Functional Analysis of an Essential GSP1/Ran Ortholog Gene, CpRan1, from the Chestnut Blight Fungus Cryphonectria parasitica Using a Heterokaryon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth
2.2. Quantitative Analysis of Transcript Accumulation Using Real-Time RT-PCR
2.3. Statistical Analysis
2.4. Cloning and Characterization of a GSP1/Ran Like Gene, CpRan1
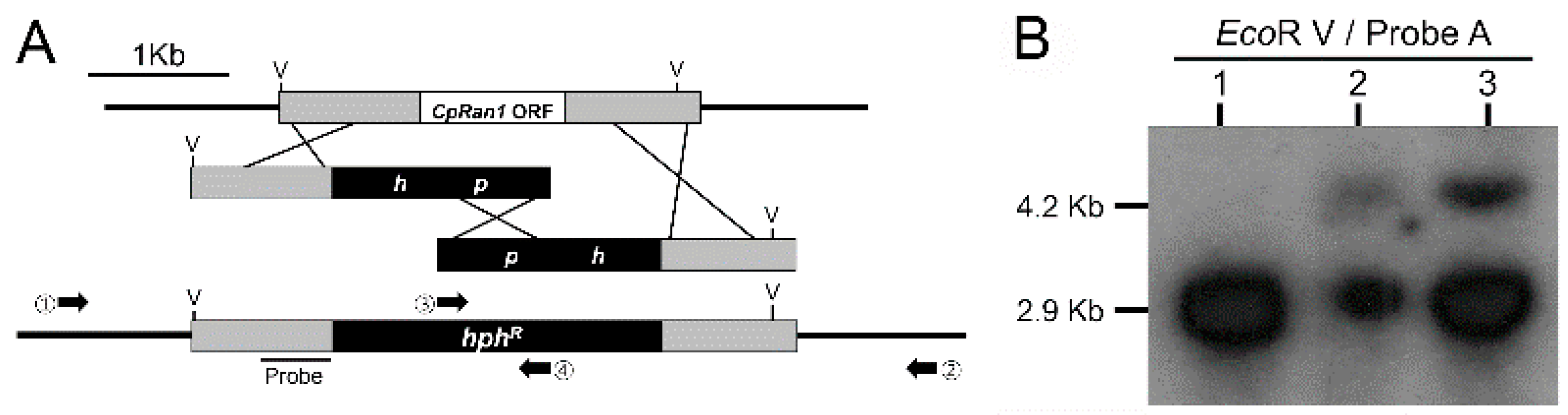
2.5. Southern and Northern Blot Analysis
2.6. Target Gene Replacement Vectors and Fungal Transformation
2.7. Staining and Microscopy
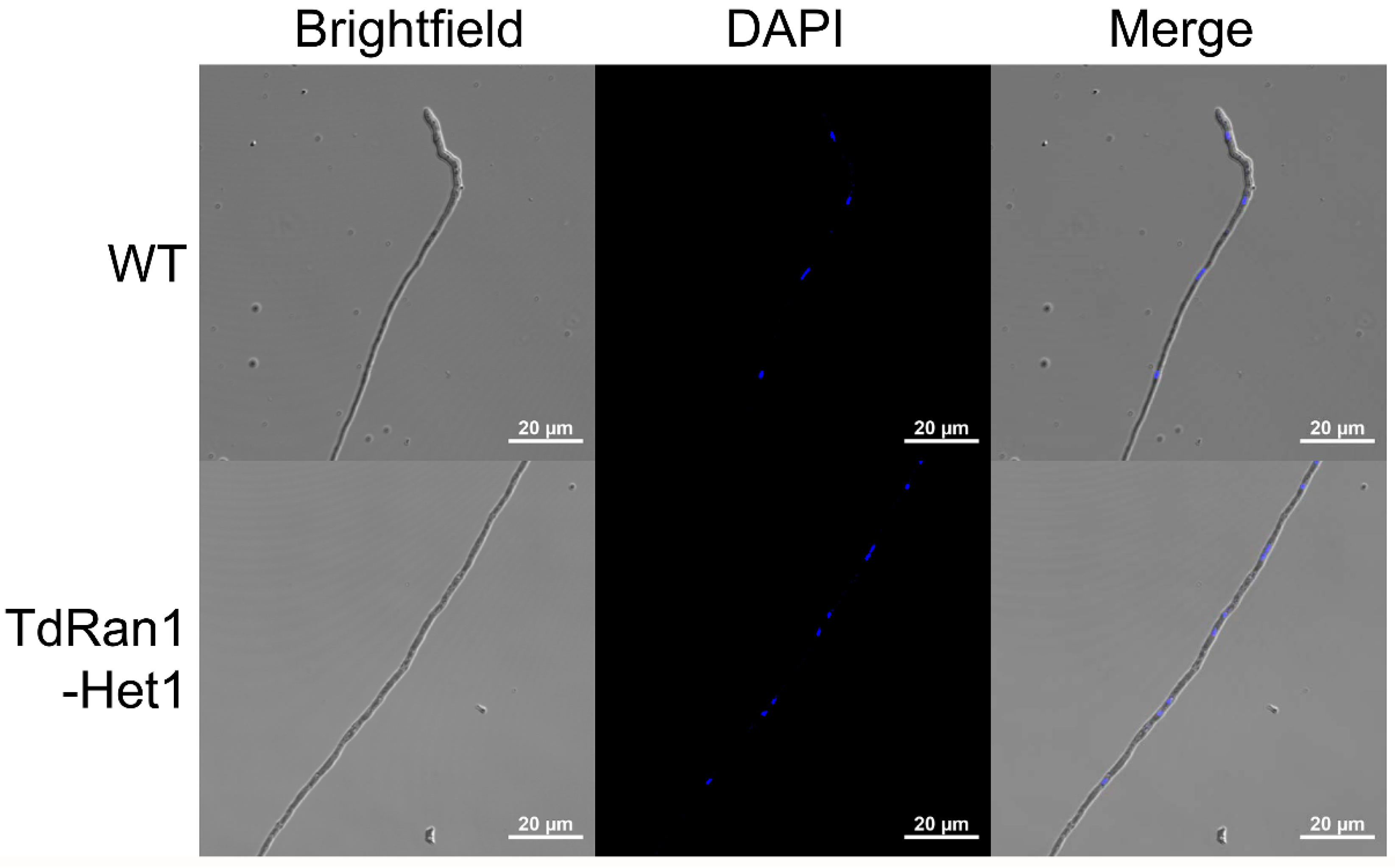
2.8. Ratio of Different Types of Nuclei
2.9. Construction of Complementary Vectors
3. Results
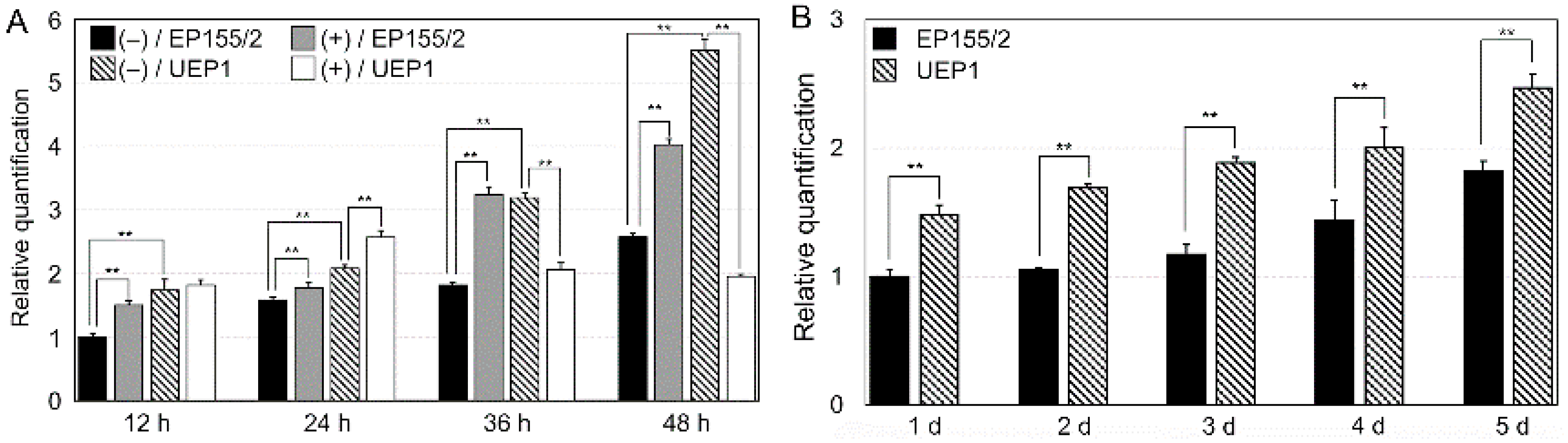
3.1. Characterization of a Tannic Acid-Responsive and Hypoviral-Regulated CpRan1 Gene
3.2. Expression of CpRan1
3.3. Screening and Identification of a CpRan1-Null Mutant
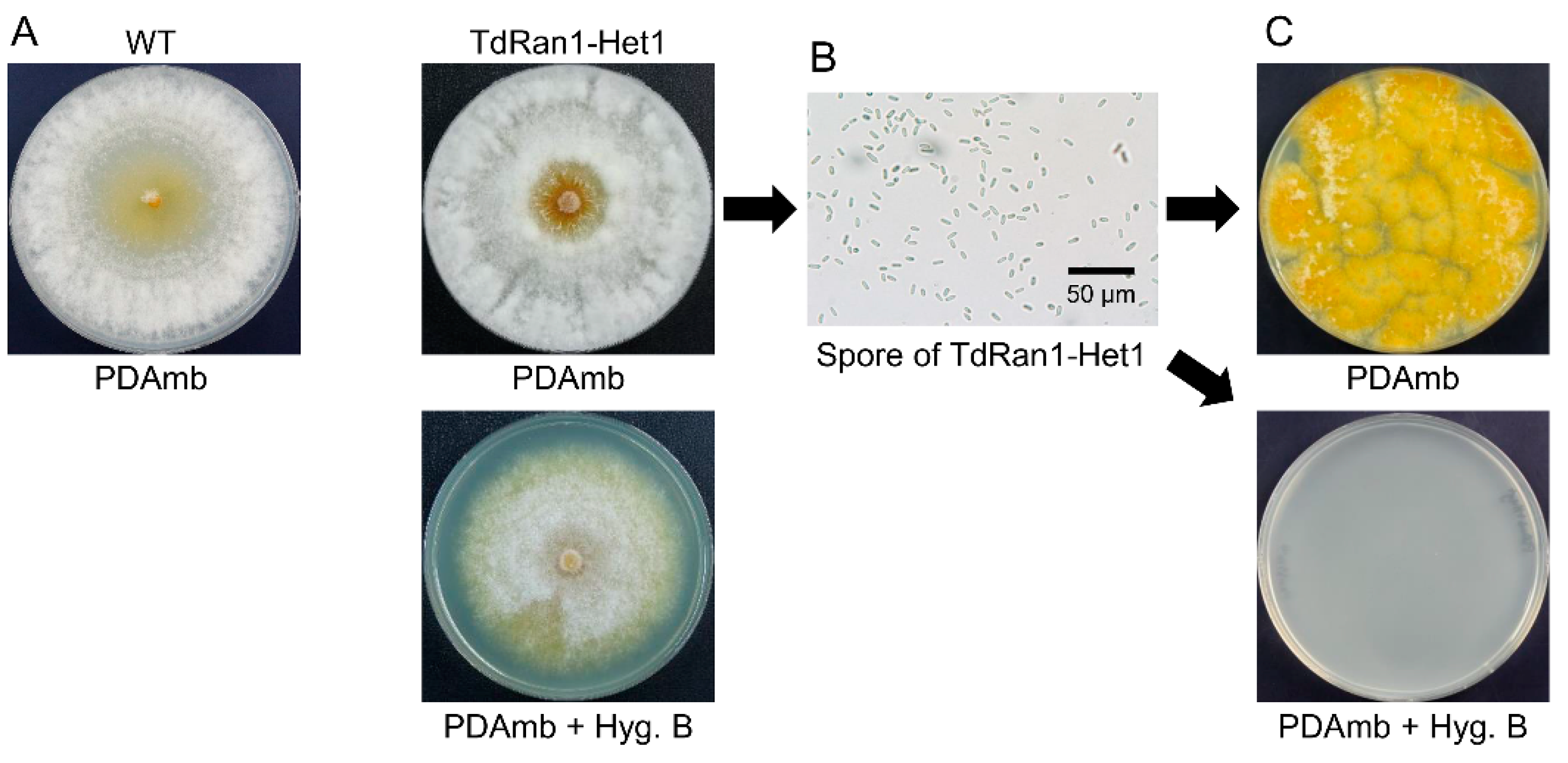
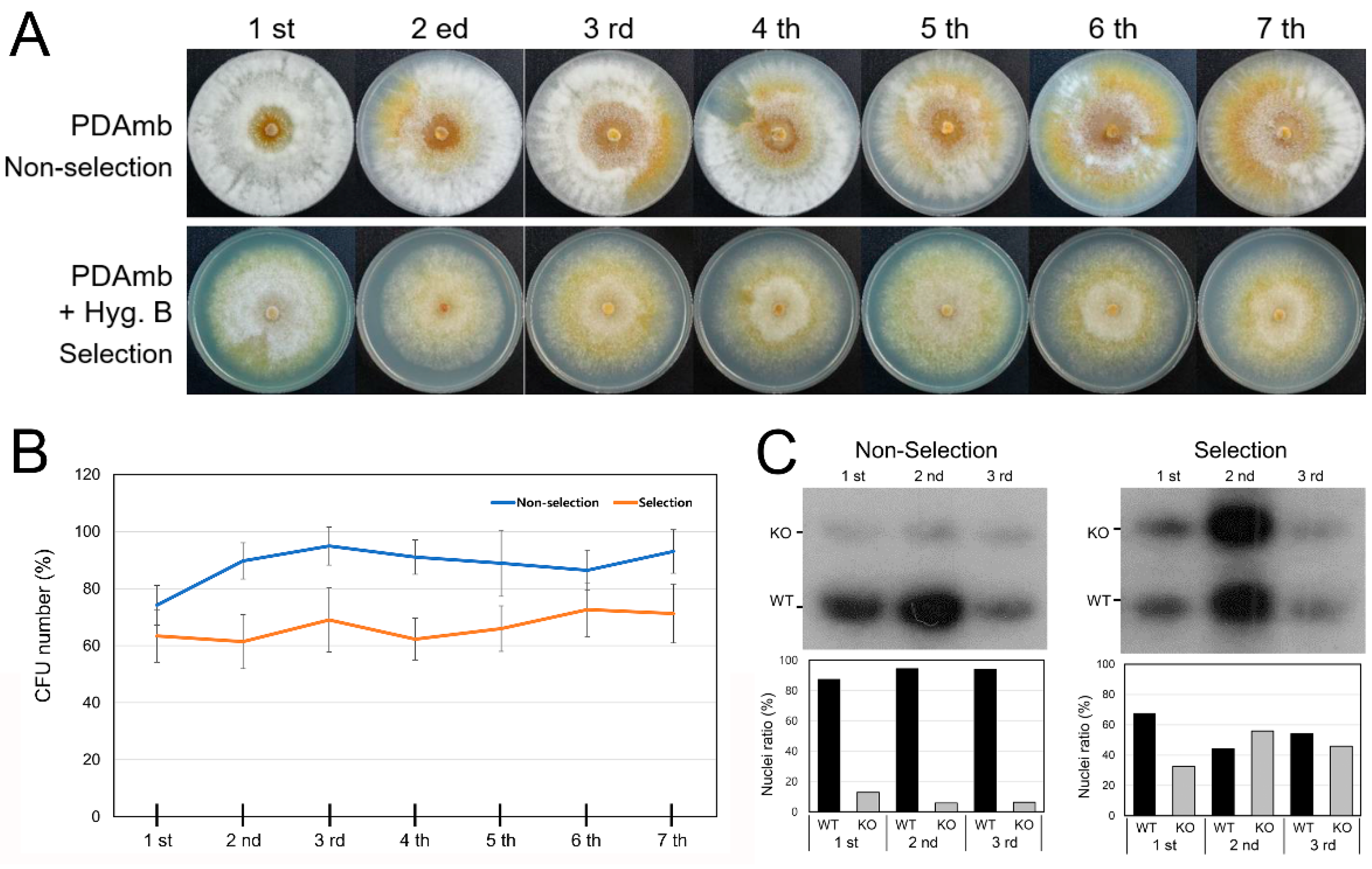
3.4. Heterokaryon Analysis of the Putative CpRan1-Null Mutant
3.5. Characteristics of Heterokaryons
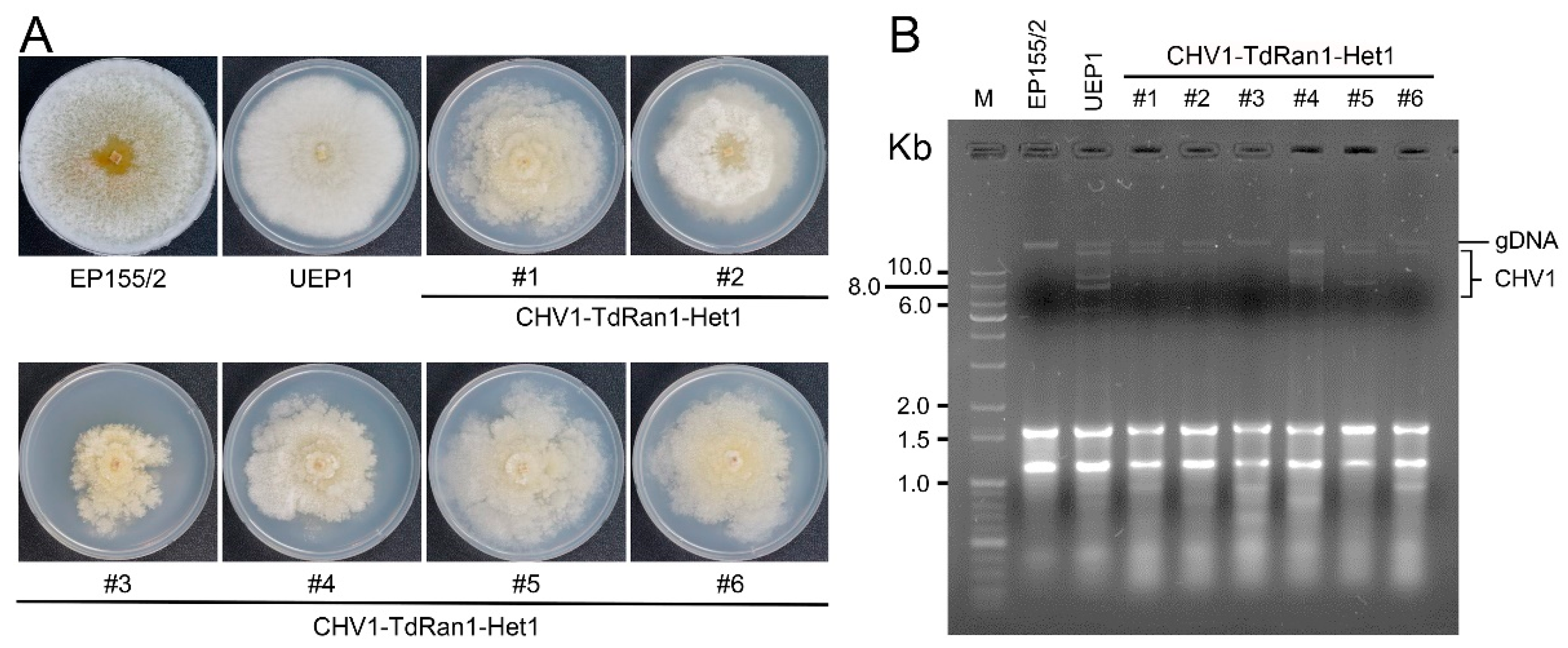
3.6. Functional Analysis of the Essential CpRan1 Gene Using Heterokaryons
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Alfen, N.K. Biology and potential for disease control of hypovirulence of Endothia parasitica. Ann. Rev. Phytopathol. 1982, 20, 349–362. [Google Scholar] [CrossRef]
- Van Alfen, N.K.; Jaynes, R.A.; Anagnostakis, S.L.; Day, P.R. Chestnut blight: Biological control by transmissible hypovirulence in Endothia parasitica. Science 1975, 189, 890–891. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Biological control of chestnut blight. Science 1982, 215, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Biological control of chestnut blight: An example of cirus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 1992, 56, 561–576. [Google Scholar] [CrossRef]
- Havir, E.A.; Anagnostakis, S.L. Oxalate production by virulent but not by hypovirulent strains of Endothia parasitica. Physiol. Plant Pathol. 1983, 23, 369–376. [Google Scholar] [CrossRef]
- Elliston, J.E. Characteristics of dsRNA-free and dsRNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology 1985, 74, 151–158. [Google Scholar] [CrossRef]
- Rigling, D.; Heiniger, U.; Hohl, H.R. Reduction of Laccse activity in dsRNA-containing hypovirulent strains of Cryphonectria (Endothia) parasitica. Phytopathology 1988, 79, 219–223. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Cornejo, C.; Aulia, A.; Shahi, S.; Hillman, B.; Rigling, D. In-Tree behavior of diverse viruses harbored in the chestnut blight fungus, Cryphonectria parasitica. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Kazmierczak, P.; Pfeiffer, P.; Zhang, L.; Van Alfen, N.K. Transcriptional repression of specific host genes by the mycovirus Cryphonectria hypovirus 1. J. Virol. 1996, 70, 1137–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.S.; Choi, J.W.; Park, S.M.; Cha, B.J.; Yang, M.S.; Kim, D.H. Ordered differential display from Cryphonectria parasitica. J. Plant Pathol. 2000, 16, 142–146. [Google Scholar]
- Allen, T.D.; Nuss, D.L. Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 2004, 78, 4145–4155. [Google Scholar] [CrossRef] [Green Version]
- Dawe, A.L.; Segers, G.C.; Allen, T.D.; McMains, V.C.; Nuss, D.L. Microarray analysis of Cryphonectria parasitica Galpha- and Gbetagamma-signalling pathways reveals extensive modulation by hypovirus infection. Microbiology 2004, 150, 4033–4043. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Allen, T.D.; Hillma, B.I.; Nuss, D.L. Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut bright fungus Cryphonectria parasitica. Eukaryot. Cell 2007, 6, 1286–1298. [Google Scholar] [CrossRef] [Green Version]
- Dawe, A.L.; Nuss, D.L. Hypovirus molecular biology: From Koch’s postulates to host self-recognition genes that restrict virus transmission. Adv. Virus Res. 2013, 86, 109–147. [Google Scholar] [PubMed]
- Churchill, A.C.L.; Ciuffetti, L.M.; Hansen, D.R.; Van Etten, H.D.; Van Alfen, N.K. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 1990, 17, 25–31. [Google Scholar] [CrossRef]
- Chen, B.; Choi, G.H.; Nuss, D.L. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 1994, 264, 1762–1764. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Rigling, D.; Zhang, L.; Van Alfen, N.K. A new extracellular laccase of Cryphonectria parasitica is revealed by deletion of lac1. Mol. Plant Microbe Interact. 1995, 8, 259–266. [Google Scholar] [CrossRef]
- Segers, G.C.; Wezel, R.V.; Zhang, X.; Hong, Y.; Nuss, D.L. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 2006, 5, 896–904. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.M.; Park, J.A.; Kim, D.H. Comparative proteomic analysis of chestnut blight fungus, Cryphonectria parasitica, under tannic-acid-inducing and hypovirus-regulating conditions. Can. J. Microbiol. 2012, 58, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Demeter, J.; Morphew, M.; Sazer, S. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc. Natl. Acad. USA 1995, 92, 1436–1440. [Google Scholar] [CrossRef] [Green Version]
- Ryan, K.J.; Michael McCaffery, J.; Wente, S.R. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 2003, 160, 1041–1053. [Google Scholar] [CrossRef] [Green Version]
- Hetzer, M.W.; Walther, T.C.; Mattaj, I.W. Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005, 21, 347–380. [Google Scholar] [CrossRef]
- Liu, H.; Söderhäll, K.; Jiravanichpaisal, P. Antiviral immunity in crustaceans. Fish Shellfish Immunol. 2009, 27, 77–88. [Google Scholar] [CrossRef]
- Mirallas, O.; Ballega, E.; Samper-Martín, B.; García-Márquez, S.; Carballar, R.; Ricco, N.; Jiménez, J.; Clotet, J. Intertwined control of the cell cycle and nucleocytoplasmic transport by the cyclin-dependent kinase Pho85 and RanGTPase Gsp1 in Saccharomyces cerevisiae. Microbiol. Res. 2018, 206, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Kutay, U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999, 15, 607–660. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Intracellular Compartments and Protein Sorting. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Ko, Y.H.; Choi, S.Y.; So, K.K.; Kim, J.M.; Chun, J.; Kim, D.H. Functional analysis of an essential Ran-binding protein gene, CpRbp1, from the chestnut blight fungus Cryphonectria parasitica using heterokaryon rescue. Sci. Rep. 2020, 10, 8111. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. A stable heterokaryon of Endothia parasitica. Mycologia 1981, 73, 570–576. [Google Scholar] [CrossRef]
- Kim, M.J.; Park, S.M.; Kim, Y.H.; Cha, B.J.; Yang, M.S.; Kim, D.H. Deletion of a hypoviral-regulated cppk1 gene in a chestnut blight fungus, Cryphonectria parasitica, results in microcolonies. Fungal Genet. Biol. 2004, 41, 482–492. [Google Scholar] [CrossRef]
- Ko, Y.H.; So, K.K.; Kim, J.M.; Kim, D.H. Heterokaryon analysis of a Cdc48-like gene, CpCdc48, from the chestnut blight fungus Cryphonectria parasitica demonstrates it is essential for cell division and growth. Fungal Genet. Biol. 2016, 88, 1–12. [Google Scholar] [CrossRef]
- Puhalla, J.E.; Anagnostakis, S.L. Genetics and nutritional requirements of Endothia parasitica. Phytopathology 1971, 61, 169–173. [Google Scholar] [CrossRef]
- Rigling, D.; Van Alfen, N.K. Extra- and Intracellular laccases of the chestnut blight fungus, Cryphonectria parasitica. Appl. Environ. Microbiol. 1993, 59, 3634–3639. [Google Scholar] [CrossRef] [Green Version]
- Powell, W.A.J.; Van Alfen, N.K. Two nonhomologus viruses of Cryphonectria (Endothia) parasitica reduce accumulation of specific virulence-associated polypeptides. J. Bacteriol. 1987, 169, 5324–5326. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Choi, J.W.; Park, S.M.; Cha, B.J.; Yang, M.S.; Kim, D.H. Characterization of a fungal protein kinase from Cryphonectria parasitica and its transcriptional upregulation by hypovirus. Mol. Microbiol. 2002, 45, 933–941. [Google Scholar] [CrossRef]
- Choi, G.H.; Nuss, D.L. Nucleotide sequence of the glyceraldehyde-3-phosphate dehydrogenase gene from Cryphonectria parasitica. Nucleic Acids Res. 1990, 18, 5566. [Google Scholar] [CrossRef] [Green Version]
- Goswami, R.S. Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol. 2012, 835, 255–269. [Google Scholar]
- Nguyen, Q.B.; Kadotani, N.; Kasahara, S.; Tosa, Y.; Mayama, S.; Nakayashiki, H. Systematic functional analysis of calcium signalling proteins in the genome of the riceblast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol. Microbiol. 2008, 68, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Osherov, N.; May, G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 2000, 155, 647–656. [Google Scholar]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]


| Vector Constructs in Figure 8 | Number of Transformant | CFU | |||
|---|---|---|---|---|---|
| PDAmb + Geneticin | PDAmb + Hyg. B | PDAmb | PDAmb + Geneticin | PDAmb + Hyg. B | |
| gDNA | 280 | 154 | 23 | 23 | 17 |
| cDNA | 68 | 8 | 8 | 8 | 6 |
| K21A | 60 | 0 | - | - | - |
| K97A | 74 | 7 | 7 | 7 | 5 |
| K121A | 65 | 0 | - | - | - |
| S151A | 60 | 0 | - | - | - |
| Mouse | 80 | 9 | 9 | 9 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, Y.-H.; Chun, J.; Kim, D.-H. Functional Analysis of an Essential GSP1/Ran Ortholog Gene, CpRan1, from the Chestnut Blight Fungus Cryphonectria parasitica Using a Heterokaryon. J. Fungi 2021, 7, 332. https://doi.org/10.3390/jof7050332
Ko Y-H, Chun J, Kim D-H. Functional Analysis of an Essential GSP1/Ran Ortholog Gene, CpRan1, from the Chestnut Blight Fungus Cryphonectria parasitica Using a Heterokaryon. Journal of Fungi. 2021; 7(5):332. https://doi.org/10.3390/jof7050332
Chicago/Turabian StyleKo, Yo-Han, Jeesun Chun, and Dae-Hyuk Kim. 2021. "Functional Analysis of an Essential GSP1/Ran Ortholog Gene, CpRan1, from the Chestnut Blight Fungus Cryphonectria parasitica Using a Heterokaryon" Journal of Fungi 7, no. 5: 332. https://doi.org/10.3390/jof7050332





