Vacuolar Protein-Sorting Receptor MoVps13 Regulates Conidiation and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Deletion and Complement Strategy
2.2. Strains, Growth Conditions, and Phenotypic Analyses
2.3. Growth Stress Assay
2.4. Autophagy Assays
2.5. Immunoblotting Analysis
3. Results
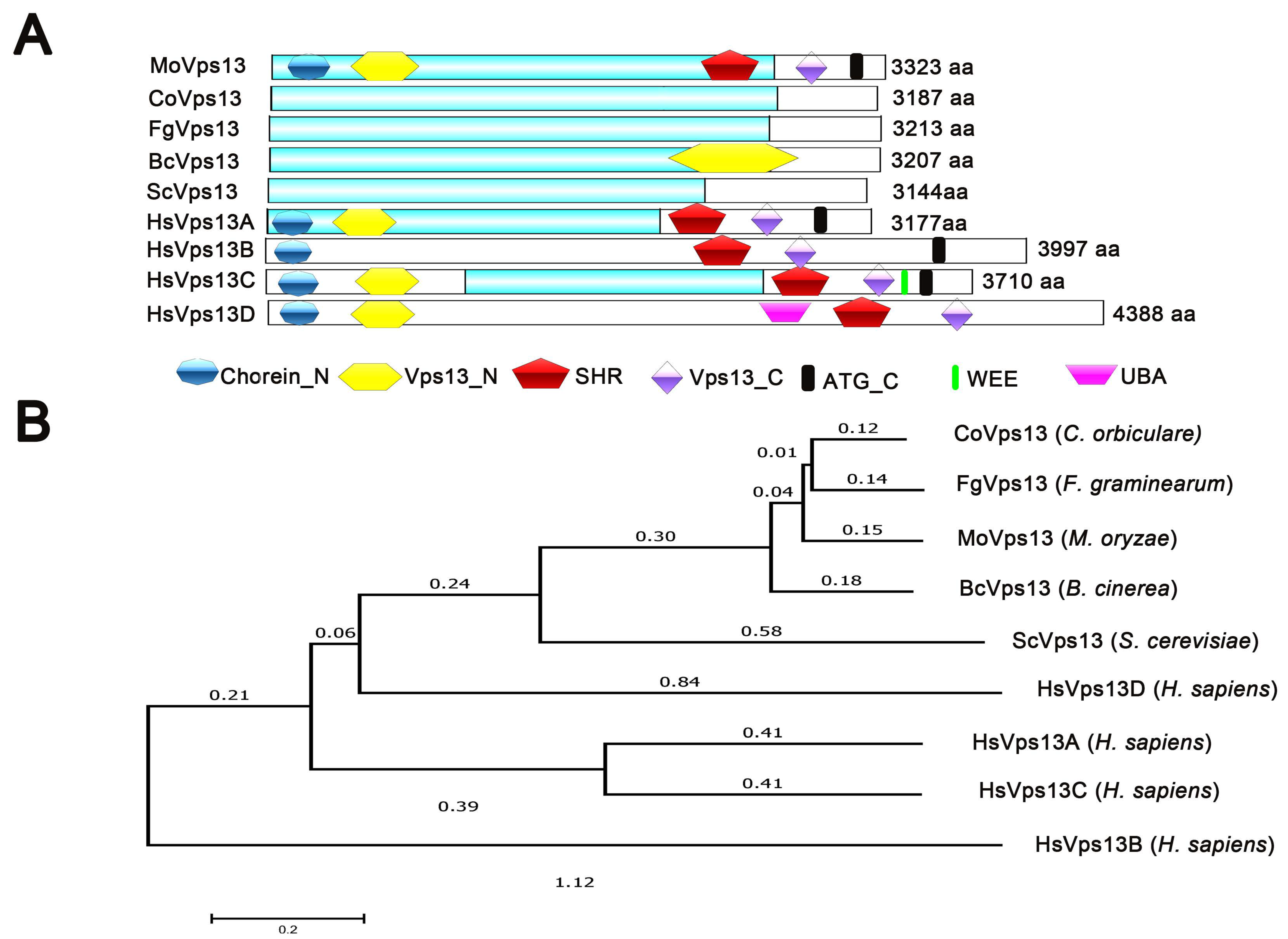
3.1. Identification of VPS13 Protein in M. oryzae
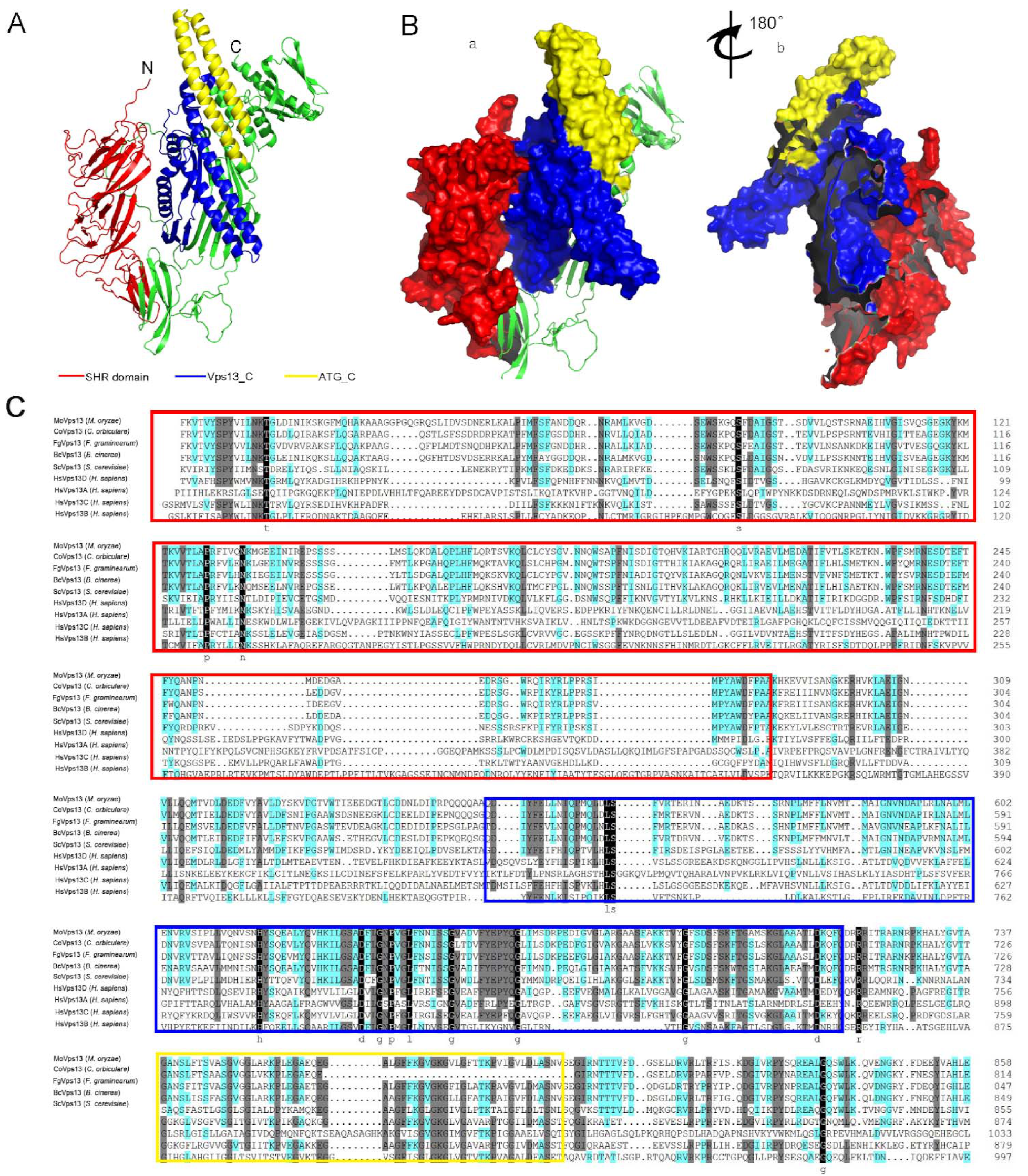
3.2. Three-Dimensional Structure of MoVps13 Protein in M. oryzae
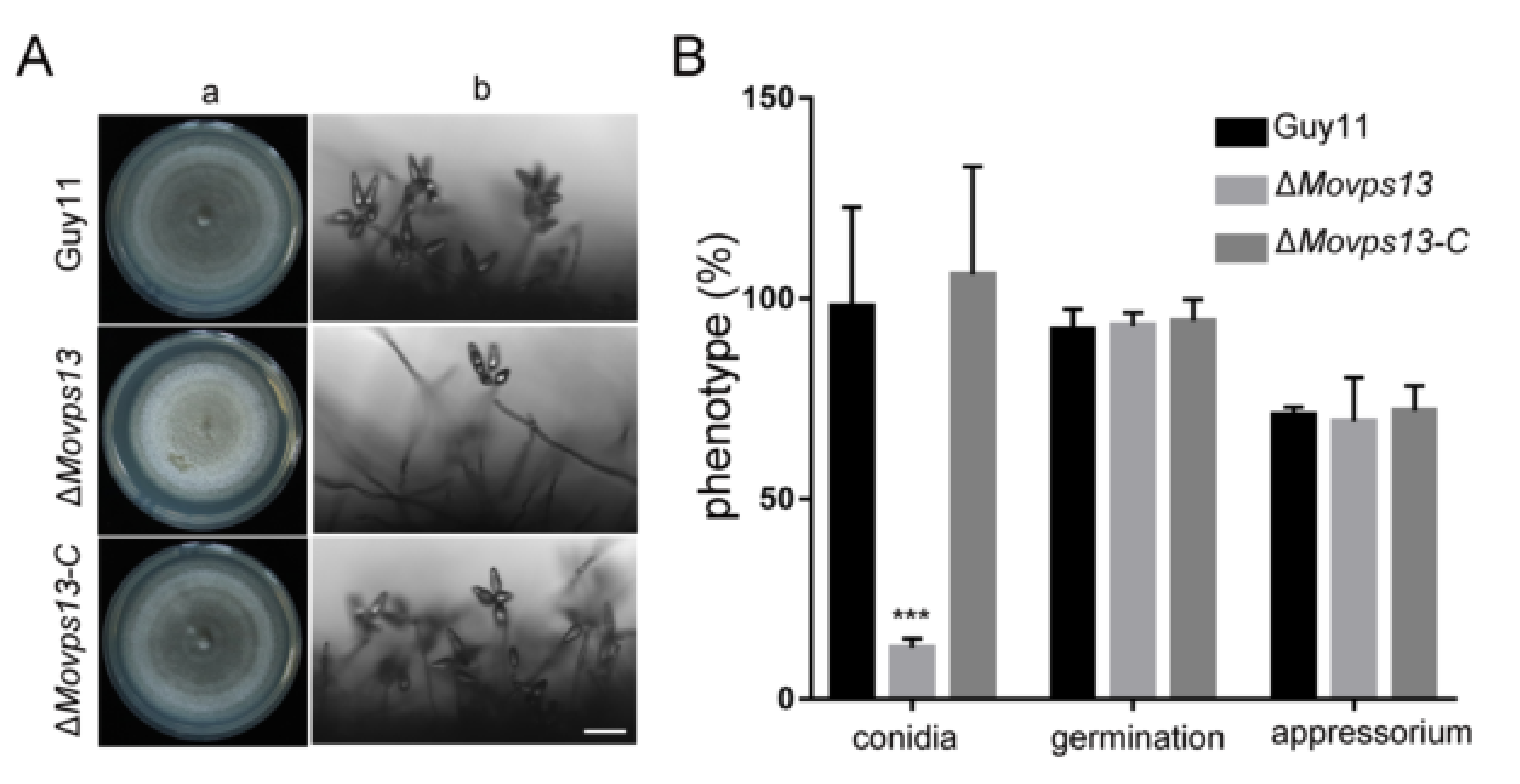
3.3. MoVps13 Is Involved in Conidiation and Virulence
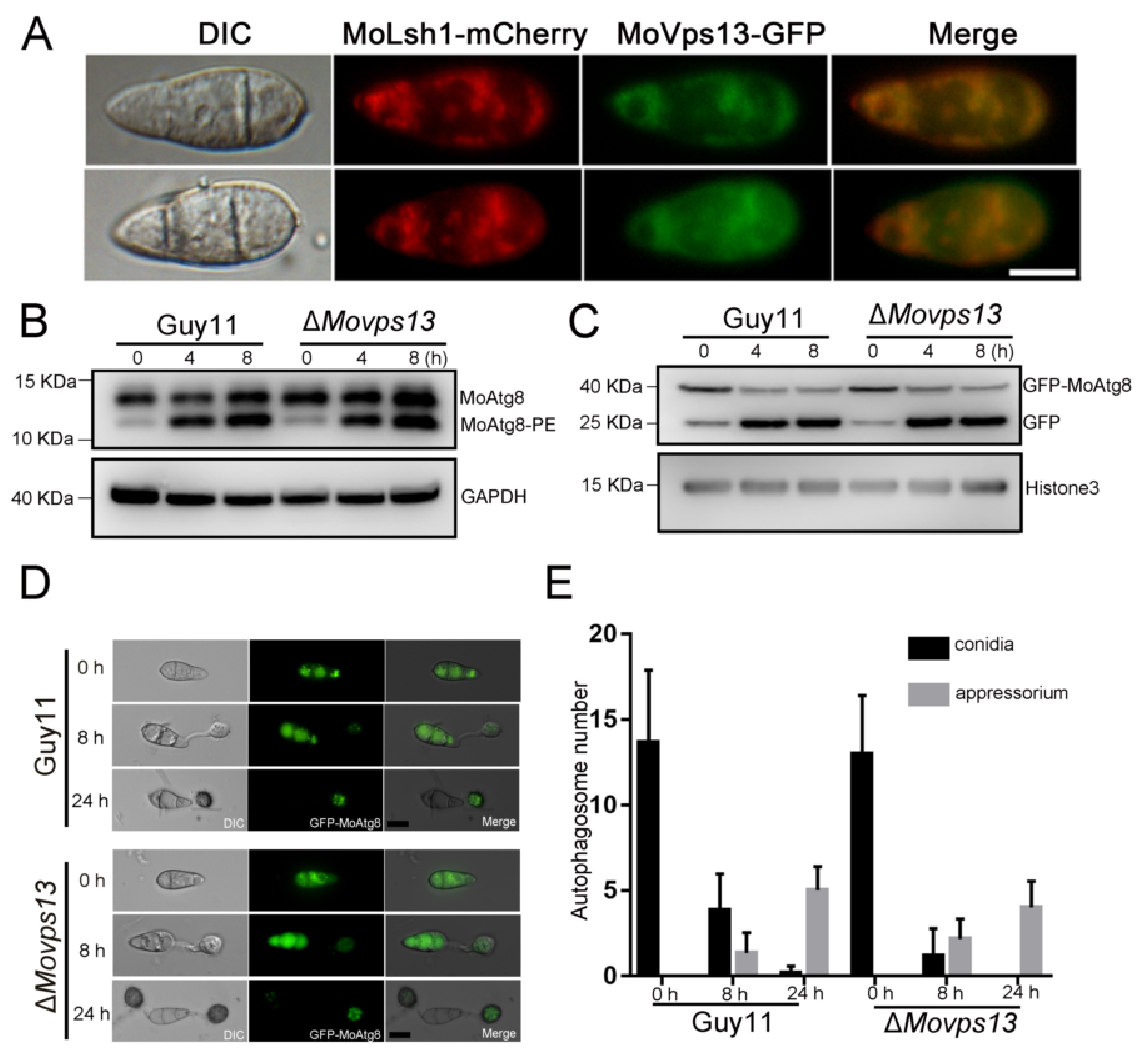
3.4. MoVps13 Localized in ER and Involved in ER-Phagy

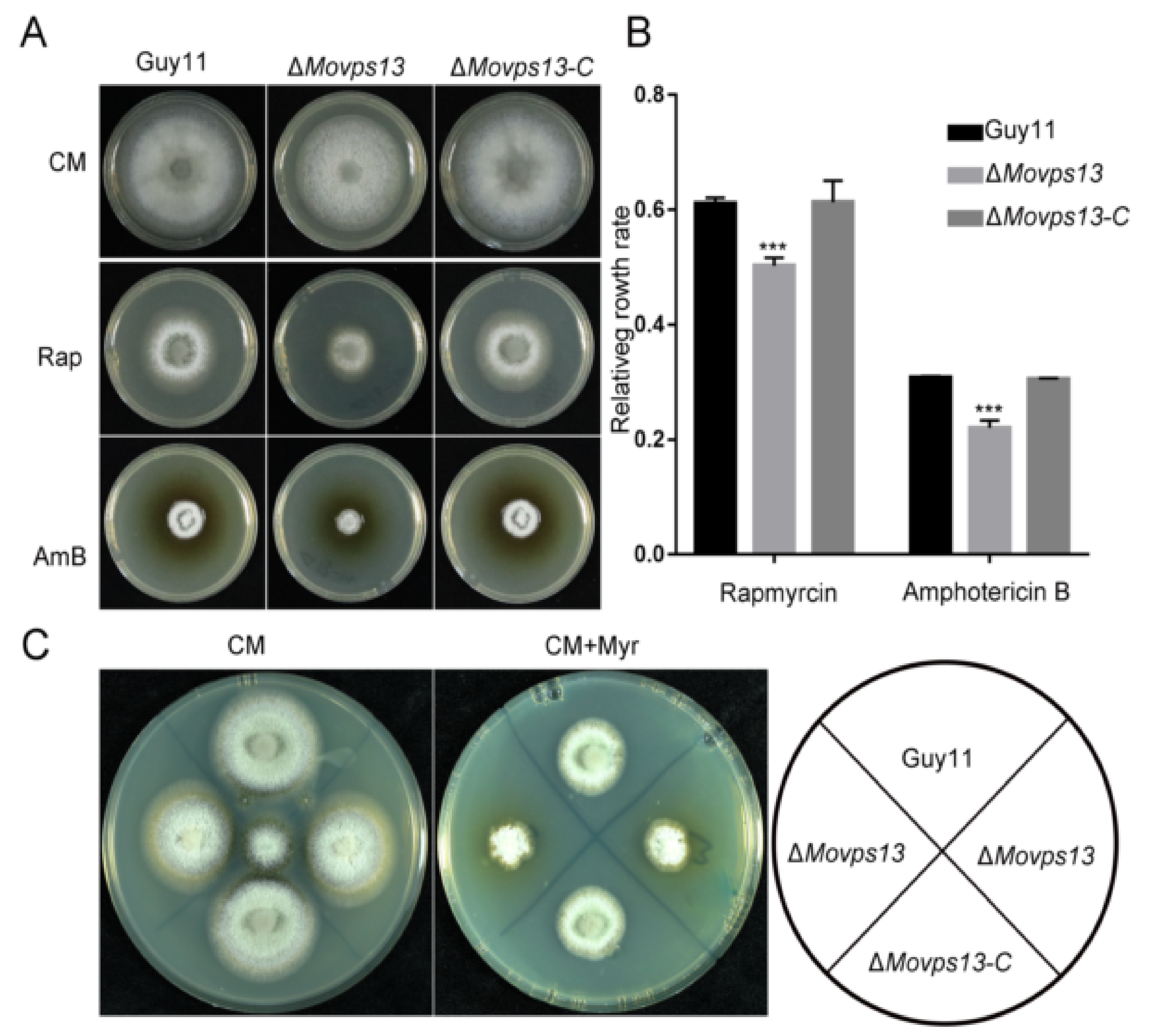
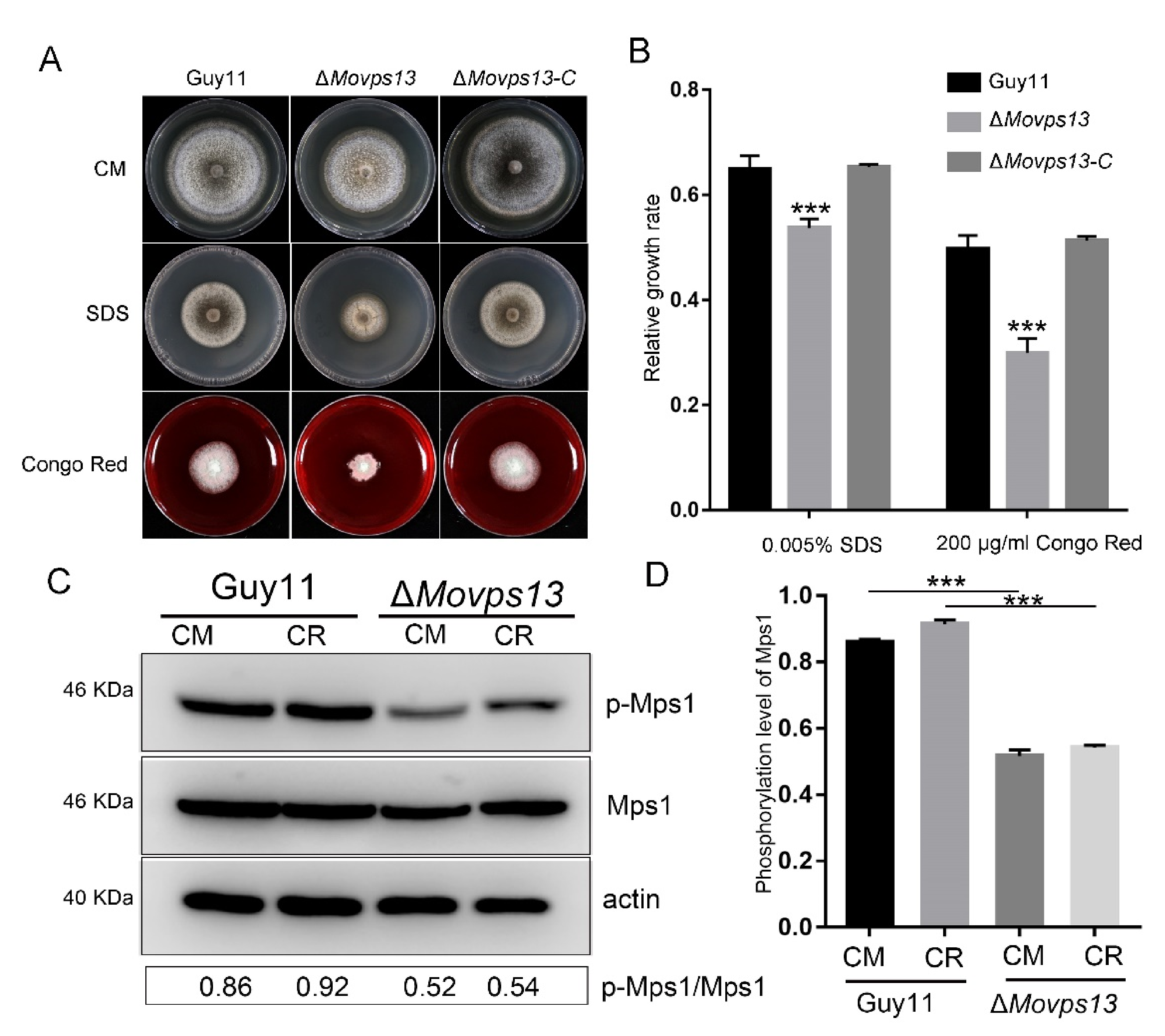
3.5. MoVps13 Is Hypersensitive to Sphingolipid Synthesis Inhibitors and Displays Cell Wall Defects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, R.A. Magnaporthe oryzae. Trends Microbiol. 2021, 29, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Huang, P.; Yan, Y.; Shi, Y.; Dong, B.; Liu, X.; Ye, L.; Lin, F.; Lu, J. The basic helix-loop-helix transcription factor Crf1 is required for development and pathogenicity of the rice blast fungus by regulating carbohydrate and lipid metabolism. Environ. Microbiol. 2018, 20, 3427–3441. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, A.; Talbot, N.J. Fungal pathogenesis: Combatting the oxidative burst. Nat. Microbiol. 2017, 2, 17095. [Google Scholar] [CrossRef] [PubMed]
- Hamer, J.E.; Howard, R.J.; Chumley, F.G.; Valent, B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 1988, 239, 288–290. [Google Scholar] [CrossRef]
- Fernandez, J.; Orth, K. Rise of a Cereal Killer: The Biology of Magnaporthe oryzae Biotrophic Growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef]
- Rocha, R.O.; Elowsky, C.; Pham, N.T.T.; Wilson, R.A. Spermine-mediated tight sealing of the Magnaporthe oryzae appressorial pore–rice leaf surface interface. Nat. Microbiol. 2020, 5, 1472–1480. [Google Scholar] [CrossRef]
- Choi, J.; Kim, Y.; Kim, S.; Park, J.; Lee, Y.-H. MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 2009, 46, 243–254. [Google Scholar] [CrossRef]
- Ryder, L.S.; Talbot, N.J. Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant. Biol. 2015, 26, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Foster, A.J.; Ryder, L.S.; Kershaw, M.J.; Talbot, N.J. The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 1008–1016. [Google Scholar] [CrossRef] [Green Version]
- Talbot, N.J. Appressoria. Curr. Biol. 2019, 29, R144–R146. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.-M.; Liang, S.; Shi, H.-B.; Lu, J.-P.; Dong, B.; Liao, Q.-S.; Lin, F.-C.; Liu, X.-H. VPS9 domain-containing proteins are essential for autophagy and endocytosis in Pyricularia oryzae. Environ. Microbiol. 2018, 20, 1516–1530. [Google Scholar] [CrossRef]
- Chen, Y.; Zhai, S.; Zhang, H.; Zuo, R.; Wang, J.; Guo, M.; Zheng, X.; Wang, P.; Zhang, Z. Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity ofMagnaporthe oryzae. Environ. Microbiol. 2014, 16, 788–801. [Google Scholar] [CrossRef]
- Feng, W.; Yin, Z.; Wu, H.; Liu, P.; Liu, X.; Liu, M.; Yu, R.; Gao, C.; Zhang, H.; Zheng, X.; et al. Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PLoS Pathog. 2021, 17, e1009080. [Google Scholar] [CrossRef]
- Nie, H.-Z.; Zhang, L.; Zhuang, H.-Q.; Shi, W.-J.; Yang, X.-F.; Qiu, D.-W.; Zeng, H.-M. The Secreted Protein MoHrip1 Is Necessary for the Virulence of Magnaporthe oryzae. Int. J. Mol. Sci. 2019, 20, 1643. [Google Scholar] [CrossRef] [Green Version]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Yang, H. VPS13: A lipid transfer protein making contacts at multiple cellular locations. J. Cell Biol. 2018, 217, 3322–3324. [Google Scholar] [CrossRef] [Green Version]
- Rzepnikowska, W.; Flis, K.; Muñoz-Braceras, S.; Menezes, R.; Escalante, R.; Zoladek, T. Yeast and other lower eukaryotic organisms for studies of Vps13 proteins in health and disease. Traffic 2017, 18, 711–719. [Google Scholar] [CrossRef] [Green Version]
- Bean, B.D.M.; Dziurdzik, S.K.; Kolehmainen, K.L.; Fowler, C.M.S.; Kwong, W.K.; Grad, L.I.; Davey, M.; Schluter, C.; Conibear, E. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 2018, 217, 3593–3607. [Google Scholar] [CrossRef] [Green Version]
- Velayos-Baeza, A.; Vettori, A.; Copley, R.R.; Dobson-Stone, C.; Monaco, A.P. Analysis of the human VPS13 gene family. Genomics 2004, 84, 536–549. [Google Scholar] [CrossRef]
- Hook, S.C.; Chadt, A.; Heesom, K.J.; Kishida, S.; Al-Hasani, H.; Tavare, J.M.; Thomas, E.C. TBC1D1 interacting proteins, VPS13A and VPS13C, regulate GLUT4 homeostasis in C2C12 myotubes. Sci. Rep. 2020, 10, 17953. [Google Scholar] [CrossRef]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, E.; Nakamura, M.; Agemura, A.; Kusumoto, A.; Ichiba, M.; Kurano, Y.; Muroya, S.; Sano, A. Brain-specific transcript variants of 5′ and 3′ ends of mouse VPS13A and VPS13C. Biochem. Biophys. Res. Commun. 2007, 353, 902–907. [Google Scholar] [CrossRef]
- Walker, R.H. Untangling the Thorns: Advances in the Neuroacanthocytosis Syndromes. J. Mov. Disord. 2015, 8, 41–54. [Google Scholar] [CrossRef]
- Douzgou, S.; Petersen, M.B. Clinical variability of genetic isolates of Cohen syndrome. Clin. Genet. 2011, 79, 501–506. [Google Scholar] [CrossRef]
- Momtazmanesh, S.; Rayzan, E.; Shahkarami, S.; Rohlfs, M.; Klein, C.; Rezaei, N. A novel VPS13B mutation in Cohen syndrome: A case report and review of literature. BMC Med. Genet. 2020, 21, 140. [Google Scholar] [CrossRef]
- Rodrigues, J.M.; Fernandes, H.D.; Caruthers, C.; Braddock, S.R.; Knutsen, A.P. Cohen Syndrome: Review of the Literature. Cureus 2018, 10, e3330. [Google Scholar] [CrossRef] [Green Version]
- Windholz, J.; Kovacs, P.; Tonjes, A.; Dittrich, K.; Bluher, S.; Kiess, W.; Stumvoll, M.; Korner, A. Effects of genetic variants in ADCY5, GIPR, GCKR and VPS13C on early impairment of glucose and insulin metabolism in children. PLoS ONE 2011, 6, e22101. [Google Scholar] [CrossRef] [Green Version]
- Nakada, T.A.; Boyd, J.H.; Russell, J.A.; Aguirre-Hernandez, R.; Wilkinson, M.D.; Thair, S.A.; Nakada, E.; McConechy, M.K.; Fjell, C.D.; Walley, K.R. VPS13D Gene Variant Is Associated with Altered IL-6 Production and Mortality in Septic Shock. J. Innate Immun. 2015, 7, 545–553. [Google Scholar] [CrossRef]
- John Peter, A.T.; Herrmann, B.; Antunes, D.; Rapaport, D.; Dimmer, K.S.; Kornmann, B. Vps13-Mcp1 interact at vacuole–mitochondria interfaces and bypass ER–mitochondria contact sites. J. Cell Biol. 2017, 216, 3219–3229. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539. [Google Scholar] [CrossRef]
- Bouatta, N.; Sorger, P.; Alquraishi, M. Protein structure prediction by AlphaFold2: Are attention and symmetries all you need? Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, T.; Wang, P.; Liu, F.; Tai, G.; Zhou, Y. The water network in galectin-3 ligand binding site guides inhibitor design. Acta Biochim. Biophys. Sin. 2015, 47, 192–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zhu, S.; Yan, Y.; Qu, Y.; Wang, J.; Feng, X.; Liu, X.; Lin, F.; Lu, J. Role refinement of melanin synthesis genes by gene knockout reveals their functional diversity in Pyricularia oryzae strains. Microbiol. Res. 2021, 242, 126620. [Google Scholar] [CrossRef]
- Talbot, N.J.; Ebbole, D.J.; Hamer, J.E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 1993, 5, 1575–1590. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.-Y.; Liang, S.; Zhang, Y.-R.; Lu, J.-P.; Lin, F.-C.; Liu, X.-H. MoSec61β, the beta subunit of Sec61, is involved in fungal development and pathogenicity, plant immunity, and ER-phagy in Magnaporthe oryzae. Virulence 2020, 11, 1685–1700. [Google Scholar] [CrossRef]
- Lu, J.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic Analysis of Zn2Cys6 Transcription Factors Required for Development and Pathogenicity by High-Throughput Gene Knockout in the Rice Blast Fungus. PLoS Pathog. 2014, 10, e1004432. [Google Scholar] [CrossRef]
- Muñoz-Braceras, S.; Calvo, R.; Escalante, R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy inDictyosteliumand human HeLa cells. Autophagy 2015, 11, 918–927. [Google Scholar] [CrossRef] [Green Version]
- Funato, K.; Riezman, H.; Muniz, M. Vesicular and non-vesicular lipid export from the ER to the secretory pathway. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158453. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, J.; Zhu, X.; Dong, B.; Liu, X.; Lu, J.; Lin, F. The P5-type ATPase Spf1 is required for development and virulence of the rice blast fungus Pyricularia oryzae. Curr. Genet. 2020, 66, 385–395. [Google Scholar] [CrossRef]
- De, M.; Oleskie, A.N.; Ayyash, M.; Dutta, S.; Mancour, L.; Abazeed, M.E.; Brace, E.J.; Skiniotis, G.; Fuller, R.S. The Vps13p–Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J. Cell Biol. 2017, 216, 425–439. [Google Scholar] [CrossRef]
- Zhu, X.-M.; Li, L.; Cai, Y.-Y.; Wu, X.-Y.; Shi, H.-B.; Liang, S.; Qu, Y.-M.; Naqvi, N.I.; Del Poeta, M.; Dong, B.; et al. A VASt-domain protein regulates autophagy, membrane tension, and sterol homeostasis in rice blast fungus. Autophagy 2020, 17, 1–23. [Google Scholar] [CrossRef]
- Zhu, X.-M.; Li, L.; Wu, M.; Liang, S.; Shi, H.-B.; Liu, X.-H.; Lin, F.-C. Current opinions on autophagy in pathogenicity of fungi. Virulence 2019, 10, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Li, L.; Zhu, X.-M.; Su, Z.-Z.; Del Poeta, M.; Liu, X.-H.; Lin, F.-C. Insights of roles played by septins in pathogenic fungi. Virulence 2021, 12, 1550–1562. [Google Scholar] [CrossRef]
- Nadal, M.; Gold, S.E. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol. Plant Pathol. 2010, 11, 463–478. [Google Scholar] [CrossRef]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 7, 11062. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, S. Emerging Principles of Selective ER Autophagy. J. Mol. Biol. 2020, 432, 185–205. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Martens, S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016, 428, 1714–1724. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-S.; Neiman, A.M. VPS13 Regulates Membrane Morphogenesis During Sporulation in Saccharomyces cerevisiae. J. Cell Sci. 2012, 125, 3004–3011. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.X.; Qian, H.; Liu, M.Y.; Wu, M.H.; Wei, Y.Y.; Zhu, X.M.; Lu, J.P.; Lin, F.C.; Liu, X.H. Endosomal sorting complexes required for transport-0 (ESCRT-0) are essential for fungal development, pathogenicity, autophagy and ER-phagy in Magnaporthe oryzae. Environ. Microbiol. 2021. [Google Scholar] [CrossRef]
- Liu, X.-H.; Chen, S.-M.; Gao, H.-M.; Ning, G.-A.; Shi, H.-B.; Wang, Y.; Dong, B.; Qi, Y.-Y.; Zhang, D.-M.; Lu, G.-D.; et al. The small GTPase MoYpt7 is required for membrane fusion in autophagy and pathogenicity ofMagnaporthe oryzae. Environ. Microbiol. 2015, 17, 4495–4510. [Google Scholar] [CrossRef]
- Sakulkoo, W.; Osés-Ruiz, M.; Oliveira Garcia, E.; Soanes, D.M.; Littlejohn, G.R.; Hacker, C.; Correia, A.; Valent, B.; Talbot, N.J. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 2018, 359, 1399–1403. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Bian, Z.; Xu, J.-R. Assays for MAP Kinase Activation in Magnaporthe oryzae and Other Plant Pathogenic Fungi. In Plant Pathogenic Fungi and Oomycetes; Springer: New York, NY, USA, 2018; pp. 93–101. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.-R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.R.; Staiger, C.J.; Hamer, J.E. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 1998, 95, 12713–12718. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Dagdas, Y.F.; Zhu, X.; Zheng, S.; Chen, L.; Cartwright, Z.; Talbot, N.J.; Wang, Z. The glycogen synthase kinase MoGsk1, regulated by Mps1 MAP kinase, is required for fungal development and pathogenicity in Magnaporthe oryzae. Sci. Rep. 2017, 7, 945. [Google Scholar] [CrossRef]
- Sun, G.; Elowsky, C.; Li, G.; Wilson, R.A. TOR-autophagy branch signaling via Imp1 dictates plant-microbe biotrophic interface longevity. PLoS Genet. 2018, 14, e1007814. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Liu, X.; Ye, Z.; Zhou, Q.; Liu, P.; Yin, Z.; Wang, W.; Zheng, X.; Zhang, H.; Zhang, Z. Phosphatase-associated protein MoTip41 interacts with the phosphatase MoPpe1 to mediate crosstalk between TOR and cell wall integrity signalling during infection by the rice blast fun. Environ. Microbiol. 2021, 23, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.; Heisenberg, C.P. Biology and physics of cell shape changes in development. Curr. Biol. 2009, 19, R790–R799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafesse, F.G.; Holthuis, J.C. Cell biology: A brake on lipid synthesis. Nature 2010, 463, 1028–1029. [Google Scholar] [CrossRef]
- Gonzalez-Solis, A.; Han, G.; Gan, L.; Li, Y.; Markham, J.E.; Cahoon, R.E.; Dunn, T.M.; Cahoon, E.B. Unregulated Sphingolipid Biosynthesis in Gene-Edited Arabidopsis ORM Mutants Results in Nonviable Seeds with Strongly Reduced Oil Content. Plant Cell 2020, 32, 2474–2490. [Google Scholar] [CrossRef]
- Guan, X.L.; Souza, C.M.; Pichler, H.; Dewhurst, G.; Schaad, O.; Kajiwara, K.; Wakabayashi, H.; Ivanova, T.; Castillon, G.A.; Piccolis, M.; et al. Functional Interactions between Sphingolipids and Sterols in Biological Membranes Regulating Cell Physiology. Mol. Biol. Cell 2009, 20, 2083–2095. [Google Scholar] [CrossRef] [Green Version]
- Hurst, L.R.; Fratti, R.A. Lipid Rafts, Sphingolipids, and Ergosterol in Yeast Vacuole Fusion and Maturation. Front. Cell Dev. Biol. 2020, 8, 539. [Google Scholar] [CrossRef]
- Yang, H.; Tong, J.; Lee, C.W.; Ha, S.; Eom, S.H.; Im, Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015, 6, 6129. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Su, J.; Xu, Y.; Chen, J.; Chern, M.; Lei, M.; Qi, T.; Wang, Z.; Ryder, L.S.; Tang, B.; et al. Discovery of broad-spectrum fungicides that block septin-dependent infection processes of pathogenic fungi. Nat. Microbiol. 2020, 5, 1565–1575. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Li, L.; Wang, J.; Zhao, L.; Shi, H.; Bao, J.; Su, Z.; Liu, X.; Lin, F. Vacuolar Protein-Sorting Receptor MoVps13 Regulates Conidiation and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae. J. Fungi 2021, 7, 1084. https://doi.org/10.3390/jof7121084
Zhu X, Li L, Wang J, Zhao L, Shi H, Bao J, Su Z, Liu X, Lin F. Vacuolar Protein-Sorting Receptor MoVps13 Regulates Conidiation and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae. Journal of Fungi. 2021; 7(12):1084. https://doi.org/10.3390/jof7121084
Chicago/Turabian StyleZhu, Xueming, Lin Li, Jiaoyu Wang, Lili Zhao, Huanbin Shi, Jiandong Bao, Zhenzhu Su, Xiaohong Liu, and Fucheng Lin. 2021. "Vacuolar Protein-Sorting Receptor MoVps13 Regulates Conidiation and Pathogenicity in Rice Blast Fungus Magnaporthe oryzae" Journal of Fungi 7, no. 12: 1084. https://doi.org/10.3390/jof7121084





