are involved in key developmental stages of the fungus and that have demonstrated pharmacological and therapeutic potentials [6,7,8].
Rehner et al. [2] identified phylogenetic relationships within morphologically classified species of Beauveria based on the multisequence of RNA polymerase II largest subunit (RBP1), RNA polymerase II second largest subunit (RBP2), translation elongation factor alpha (TEF-α), and Bloc nuclear intergenic region. Including a few new taxa described since that phylogenetically based revision of the genus, 14 well-supported terminal lineages are now formally recognized: B. amorpha, B. bassiana, B. brongniartii, B. caledonica, B. malawiensis, B. vermiconia, B. asiatica, B. australis, B. kipukae, B. pseudobassiana, B. sungii, B. varroae, B. lii, and B. hoplocheli [2,9,10].
In Lebanon, there were no studies on the taxonomy of entomopathogenic fungi (EPF) until the identification of LTB01, an entomopathogenic strain of Beauveria infecting larvae of cedar sawfly, Cephalcia tannourinensis Chevin (Hymenoptera: Pamphiliidae), from a Tannourine Cedar Forest Nature Reserve [11]. During the last three decades and until now, the endemic C. tannourinensis, the first Cephalcia species reported on Cedrus libani (Pinales: Pinaceae), has been considered the principal defoliator insect of the emblematic cedars in Lebanon. The infestation affects about 70% of the forest, with the major concern of its rapid spread and the subsequent infestation of the remaining cedar forests within the Mediterranean region [12]. The most prevalent natural pathogen of C. tannourinensis is Beauveria, which infects all developmental stages of this pest [11]. During its life cycle, Cephalcia passes most of its time as prepupae in diapause inside an earth-walled chamber in the mineral layer of the soil and, hence, is directly affected by soil entomopathogens [13]. Even though Beauveria is the first and only identified natural enemy of C. tannourinensis, there is still considerable ambiguity with regard to the occurrence, as well as the phylogeny, of this natural pest regulator. The objectives of this study were to collect and to characterize Beauveria isolates from the soil of three Lebanese forests where outbreaks of C. tannourinensis caused substantial damage to the cedars.
2. Materials and Methods
2.1. Study Sites and Collection of Soil Samples
Three Lebanese forests were chosen for the collection of soil samples: the Tannourine Cedar Forest Nature Reserve (1600–1800 masl, precipitation: 850 mm; average temperature per year: 14 °C; area: 625 hectares), the Horch Ehden natural reserve (1400–1600 masl; precipitation: 1095 mm; average temperature per year: 18.6 °C; area: 200 hectares), and the Bcharre cedar forest (1950 masl; precipitation: 950 mm; average temperature per year: 13 °C; area: 11 hectares). The average age of cedar trees in the three forests is over 300 years with an exception to the old cedar forest in Bcharre where a few specimens are more than 1000 years old. Different plots were selected for each forest based on their soil structure and texture, forest structure, and surface of the natural reserve. Three plots were selected from Tannourine Cedar Forest Nature Reserve and were numbered T1 (N 34°12′, 517; E 35°55′, 699), T2 (N 34°12′, 485′; E 35°55′, 510), and T3 (N 34°12′, 311; E 35°56′, 125). Ehden Cedar Forest Nature Reserve was divided into two plots E1 (N 34°18′, 480; E 35°59′, 510) and E2 (N 34°18′, 550; E 35°59′, 721)), and Bcharre cedar forest was considered as one plot B1 (N 34°14′, 571; E 36°02′, 905). For each plot, 10 random collection sites were selected for soil sampling. Therefore, a total of 60 soil samples were collected from the six different plots (T1, T2, T3, E1, E2, and B1). This collection was repeated twice (two biological replicates), during May 2017 and May 2018, and resulted in a total of 120 soil samples. Soil weighing approximately 1 kg was collected for each sample from a depth of up to 15 cm using small shovels. Tools were disinfected with 70% ethanol between each sample collected, in order to avoid cross-contamination. The collected samples were placed in plastic bags and carried to the entomology laboratory of the Faculty of Agricultural and Food Sciences at Holy Spirit University of Kaslik for further treatment. Gravel, stones, and plant debris were eliminated by passing the soil through a 2 mm pore sieve. Sieves were sterilized with 70% ethanol and 1% sodium hypochlorite for 10 s and 3 min respectively, then rinsed twice with sterile distilled water between soil samples. Sieved soil samples were placed separately in new plastic bags, sealed, and stored at 4 °C until their analysis within no longer than 6 months.
2.2. Isolation of EPF from Soil by Means of Insect Bait
EPF were isolated from soil samples using the third instar larvae stages of Galleria mellonella [14] (Lepidoptera: Pyralidae), prepared as described by Meyling and Eilenberg [15], and naturally collected prenymphal stages of C. tannourinensis as baits. Soil samples were moistened with sterile distilled water. From each soil sample, sub-samples of 60 g each were deposited into new sterile plastic containers. Two repetitions (two plastic containers) were made for each bait method. The top 2 cm of each plastic container were intentionally left empty to allow for proper ventilation. Two individuals of each bait kind (two third instar larval stages of G. mellonella or two prenymphal stages of C. tannourinensis) were placed on the soil in each plastic container, making it a total of four insects for each baiting method utilized per soil sample. The plastic containers were closed and incubated in dark conditions for fungal growth (25 °C, 90% RH), and inverted daily to maximize the interaction between spores and insects. Infection of insects was recorded for 3 weeks. This test was repeated in two biological replicates (two different soil collections from May 2017 and May 2018), making it a total of 8 insects for each baiting method utilized. Dead insects were isolated from containers, placed in 70% ethanol and 1% sodium hypochlorite for 10 s and 15 s respectively, then rinsed twice with sterile distilled water. Each disinfected larva was placed in a sterile Petri dish containing filter paper Whatman No. 1 and incubated at 25 °C with 90% RH. Once sporulation occurred on the infected subjects, a sample of conidia was inoculated onto potato dextrose agar (PDA) medium culture using a sterile loop. Potato dextrose agar cultures were incubated at 25 °C, and this was considered as an isolate. Discs (6 mm) from the vigorously growing cultures of nonsporulating Beauveria cultures were placed in sterile cryotubes containing 10% glycerol in water and maintained at −80 °C at the Plant Pathology Fungal Collection of the Post Herbarium at the American University of Beirut.
2.3. Isolation of EPF from Soil by Means of Selective Media
Various media were prepared for isolation of EPF directly from soil: (i) dodine-based medium originally described by Beilharzet al. [16] to isolate Beauveria spp. and confirmed by several studies [17,18,19,20]; (ii) CTAB-based medium [21]; and (iii) Low sugar-content medium (DOC2) [22]. For soil homogenization, 10 g of soil were placed into 90 mL of sterile water and shaken at 150 rpm for 15 min to release fungal propagules from the soil matrix. Using a sterile pipette, 1 mL of aliquots were removed and spread on the three different selective media using a sterile loop. Cultures were incubated in total darkness to promote fungal growth (25 °C, RH 90%) for 7 days. Three repetitions were made for each aliquot sample (three Petri dishes of each selective medium). This test was repeated in two biological replicates (two different soil collections from May 2017 and May 2018), making it a total of six Petri dishes of each medium.
2.4. Morphological Identification of Fungal Isolates
The morphological examination of each isolate was determined based on macro- and microscopic identification criteria [23]. Mycelium was removed from an in vitro culture using a sterile loop, placed onto a glass slide in a drop of lactophenol and covered with a coverslip. Microscopic measurements of conidiogenous cells and conidia were taken from the agar cultures at 15 days post-inoculation.
Conidia were suspended in 0.01% Tween and mixed with an equal volume of molten (70 °C) 0.1% Nusieve GTG agarose. A transparent adhesive tape preparation stained with lactophenol was performed for the observation of fungal cells. Thirty conidia from 15-day old cultures were measured for each sample. This experiment was done in triplicates, making it a total of 90 conidia measured for each isolate.
2.5. Molecular Characterization
For the phylogenetic analysis, a subsample of isolates was used. Fifteen soil samples (out of 60) were positive for Beauveria spp. Only one isolate from each of the 15 positive soil samples was randomly selected for further analysis. Potato dextrose broth (PDB) was used to grow isolates for DNA extraction. Erlenmeyer flasks containing 25 mL of PDB were inoculated with 0.5 cm plugs of fungus from 15-day-old cultures described above and incubated at 25 °C for 7 days to promote fungal growth. The mycelium was harvested by centrifugation of the medium on 8000 rpm for 10 min. To remove any residual medium, the mycelium was washed twice with sterile distilled water. The mycelium was frozen at −20 °C until further use. DNA extraction was performed using DNeasy kit (QIAGEN, GmbH, Hilden, Germany) following the manufacturer’s instructions. Sequences of two protein-coding genes (RBP2 and TEF-α) were used because of their demonstrated utility in phylogenetic placement of Beauveria spp [24], and the nuclear intergenic region Bloc for cryptic speciation review within B. bassiana [2,25]. For all loci, PCR amplifications were done in a total volume of 20 µL, which included 10 µL PCR Master Mix 2x Thermo Scientific, MA, USA, Taq DNA polymerase (0.05 U/µL), reaction buffer, 4 mM MgCl2, 0.4 mM of each dNTP, nuclease-free water), 20 ng of the genomic DNA, 10 pmol each of the opposing amplification primers (Table 1), and 7 µL of distilled water. Amplification cycles, using CFX96 Real-Time System (BIO-RAD, CA, USA), were performed according to Rehner et al. [2]. All PCR reaction volumes were separated on a 1.5% agarose gel to check the success of amplification reactions. All PCR products were purified using the QIAquick PCR Purification kit (QIAGEN, GmbH, Hilden, Germany) following the manufacturer’s instructions. The purified amplicons were quantified, and Sanger sequencing with either the forward or reverse primers were undertaken at the molecular core facility at the American University of Beirut using ABI3500 instrument as previously described [26].

Table 1.
Primers for PCR amplification and sequencing.
2.6. Phylogenetic Analysis
Phylogenetic analyses were performed on sequences representing 14 species of Beauveria and the outgroup taxon Cordyceps cicadae. The three-locus datasets (Bloc, TEF-α, RPB2) DNA sequences were edited and aligned with Muscle version 3.2 [30] using MEGA version 7 (Molecular Evolutionary Genetic Analysis) [31]. All three sequences were concatenated using sequenceMatrix [32], and the resulting concatenated sequences with 5814 nucleotide sites were explored in the subsequent phylogenetic analysis (TreeBASE ID: 24955). Maximum-likelihood (ML) trees were computed with IQTREE (version 1.6.12), which relies on novel and improved algorithms to estimate maximum likelihood phylogenies. Briefly, it takes a multiple sequence alignment as input, and the appropriate model of DNA substitution was obtained with ModelFinder [33] from the Bayesian Information Criterion (BIC). The TPM2u+F+I+G4 was selected as best fitting model for the concatenated dataset. Finally, the ML tree was constructed from the best fitting model and analyses were conducted with 1000 bootstrap replicates [34]. Maximum parsimony (MP) and non-parametric bootstrapping (MP BS) (1000 replicates), to assess clade support, were executed with PAUP* 4.0a [35]. The gaps were excluded, and all nucleotides were unordered and equally weighted. All MP searches were performed using 1000 random taxon-addition sequence replicates with tree-bisection and reconnection branch-swapping (TBR). To find out if data partition could be combined, a conditional combination criterion was utilized [36]. Data were combined if (i) monophyly statement were congruent or (ii) >70% bootstrapping score was recorded with one or both monophyletic groups. A Bayesian approach (MB) was also performed using MrBayes 3.2.7 [37] with two runs of Markov chains (diagnostic calculated every 200 generations) for 30 × 106 generations (standard deviation < 0.01). The lambda exponential rate parameter for the branch-length prior was calculated as described by Brown et al. [38]. Traces were examined graphically with Tracer 1.6 [39], and trees generated before stabilization of ML Scores were discarded. The paired sites test, Shimodaira and Hasegawa test, was conducted using PAUP* 4.0a to compare topologies obtained with the three different statistical approaches used in this study [40]. The significant difference in the sum of site-wise log-likelihoods for all trees was evaluated by bootstrap sampling of site scores (RELL sampling) with 1000 replicates [41].
2.7. Statistical Analysis
Data relevant to the presence/absence of Beauveria using the different extraction methods was calculated as percentages in each forest. A chi-square of homogeneity test was applied to determine if the presence or absence of Beauveria differed between the various sampled forest habitats after adjusting the computing p values by the Bonferroni method and using the Z-test to compare the percentages. To check for the best selective method of extraction, a Chi-Square Goodness of fit was performed after computing the frequencies of Beauveria isolated by the four methods. Then, a multiple binomial test was executed after adjusting the computed p-values with the Bonferroni method to detect the significance between pairs of extraction methods. Data relevant to the species of Beauveria using the different extraction methods were analyzed by the statistical comparison test of means (ANOVA). The Tukey test was used at the 5% threshold for the separation of means.
3. Results
3.1. Occurrence of EPF
We used two insect baits and three selective media to isolate EPF from the soil in three different locations in Lebanon. The results obtained demonstrated the wide presence of EPF strains including Beauveria spp., Cordyceps fumosorosea (accession number TEF-α: MK975968) and saprophytic fungi such as Fusarium spp., Aspergillus spp., and Pythium spp. All saprophytic fungi were, however, considered as contaminants and discarded from future analysis; only Beauveria spp. were kept for analysis. The percentage of occurrence and number of detected species was significantly affected by the isolation method. (X2(3) = 12.031; p = 0.007) taking into account that the low sugar medium (DOC2) was not included in the analysis since no fungi was extracted by this medium. Overall, a total of 249 isolates was recorded from the six different locations. Based on the number of isolates, the occurrence of Beauveria isolates in sampling locations indicated that Beauveria was undetectable in plot T3 in Tannourine Cedar Forest Nature Reserve and Bcharre Cedar Forest regardless of the method of isolation used.
The isolation of Beauveria with the G. mellonella method, the C. tannourinensis method, the dodine medium method, as well as the CTAB medium method was highly statistically dependent on the site location (Tannourine, Ehden, and Bcharre) (Table 2). DOC2 isolation medium method was not successful in isolating Beauveria from any sites. The chi square test of homogeneity for Tannourine Cedar Forest indicated that the presence or absence of Beauveria is very highly significantly dependent on the considered plot in the forest for each of the selected isolation methods. Thus, each extraction method was affected by the habitat (plot) whereby plot 1 and plot 2 harbored more Beauveria than plot 3 irrespective of the extraction method used (C. tannourinensis: X2 = 19.46, df = 2, p < 0.05; G. mellonella: X2 = 26.97, df = 2, p < 0.05; dodine: X2 = 37.6, df = 2, p < 0.05; CTAB: X2 = 22.7, df = 2, p < 0.05). The same applies for the cedar forest of Ehden which also indicated that the presence or absence of the EPF is significantly dependent of the considered habitat (plot), for each of the selected isolation methods (C. tannourinensis: X2 = 3.93, df = 2, p < 0.05; G. mellonella: X2 = 9.26, df = 2, p < 0.05; dodine: X2 = 16.88, df = 2, p < 0.05; CTAB: X2 = 9.22, df = 2, p < 0.05).

Table 2.
Occurrence frequency of Beauveria EPF recovered from soil samples.
To determine the best selective method of extraction in all cedar forests, results showed that there is a high statistically significant difference between levels of Beauveria extracted by the four methods (C. tannourinensis, G. mellonella, dodine, and CTAB) with a chi-square goodness of fit . There was a significant difference between Beauveria frequencies detected by C. tannourinensis (lowest one) and dodine (highest one) extraction methods. No significant differences were detected between all the other combinations of methods of extraction.
There was no significant difference between the occurrence of B. bassiana and B. pseudobassiana using G. mellonella (F = 0.85, df = 1, p > 0.05), dodine (F = 0.461, df = 1, p > 0.05), and CTAB (F = 0.405, df = 1, p > 0.05). This result was significant exclusively using the C. tannourinensis bait method for the isolation of Beauveria spp. (F = 8.013, df = 1, p < 0.05) (Figure 1).
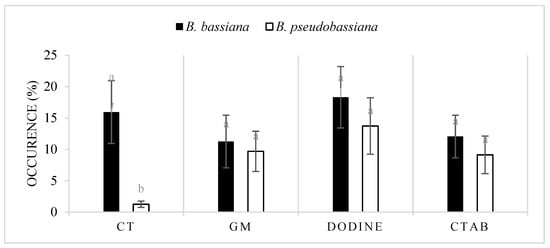
Figure 1.
Occurrence rate of B. bassiana and B. pseudobassiana in the Lebanese soils using different isolation methods (CT: Cephalcia tannourinensis bait method, GM: Galleria mellonella bait method, dodine: dodine-based selective medium, and CTAB: CTAB-based selective medium). The values with different superscript letters in a column are significantly different (p < 0.05).
3.2. Morphological Characterization of Beauveria Isolates
The morphological description of ex-type cultures has been previously reported by Rehner et al. [2]. Due to its structural simplicity and lack of phenotypical differences, Beauveria isolates cannot be identified to the species level by their morphologies; however, the conidial shape and size can be considered as key morphological features shared by a few of the species in this genus. Beauveria bassiana conidia averaged 1.98 × 1.33 µm (1.82–2.23 × 0.9–1.56 µm), mostly ellipsoidal and rarely forming spherical spore clusters among aerial hyphae, and colorless in mass. Beauveria pseudobassiana conidia averaged 2.00 × 1.90 µm (1.93–2.21 × 1.85–2.00 µm) with mostly globose spore clusters among aerial hyphae, and colorless in mass.
3.3. Phylogenetic Placement of Isolates
In order to further characterize the isolates, we carried Sanger sequencing of the TEF-α, RBP2, Bloc, and Internal transcribed spacer “rDNA ITS1-5.8S-ITS2 (ITS barcode)” loci. The sequences were uploaded to the NCBI portal (Table 3), and phylogenetic analyses were undertaken to place them on an evolutionary tree. The ITS sequences were not included in the phylogenetic analysis because of their limited informative value within hypocrealean ingroups [2].

Table 3.
Collection site and accession numbers (TEF-α, RBP2, Bloc, and ITS genes) of the representative strains.
The SH test showed that all approaches utilized in this study produced congruent trees (p < 0.05). The phylogenetic tree, built upon the ML approach, and generated from the concatenated dataset clustered Beauveria species into 16 distinct and robust groups independent of approach (ML, MP, and MB), supported by ML and MP bootstrap percentages in addition to MB posterior probabilities. Fourteen out of 16 groups corresponded to the Beauveria spp. previously described [2,9,10]. The two remaining groups corresponded to the two distinct indigenous groups comprising 10 (LTB01, LTB02, LTB03, LTB04, LTB05, LTB06, LTB07, LTB08, LTB09, LTB10) and 6 (LTB11, LTB12, LTB13, LTB14, LTB15, LTB16) isolates, clustered distinctly from the other Beauveria groups, although these two groups were related to B. bassiana and B. pseudobassiana, respectively, according to the ML tree (Figure 2).
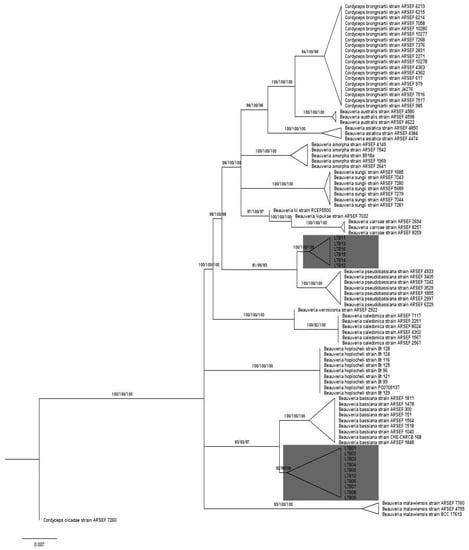
Figure 2.
Phylogenetic tree of the Beauveria species showing the relationships between isolates from Lebanon and species described by Rehner et al. [2], Zhang et al. [9], and Robène-Soustrade et al. [10], in addition to an outgroup taxon: Cordyceps cicadea. The tree was inferred with the maximum likelihood method (TPM2u+F+I+G4 model) from the concatenated dataset including, TEF-α, RPB2, and Bloc sequences. Maximum likelihood (ML), maximum parsimony (MP), and Bayesian approach (MB) produced congruent trees (p < 0.05). The maximum likelihood (ML) method produced the corresponding cladogram from the concatenated sequences and visualized with tree application. Values next to branches indicate ML bootstrap percentage, MP bootstrap percentage, and MB posterior probabilities, respectively. Triangles represent the collapsed clades.
The branching patterns 93/93/87 and 81/95/83 within Beauveria LTB01 to LTB10 and LTB11 to LTB16, respectively, are highly supportive of new lineages. These branches were assigned with ML bootstrap percentage, MP bootstrap percentage and, MB posterior probabilities
Morphological features coupled to a detailed molecular phylogenetic analysis allowed us to describe novel indigenous species Beauveria. These species were classified as new species referred to herein as B. tannourinensis (LTB01 to LTB10) and B. ehdenensis (LTB11 to LTB16).
3.4. Taxonomy
Beauveria tannourinensis Al Khoury C, Nemer G, Humber R, Guillot J, Nemer N, sp. nov.
3.4.1. Mycobank: MB 832258
Holotype: BT0517 Lebanon: Batroun, North Lebanon (34°12′46.3″ N 35°55′44.3″ E), on a C. tannourinensis nymph (Hymenoptera: Pamphiliidae) in Tannourine Cedar Forest Nature Reserve, 14 May 2017.
Ex-type culture LTB02, deposited in the Plant Pathology Fungal Collection of the Post Herbarium at the American University of Beirut. Sequences from LTB02 strain were deposited in GenBank under accession numbers: RPB2 = MK908082, Bloc = MK884864, ITS = MK884879, and TEF-α = MK975955.
Colony characteristics on full strength PDA, 36–42 mm diam at 10 d at 23 °C, appearance densely cushion-like and up to 7 mm thick, and uncolored. The reverse is white at first, then becoming yellow at margins and brown at the center. Odor indistinct. Conidia 1.98 × 1.33 (2.23–1.82 × 1.56–0.9) µm, aggregated in mostly ellipsoidal and rarely spherical clusters among aerial hyphae and white in mass. Vegetative hyphae septate, branched, hyaline, smooth-walled, 1–2 µm wide. Conidiogenous cells are solitary in but usually in dense clusters of five or more, base subspherical to ampulliform and 3–5 µm wide, apex with an indeterminate 1 µm wide geniculate, denticulate rachis, produced laterally on aerial hyphae or from subtending cells.
Beauveria ehdenensis Al Khoury C., Nemer G, Humber R., Guillot J, Nemer N, sp. nov.
3.4.2. Mycobank: MB 832259
Holotype: BE0518 Lebanon.: Zgharta, North Lebanon (34°18′42.1″ N, 35°59′03.3″ E), on a G. mellonella third instar larva (Lepidoptera: Pyralidae) in Horsh Ehden Nature Reserve, 10 May 2018.
Ex-type culture LTB15, deposited in the Plant Pathology Fungal Collection of the Post Herbarium at the American University of Beirut. Sequences from LTB15 strain were deposited in GenBank under accession numbers: RPB2 = MK908085, Bloc = MK884867, ITS = MK884883, and TEF-α = MK975958.
Colony characteristics on full strength PDA, 48–53 mm diam at 10 d at 23 °C, appearance densely dotted-like and up to 2 mm thick, and uncolored. The reverse is white at the center and margins. Odor indistinct. Conidia 2 × 1.9 µm (1.93–2.21 × 1.85–2) µm, aggregated in mostly globose clusters among aerial hyphae and white in mass. Vegetative hyphae septate, branched, hyaline, smooth-walled, 1–2 µm wide. Conidiogenous cells solitary in but usually in dense clusters of five or more, base subspherical to ampulliform and 3–4.5 µm wide, apex with an indeterminate 1 µm wide geniculate, denticulate rachis, produced laterally on aerial hyphae or from subtending cells.
4. Discussion
4.1. Bioexploration of EPF
The present study reports the results of the first bioexploration of entomopathogenic Beauveria species in Lebanon. A total of 249 isolates were recovered from soils of three Lebanese cedar forests using different isolation methods. Similar occurrences have been previously reported by Keller et al. [17], Rath et al. [42], Bidochka and Kasperski [43], Pérez-González et al. [44], Niemczyk et al. [45], and Sánchez-Peña et al. [46]. Beauveria isolates were recovered from 25% of the soil samples. In addition, the present study has identified that Beauveria strains isolated from the cedar forests are quite different from described species on the morphological and genetical levels. An in-depth study of fungal–insect relationships in the cedars’ ecological natural vegetation might enhance our understanding of the evolution and diversity of these fungi.
The evaluation of the selective medium for culturing Beauveria spp. showed that the dodine-based medium is the most efficient among the three media tested. This result is in accordance with the studies from Keller et al. [17] Enkerli et al. [20]. Strasser et al. [47] showed that dodine-based media can be used successfully to isolate Beauveria spp. and Metarhizium spp. from soil and insect cadavers. Moreover, a low level of contamination by saprophytic fungi has been also recorded using this medium. Isolation of EPF was also successful using a CTAB-based medium. This result is in accordance with the study from Posadas et al. [21]. However, CTAB-based medium was less selective because it allowed the development of many contaminant fungi. Shimazu and Sato [22] developed a DOC2 medium based on low sugar content and high pH supplemented with CuCl2 and crystal violet for the selective isolation of Beauveria spp. This medium has been successfully used by Safavi [48] to recover a new isolate of Beauveria bassiana. Surprisingly, in the present study, the DOC2 medium did not allow the growth of any single colony of Beauveria spp. Local conditions might have affected the tolerance ability of indigenous strains, inhibiting the possibility of their development on such an unconventional medium. In addition, dodine is not readily available in many countries in contrast to the ingredients for DOC2 medium, whose ingredients are widely available. Therefore, advanced studies should be held, trying slight modifications to the DOC2 medium, making it suitable to the development of indigenous EPF.
Two insect bait methods were used in the present study. The first one included highly susceptible larvae of the wax moth, G. mellonella [15,17,49]. This traditional method was able to recover Beauveria spp. from different Lebanese soils, and B. bassiana and B. pseudobassiana were both recovered at similar rates. The present study also included the first use of web-spinning sawfly, C. tannourinensis that proved to be a highly efficient bait method in isolating Beauveria from the Tannourine Cedar Forest. After the molecular analysis, most of the Beauveria isolated by the Cephalcia bait method turned out to be closely related to B. bassiana whereas the Galleria bait method showed an equal isolation between the species closely related to B. bassiana and B. pseudobassiana. This could be due to the fact that C. tannourinensis is more vulnerable to B. bassiana than to B. pseudobassiana. This is in accordance with findings reported by Al Khoury et al. [50] who showed that local isolates of Beauveria are relatively active against the eggs of S. scabiei.
4.2. Identification and Phylogenetic Placement
Macroscopic and microscopic examination, as well as multilocus phylogenetic analysis, proved that all Beauveria isolates from the Lebanese forests were distinct from previously described Beauveria species. In spite of morphological differences when collected for the first time from Tannourine Cedar Forest Nature Reserve in Lebanon, analyses of partial sequences of ITS and TEF-α loci identified LTB01 strain as B. bassiana Clade C [11,51]. Morphological characteristics of conidia were compared to ex-type cultures previously described by Rehner et al. [2], Zhang et al. [9], Robène-Soustrade et al. [10], Rehner and Buckley [24], and Rehner et al. [25] to confirm species assignments. The isolate LTB02 (representing the first lineage of isolates) produced mostly ellipsoidal conidia. Similar morphology was previously observed [11]. The ellipsoidal conidial shape has been previously reported for B. brongniartii (ARSEF 1431), B. bassiana (ARSEF 678), B. asiatica (ARSEF 4384), and B. sungii (ARSEF 1685) but the conidia produced by the indigenous Lebanese isolates were smaller. Furthermore, based on concatenated data of TEF-α, RPB2, and Bloc, B. brongniartii, B. bassiana, B. asiatica, and B. sungii represented four well supported terminal lineages, as described by Robène-Soustrade et al. [10] and Rehner et al. [2] and therefore were clearly separated from our local isolates. LTB15 isolate (representing the second lineage of isolates) produced mostly globose conidia with an average size of 2 × 1.9 µm. Although B. pseudobassiana is known to produce spherical conidia with an average size 2–3 × 1.5–2.5 µm, Robène-Soustrade et al. [10] reported B. pseudobassiana strain producing globose conidia. However, the latter strain (ARSEF 152) produced conidia with a larger size (3.5 × 3 µm) when compared to the conidia produced by the indigenous isolate (strain LTB15). Moreover, a study conducted by Al Khoury et al. [52] showed a significant difference in the expression of genes involved in the virulence of the local strain (LTB02) when compared to a commercially available strain of B. bassiana. This is also in good agreement with results obtained by Al Khoury et al. [53] demonstrating a significantly higher efficacy of the local strain against all developmental stages of Tetranychus urticae Koch (Acari: Trombidiformes) than to the commercial one (Tracer, Krishi Rasayan, India). These differences in the genetic expression, as well as bioactivities, could be considered as additional support that the local isolates of Beauveria are separated from previously described strains.
4.3. Occurence of EPF
A high incidence of EPF was recorded in the soil of the Tannourine Cedar Forest Nature Reserve. This forest may offer better environmental conditions for the development of EPF and/or have appropriate hosts for entomopathogenic fungal development exemplified by the massive presence of the diapausing stages of C. tannourinensis present in the soil. A higher incidence of EPF was recorded in the first and second plots (T1 and T2) than in the third plot (T3). A study conducted by Nemer et al. [12] showed that the soil samples from the T1 and T2 plots were of a clayey type and can be further classified into Red Mediterranean soil. It is possible that the soil’s light sandy structure in T3 is not suitable for the development of either EPF or for C. tannourinensis. This finding is consistent with previous findings showing that larvae of C. tannourinensis cannot survive in sandy soils [12]. Furthermore, these results are in accordance with the study from Strasser et al. [54] and Niemczyk et al. [45] suggesting that heavy clay soils could enhance the occurrence of EPF. Quesada-Moraga et al. [55], and Abdullah et al. [56] demonstrated that soils with higher organic matter content were favored by entomopathogenic fungal populations. A significant difference of fungal occurrence was observed between the two plots at Horch Ehden Natural Reserve (Table 2). Despite having the same environmental conditions, the occurrence of Beauveria spp. was higher in E1 where the habitat is composed mainly of cedars. Therefore, the high density of insect hosts in which the fungi can multiply has favored its occurrence at the location where Cedar trees and their pest are available. Even though these results differ from those reported by Bueno-Pallero [57] who hypothesized that vegetation type has no effect on EPF occurrence, they are consistent with those from Carrillo-Benitez et al. [58] and Pérez-Gonzalez et al. [44] who suggested that different Beauveria species may infect different insect hosts and adapt to specific pests and environments.
Several studies previously reported possible factors affecting the occurrence of Beauveria within different areas. Vega et al. [59] suggested that differences in environmental factors were the main reason for this fungal variation. Quesada-Moraga et al. [55] demonstrated the significant effect of soil properties on the isolation rate of EPF. In the present study, we demonstrated that the occurrence of EPF could also be affected significantly by using different isolation methods. These results are in line with those obtained by Bueno-Pallero et al. [57] who demonstrated that the utilization of selective media results in higher EPF occurrence rates relative to the baiting method. Our tests revealed that the best mass extraction method for Beauveria from the soil is the use of the dodine medium; however, for a species-specific isolation, baiting with a specific insect host would be recommended.
Since no bioexploration has been undertaken for EPF in Lebanon, this study could be considered as key for future bioexploration to elucidate the behavior of EPF in soil habitats. During the last three decades, in northern Lebanon, where more than 70% of the cedar forests occur these trees have been under threats from the defoliating insect, C. tannourinensis [13,60,61]. The only available control method is the use of endemic entomopathogens and in order to be able to integrate their usage within the cedars forest ecosystems, this study demonstrated that the presence of Beauveria species in the soil depends on complex interactions between biotic, abiotic, and edaphic factors. In their investigations into the utilization of Beauveria inoculum to control pest outbreak, Tartanus et al. [62] showed that the metabolic activity/versatility of fungi may also play a fundamental role in the soil abundance.
In conclusion, multiple factors affected the occurrence of EPF in this study beginning with the soil habitats, to the forest structure, the hosts, and the isolation methods. Combined results from morphological characteristics and multigene phylogenetic analysis successfully recovered two novel lineages that have been described here as the new species Beauveria tannourinensis and Beauveria ehdenensis. Much work remains to be conducted to study the behavior, survival, growth, and sporulation of these new entomopathogenic Beauveria species. Because the pest C. tannourinensis is being monitored regularly, a close monitoring of these entomopathogens is important in order to predict future outbreaks in these threatened cedar forest ecosystems.
Author Contributions
Conceptualization, C.A.K., G.N., J.G. and N.N.; Methodology, C.A.K., R.C., E.N. and Y.E.K.; Software, C.A.K., N.E.-H. and G.N.; Validation, C.A.K., R.H. and E.N.; Formal analysis, C.A.K., N.E. and W.S.; Investigation, C.A.K., R.C. and Y.E.K.; Resources, C.A.K., G.N., J.G. and N.N.; Data curation, C.A.K., G.N., J.G., R.H. and N.N.; Writing—original draft preparation, C.A.K.; Writing—review and editing, C.A.K., G.N., J.G., R.H. and N.N.; Visualization, G.N., J.G., R.H. and N.N.; Supervision, G.N., J.G., R.H. and N.N.; Project administration, C.A.K., G.N., J.G., R.H. and N.N.; Funding acquisition, C.A.K., G.N., J.G. and N.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Council for Scientific Research of Lebanon, grant name (CNRS-USEK). This research was also funded by the “Coopération pour l’évaluation et le développement de la recherche” (CEDRE) Program; grant no. 37349SA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors would like to thank Stephen Rehner for his insightful comments regarding the phylogenetic placement of Beauveria. The authors would also like to thank Cynthia Elia and Tamar Zaidan for proofreading this study. The authors would like to thank the insightful comments of the anonymous reviewers of this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Keller, S.; Zimmermann, G. Mycopathogens of soil insects. In Insect-Fungus Interactions; Wilding, N., Collins, N.M., Hammond, P.M., Webber, J.F., Eds.; Academic Press: London, UK, 1989; pp. 239–270. [Google Scholar]
- Rehner, S.A.; Minnis, A.M.; Sung, G.; Luangsa-ard, J.J.; Devotto, L.; Humber, R.A. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 2011, 103, 1055–1073. [Google Scholar] [CrossRef]
- de Faria, M.R.; Wraight, S.P. Mycoinsecticides and mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Li, Z.Z. A list of insect hosts of Beauveria bassiana. In Study and Application of Entomogenous Fungi in China; Li, Y.W., Li, Z.Z., Liang, Z.Q., Wu, J.W., Wu, Z.K., Xi., Q.F., Eds.; Academic Periodical Press: Beijing, China, 1988; Volume 1, pp. 241–255. [Google Scholar]
- Chandler, D.; Davidson, G.; Pell, J.K.; Ball, B.V.; Shaw, K.; Sunderland, K.D. Fungal biocontrol of Acari. Biocontrol Sci. Technol. 2000, 10, 357–384. [Google Scholar] [CrossRef]
- Al Khoury, C.; Nemer, N.; Nemer, G. Beauvericin potentiates the activity of pesticides by neutralizing the ATP-binding cassette transporters in arthropods. Sci. Rep. 2021, 11, 10865. [Google Scholar] [CrossRef]
- Al Khoury, C.; Nemer, N.; Nemer, G.; Kurban, M.; Bernigaud, C.; Fischer, K.; Guillot, J. In vitro activity of beauvericin against all developmental stages of Sarcoptes scabiei. Antimicrob. Agents Chemother. 2020, 64, 2118. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Patocka, J.; Nepovimova, E.; Kuca, K. A review on the synthesis and bioactivity aspects of beauvericin, a Fusarium mycotoxin. Front. Pharmacol. 2018, 9, 1338. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; He, L.; Chen, X.; Huang, B. Beauveria lii sp. nov. isolated from Henosepilachna vigintioctopunctata. Mycotaxon 2013, 121, 199–206. [Google Scholar] [CrossRef]
- Robène-Soustrade, I.; Jouen, E.; Pastou, D.; Payet-Hoarau, M.; Goble, T.; Linderme, D.; Lefeuvre, P.; Calmès, C.; Reynaud, B.; Nibouche, S. Description and phylogenetic placement of Beauveria hoplocheli sp. nov. used in the biological control of the sugarcane white grub, Hoplochelus marginalis, on Reunion Island. Mycologia 2015, 107, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Abdo, C.; Nemer, N.; Nemer, G.; Abou Jawdah, Y.; Atamian, H.; Kawar, N.S. Isolation of Beauveria species from Lebanon and evaluation of its efficacy against the cedar web-spinning sawfly, Cephalcia tannourinensis. Biocontrol 2008, 53, 341–352. [Google Scholar] [CrossRef]
- Nemer, N.; El Beyrouthy, M.; Lahoud, C.; Mnif, W.; Bashour, I.; Kawar, N. The influence of soil properties on the development of Cephalcia tannourinensis Chevin (Hym. Pamphiliidae) infesting the cedar forests in Lebanon. Afr. J. Biotechnol. 2014, 13, 4369–4381. [Google Scholar]
- Nemer, N.; Demolin, G.; Kawar, N.; Kfoury, L.; Zakhour, E. Monitoring of the new cedar web-spinning sawfly, Cephalcia tannourinensis n. sp. in cedar forests of Lebanon. In Entomological Research in Mediterranean Forest Ecosystems; Lieutier, F., Ghaioule, D., Eds.; INRA Publications: Paris, France, 2005; pp. 247–255. [Google Scholar]
- Zimmermann, G. The ‘Galleria bait method’ for detection of entomopathogenic fungi in soil. J. Appl. Entomol. 1986, 102, 213–215. [Google Scholar] [CrossRef]
- Meyling, N.V.; Eilenberg, J. Isolation and characterisation of Beauveria bassiana isolates from phylloplanes of hedgerow vegetation. Mycol. Res. 2006, 110, 188–195. [Google Scholar] [CrossRef]
- Beilharz, V.C.; Parbery, D.G.; Swart, H.J. Dodine: A selective agent for certain soil fungi. Trans. Br. Mycol. Soc. 1982, 79, 507–511. [Google Scholar] [CrossRef]
- Keller, S.; Kessler, P.; Schweizer, C. Distribution of insect pathogenic soil fungi in Switzerland with special reference to Beauveria brongniartii and Metharhizium anisopliae. Biocontrol 2003, 48, 307–319. [Google Scholar] [CrossRef]
- Kessler, P.; Matzke, H.; Keller, S. The effect of application time and soil factors on the occurrence of Beauveria brongniartii applied as a biological control agent in soil. J. Invertebr. Pathol. 2003, 84, 15–23. [Google Scholar] [CrossRef]
- Kessler, P.; Enkerl, J.; Schweize, C.; Keller, S. Survival of Beauveria brongniartii in the soil after application as a biocontrol agent against the European cockchafer Melolontha melolontha. Biocontrol 2004, 49, 563–581. [Google Scholar] [CrossRef]
- Enkerli, J.; Widmer, F.; Keller, S. Long-term field persistence of Beauveria brongniartii strains applied as biocontrol agents against European cockchafer larvae in Switzerland. Biol. Control 2004, 29, 115–123. [Google Scholar] [CrossRef]
- Posadas, J.B.; Comerio, R.M.; Mini, J.I.; Nussenbaum, A.L.; Lecuona, R.E. A novel dodine-free selective medium based on the use of cetyl trimethyl ammonium bromide (CTAB) to isolate Beauveria bassiana, Metarhizium anisopliae sensu lato and Paecilomyces lilacinus from soil. Mycologia 2012, 104, 974–980. [Google Scholar] [CrossRef]
- Shimazu, M.; Sato, H. Media for selective isolation of an entomogenous fungus, Beauveria bassiana (Deuteromycotina: Hyphomycetes). Appl. Entomol. Zool. 1996, 31, 291–298. [Google Scholar] [CrossRef] [Green Version]
- Humber, R.A. Fungal pathogens and parasites of insects. In Applied Microbial Systematics; Priest, F.G., Goodfellow, M., Eds.; Kluwer Academic Publishers: Derdrecht, The Netherlands, 2000; pp. 203–230. [Google Scholar]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Posada, F.; Buckley, E.P.; Infante, F.; Castillo, A.; Vega, F.E. Phylogenetic origins of African and Neotropical Beauveria bassiana sl pathogens of the coffee berry borer, Hypothenemus hampei. J. Invertebr. Pathol. 2006, 93, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Nemer, G.; Fadlalah, F.; Usta, J.; Nemer, M.; Dbaibo, G.; Obeid, M.; Bitar, F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum. Mutat. 2006, 27, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony; Mac Version 3.1.1 (Computer Program and Manual); Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Mason-Gamer, R.J.; Kellogg, E.A. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst. Biol. 1996, 45, 524–545. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Hedtke, S.M.; Lemmon, A.R.; Lemmon, E.M. When trees grow too long: Investigating the causes of highly inaccurate Bayesian branch-length estimates. Syst. Biol. 2010, 59, 145–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer Version 1.6. Available online: http://tree.bio.ed.ac.uk/software/tracer/ (accessed on 14 May 2018).
- Shimodaira, H.; Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef] [Green Version]
- Kishino, H.; Hasegawa, M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 1989, 29, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.C.; Koen, T.B.; Yip, H.Y. The influence of abiotic factors on the distribution and abundance of Metarhizium anisopliae in Tasmanian pasture soils. Mycol. Res. 1992, 96, 378–384. [Google Scholar] [CrossRef]
- Bidochka, M.J.; Kasperski, J.E.; Wild, G.A. Occurrence of the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana in soils from temperate and near-northern habitats. Can. J. Bot. 1998, 76, 1198–1204. [Google Scholar]
- Pérez-González, V.H.; Guzmán-Franco, A.W.; Alatorre-Rosas, R.; Hernández-López, J.; Hernández-López, A.; Carrillo-Benítez, M.G.; Baverstock, J. Specific diversity of the entomopathogenic fungi Beauveria and Metarhizium in Mexican agricultural soils. J. Invertebr. Pathol. 2014, 119, 54–61. [Google Scholar] [CrossRef]
- Niemczyk, M.; Sierpińska, A.; Tereba, A.; Sokołowski, K.; Przybylski, P. Natural occurrence of Beauveria spp. in outbreak areas of cockchafers (Melolontha spp.) in forest soils from Poland. Biocontrol 2019, 64, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Peña, S.R.; Lara, J.S.; Medina, R.F. Occurrence of entomopathogenic fungi from agricultural and natural ecosystems in Saltillo, México, and their virulence towards thrips and whiteflies. J. Insect Sci. 2011, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Strasser, H.; Forer, A.; Schinner, F. Development of media for the selective isolation and maintenance of virulence of Beauveria brongniartii. In Proceedings of the 3rd International Workshop on Microbial Control of Soil Dwelling Pests, Lincoln, New Zealand, 21–23 February 1996; pp. 21–23. [Google Scholar]
- Safavi, S. Isolation, identification and pathogenicity assessment of a new isolate of entomopathogenic fungus, Beauveria bassiana in Iran. J. Plant Prot. Res. 2010, 50, 158–163. [Google Scholar] [CrossRef]
- Klingen, I.; Eilenberg, J.; Meadow, R. Effects of farming system, field margins and bait insect on the occurrence of insect pathogenic fungi in soils. Agric. Ecosyst. Environ. 2002, 91, 191–198. [Google Scholar] [CrossRef]
- Al Khoury, C.; Nemer, N.; Bernigaud, C.; Fischer, K.; Guillot, J. First evidence of the activity of an entomopathogenic fungus against the eggs of Sarcoptes scabiei. Vet. Parasitol. 2021, 298, 109553. [Google Scholar] [CrossRef]
- Abou-Jawdah, Y.; Atamian, H.; Nemer, G.; Kfoury, L.; Choukrallah, N.; Hanna, L.; Nemer, N. Efficacy and molecular studies of a Lebanese isolate of Beauveria for control of Thaumetopoea wilkinsoni (Lepidoptera: Thaumetopoeidae). Biocontrol Sci. Technol. 2008, 18, 573–581. [Google Scholar] [CrossRef]
- Al Khoury, C.; Nemer, G.; Guillot, J.; Nour, A.A.; Nemer, N. Expression analysis of the genes involved in the virulence of Beauveria bassiana. Agri Gene 2019, 14, 100094. [Google Scholar] [CrossRef]
- Al Khoury, C.; Guillot, J.; Nemer, N. Susceptibility and development of resistance of the mite Tetranychus urticae to aerial conidia and blastospores of the entomopathogenic fungus Beauveria bassiana. Syst. Appl. Acarol. 2020, 25, 429–443. [Google Scholar]
- Strasser, H. Use of hyphomycetous fungi for managing insect pests. In Fungi as Biocontrol Agents: Progress Problems and Potential; Butt, T.M., Jackson, C., Magan, N., Eds.; CABI: Wallingford, UK, 2001; p. 23. [Google Scholar]
- Quesada-Moraga, E.; Navas-Cortés, J.A.; Maranhao, E.A.; Ortiz-Urquiza, A.; Santiago-Álvarez, C. Factors affecting the occurrence and distribution of entomopathogenic fungi in natural and cultivated soils. Mycol. Res. 2007, 111, 947–966. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.K.; Mustafa, R.A.; Assaf, L.H. Isolation of entomopathogenic and opportunistic fungi from soil in Duhok province, Kurdistan region of Iraq by different selective isolation media. J. Biol. Agric. Healthc. 2015, 5, 73–79. [Google Scholar]
- Bueno-Pallero, F.Á.; Blanco-Pérez, R.; Vicente-Díez, I.; Rodríguez Martín, J.A.; Dionísio, L.; Campos-Herrera, R. Patterns of occurrence and activity of entomopathogenic fungi in the Algarve (Portugal) using different isolation methods. Insects 2020, 11, 352. [Google Scholar] [CrossRef]
- Carrillo-Benítez, M.G.; Guzmán-Franco, A.W.; Alatorre-Rosas, R.; Enríquez-Vara, J.N. Diversity and genetic population structure of fungal pathogens infecting white grub larvae in agricultural soils. Microb. Ecol. 2013, 65, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.E.; Meyling, N.V.; Luangsa-ard, J.J.; Blackwell, M. Fungal entomopathogens. In Insect Pathology; Academic Press: Cambridge, MA, USA, 2012; pp. 171–220. [Google Scholar]
- Bassil, S.; Kattar, S.; Navarro-Cerrillo, R.M.; Navarrete Poyatos, M.A.; Nemer, N.; Palacios Rodriguez, G. Stand structure and regeneration of Cedrus libani (A. Rich) in Tannourine Cedar Forest Reserve (Lebanon) affected by cedar web-spinning sawfly (Cephalcia tannourinensis, Hymenoptera: Pamphiliidae). iFor.-Biogeosci. For. 2018, 11, 300. [Google Scholar] [CrossRef] [Green Version]
- Rehayem, M.; Noujeim, E.; Nemer, N.; Pagès, S.; Ogier, J.; Thaler, O.; Duvic, B. New Insights in biocontrol strategy against Cephalcia tannourinensis, the principal insect defoliator of Lebanese cedars. For. Sci. 2018, 64, 383–391. [Google Scholar] [CrossRef] [Green Version]
- Tartanus, M.; Furmanczyk, E.M.; Canfora, L.; Pinzari, F.; Tkaczuk, C.; Majchrowska-Safaryan, A.; Malusá, E. Biocontrol of Melolontha spp. grubs in organic strawberry plantations by entomopathogenic fungi as affected by environmental and metabolic factors and the interaction with soil microbial biodiversity. Insects 2021, 12, 127. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).