Citrate Regulates the Saccharomyces cerevisiae Mitochondrial GDP/GTP Carrier (Ggc1p) by Triggering Unidirectional Transport of GTP
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Expression and Purification of Wild-Type and Mutant Ggc1p
2.2. Yeast Strains, Media and Growth Conditions
2.3. Reconstitution into Liposomes and Transport Assays
2.4. Comparative Modeling and Docking Investigations
3. Results
3.1. Bacterial Expression of Ggc1p
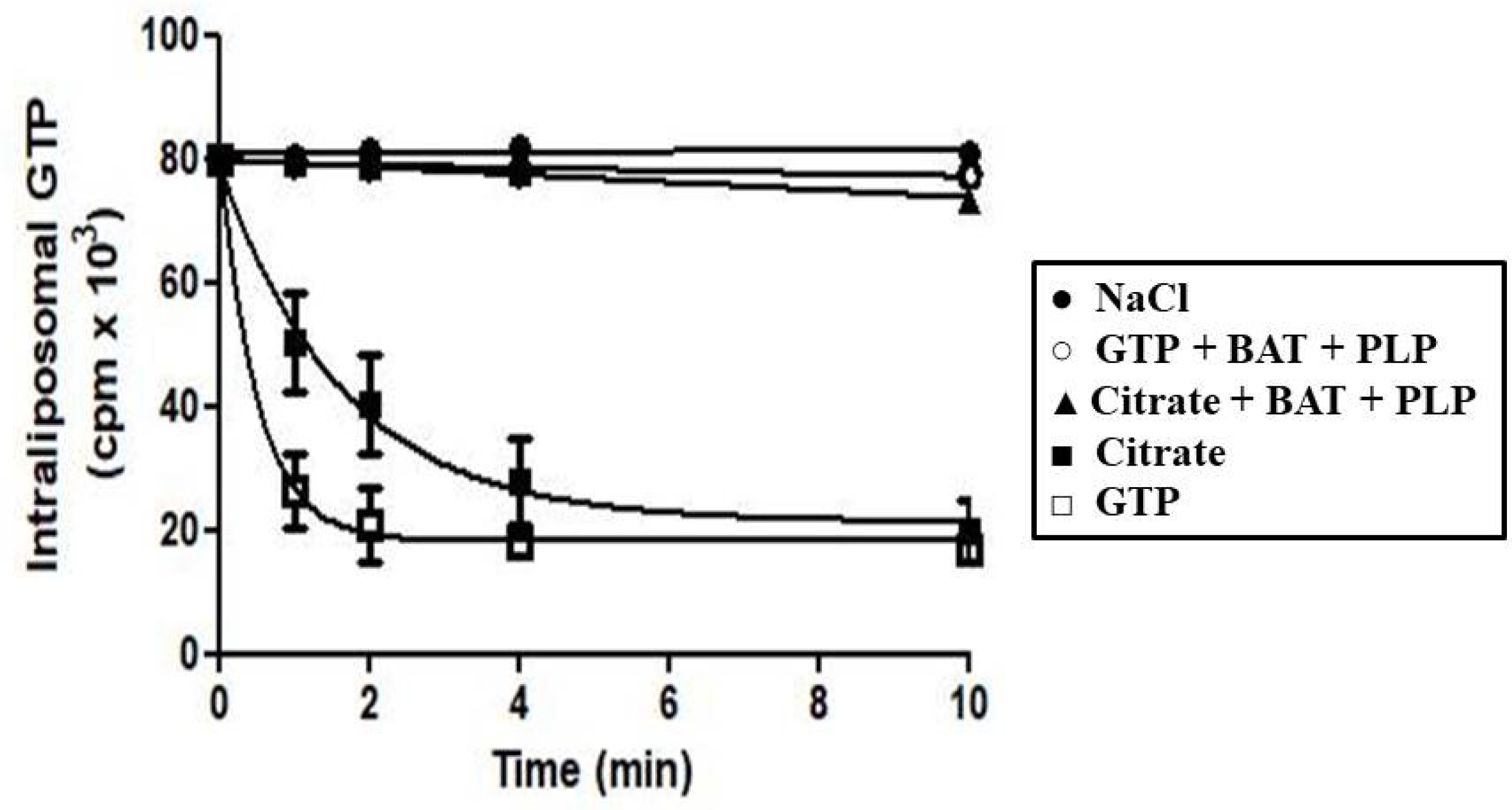
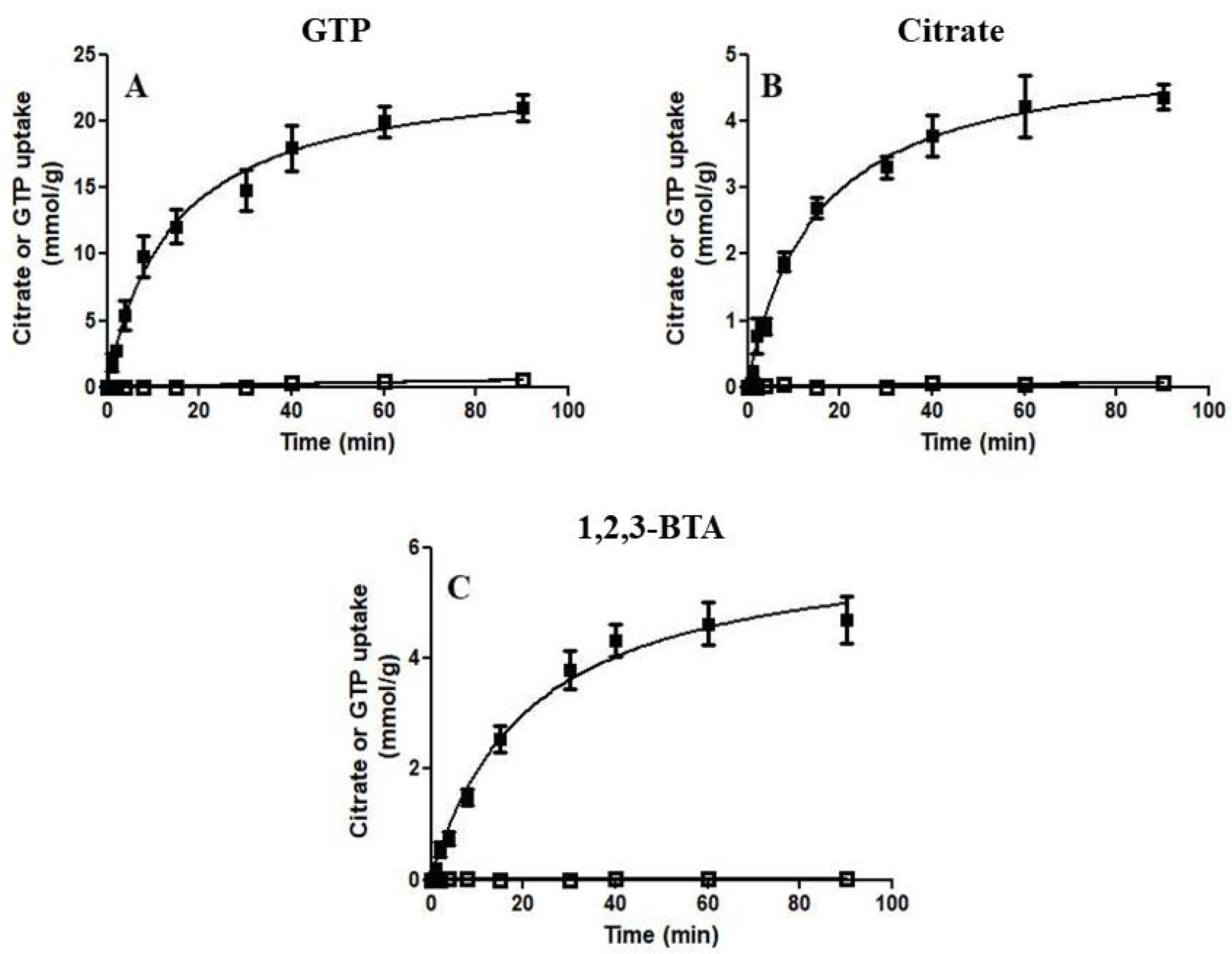
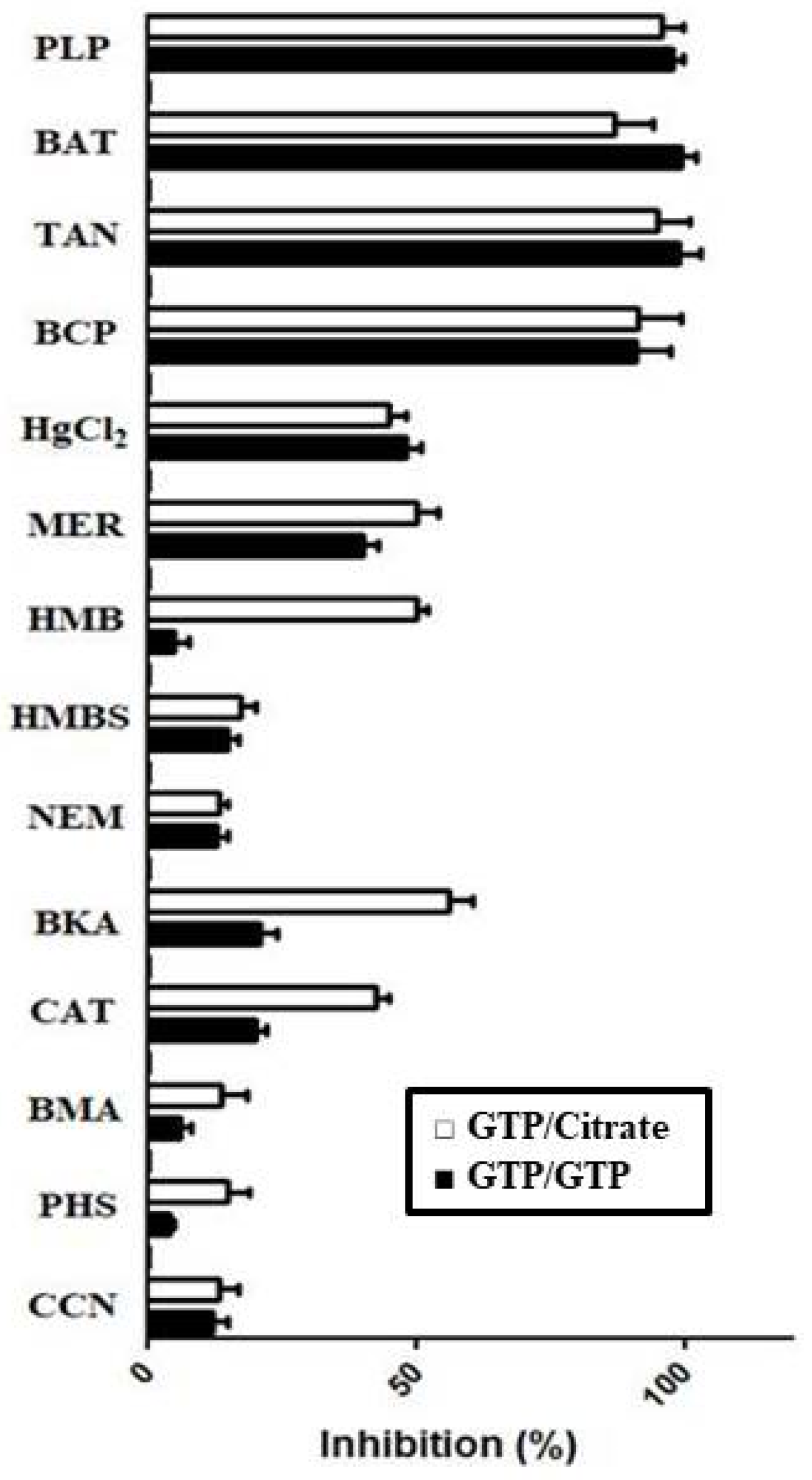
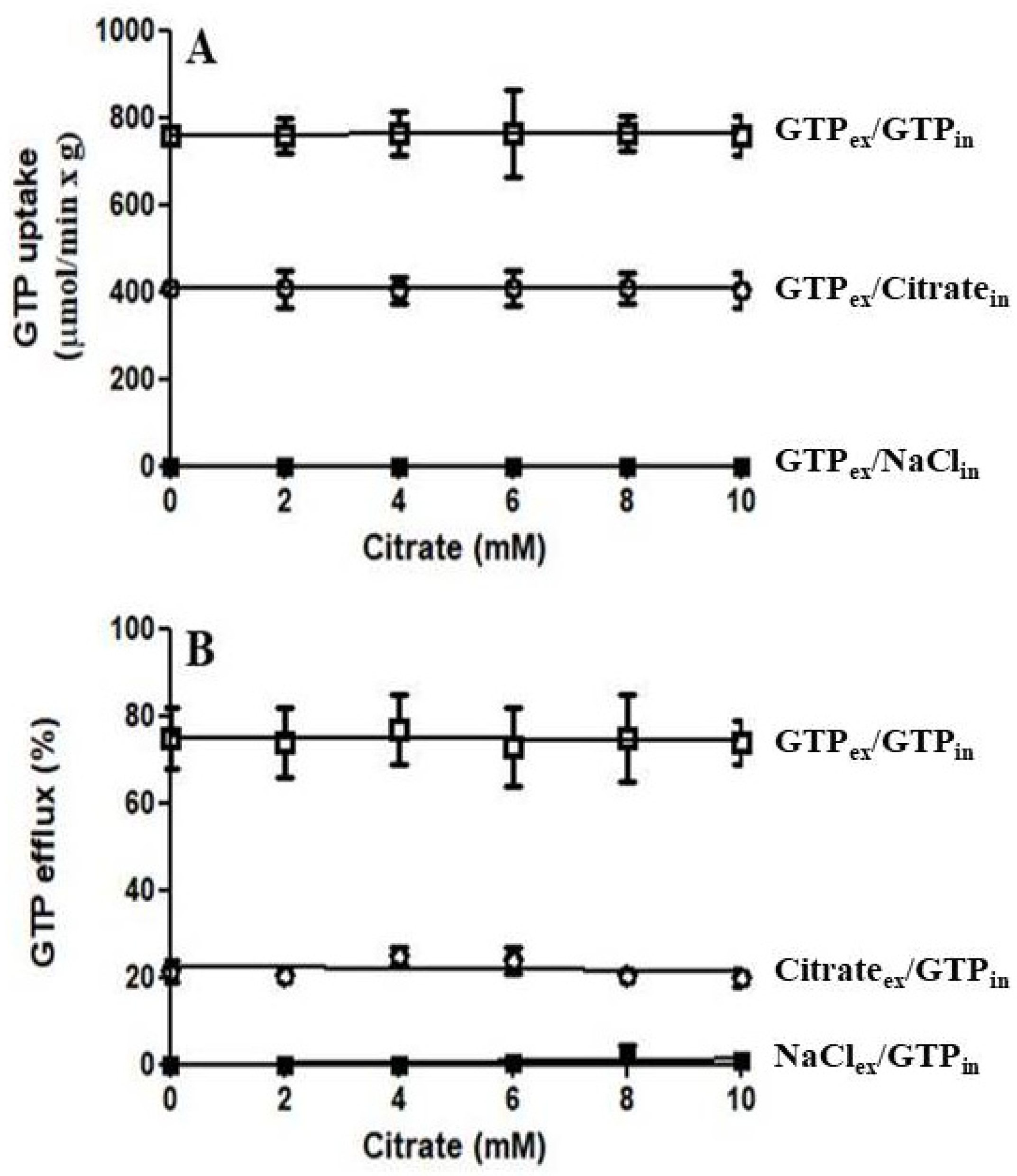
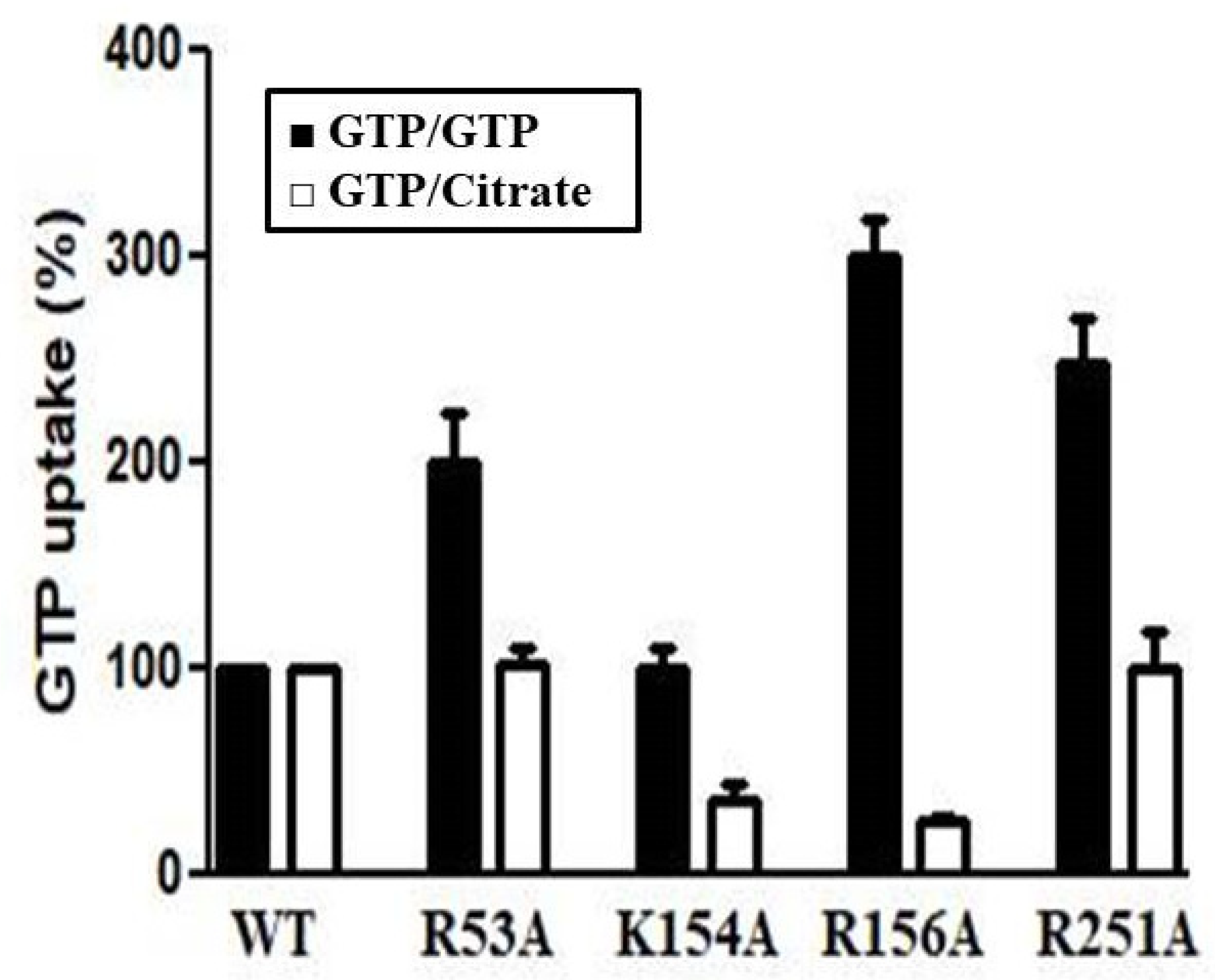
3.2. Functional Characterization of Recombinant Ggc1p
3.3. Kinetic Characteristics of Recombinant Ggc1p
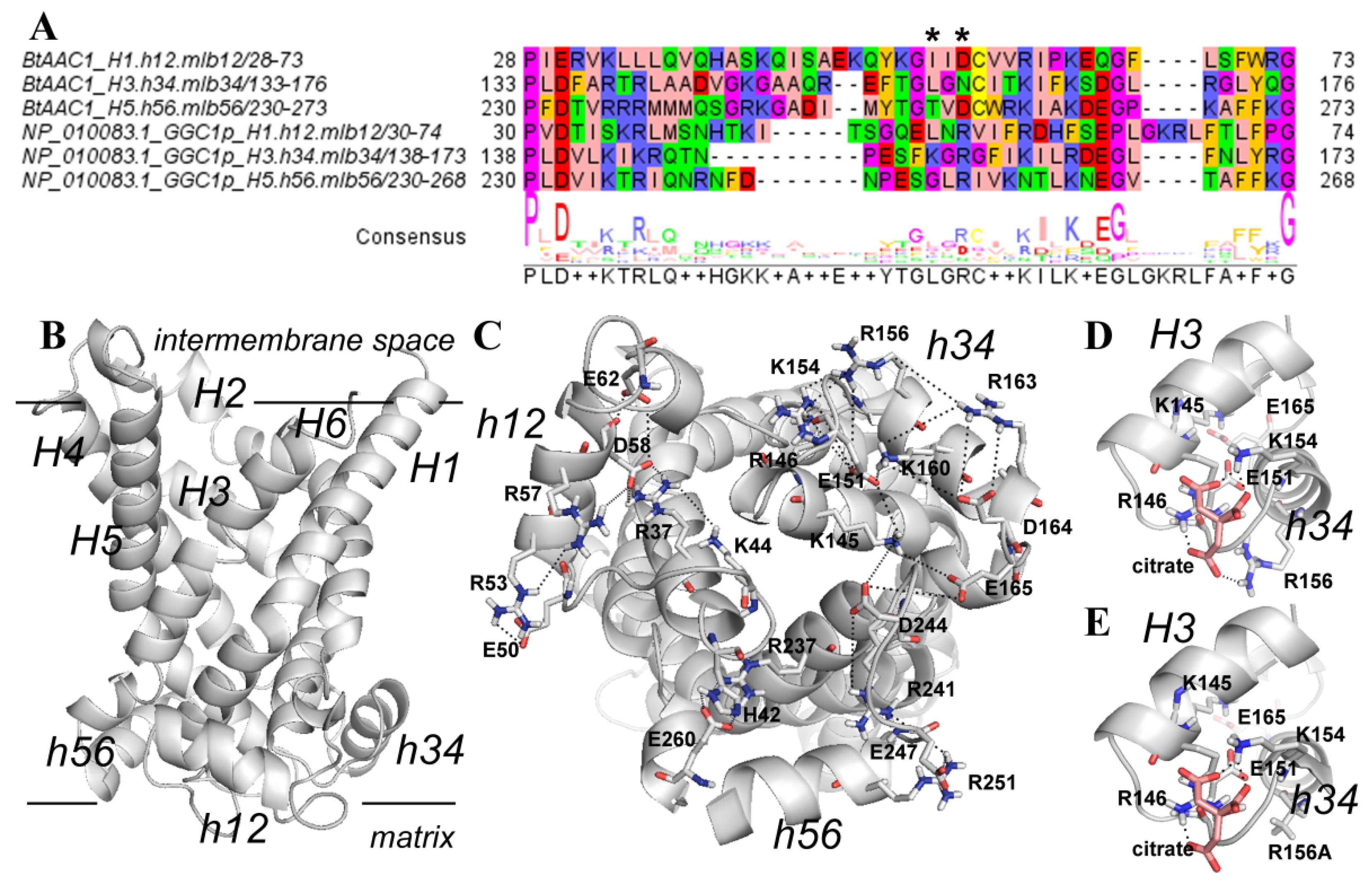
3.4. Structural Analysis of a Possible Citrate Binding Region on the Ggc1p 3D Model
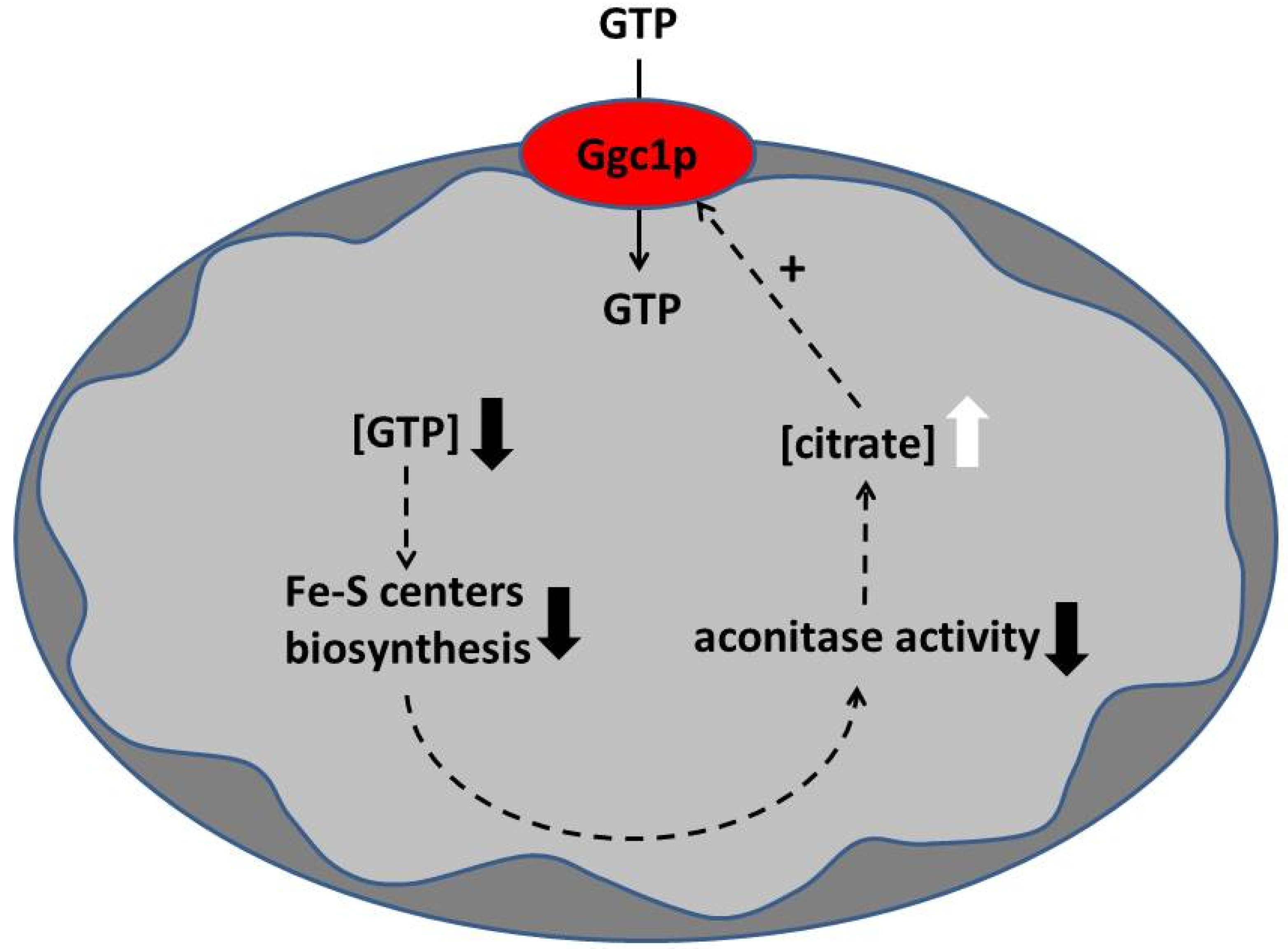
4. Discussions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomson, M. The Regulation of Mitochondrial Physiology by Organelle-Associated GTP-Binding Proteins. Cell Biochem. Funct. 2002, 20, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Sickmann, A.; Reinders, J.; Wagner, Y.; Joppich, C.; Zahedi, R.; Meyer, H.E.; Schönfisch, B.; Perschil, I.; Chacinska, A.; Guiard, B.; et al. The Proteome of Saccharomyces cerevisiae Mitochondria. Proc. Natl. Acad. Sci. USA 2003, 100, 13207–13212. [Google Scholar] [CrossRef] [Green Version]
- Newman, L.E.; Zhou, C.; Mudigonda, S.; Mattheyses, A.L.; Paradies, E.; Marobbio, C.M.T.; Kahn, R.A. The ARL2 GTPase Is Required for Mitochondrial Morphology, Motility, and Maintenance of ATP Levels. PLoS ONE 2014, 9, e99270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amutha, B.; Gordon, D.M.; Gu, Y.; Lyver, E.R.; Dancis, A.; Pain, D. GTP Is Required for Iron-Sulfur Cluster Biogenesis in Mitochondria. J. Biol. Chem. 2008, 283, 1362–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lascu, I.; Gonin, P. The Catalytic Mechanism of Nucleoside Diphosphate Kinases. J. Bioenerg. Biomembr. 2000, 32, 237–246. [Google Scholar] [CrossRef]
- Przybyla-Zawislak, B.; Gadde, D.M.; Ducharme, K.; McCammon, M.T. Genetic and Biochemical Interactions Involving Tricarboxylic Acid Cycle (TCA) Function Using a Collection of Mutants Defective in All TCA Cycle Genes. Genetics 1999, 152, 153–166. [Google Scholar] [CrossRef]
- Tsunehiro, F.; Junichi, N.; Narimichi, K.; Kazutada, W. Isolation, Overexpression and Disruption of a Saccharomyces cerevisiae YNK Gene Encoding Nucleoside Diphosphate Kinase. Gene 1993, 129, 141–146. [Google Scholar] [CrossRef]
- Amutha, B.; Pain, D. Nucleoside Diphosphate Kinase of Saccharomyces cerevisiae, Ynk1p: Localization to the Mitochondrial Intermembrane Space. Biochem. J. 2003, 370, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Vozza, A.; Blanco, E.; Palmieri, L.; Palmieri, F. Identification of the Mitochondrial GTP/GDP Transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20850–20857. [Google Scholar] [CrossRef] [Green Version]
- Kao, L.R.; Megraw, T.L.; Chae, C.B. SHM1: A Multicopy Suppressor of a Temperature-Sensitive Null Mutation in the HMG1-like Abf2 Gene. Yeast 1996, 12, 1239–1250. [Google Scholar] [CrossRef]
- Palmieri, F.; Agrimi, G.; Blanco, E.; Castegna, A.; Di Noia, M.A.; Iacobazzi, V.; Lasorsa, F.M.; Marobbio, C.M.T.; Palmieri, L.; Scarcia, P.; et al. Identification of Mitochondrial Carriers in Saccharomyces cerevisiae by Transport Assay of Reconstituted Recombinant Proteins. Biochim. Biophys. Acta (BBA)-Bioenerg. 2006, 1757, 1249–1262. [Google Scholar] [CrossRef] [Green Version]
- Fournier, M.L.; Paulson, A.; Pavelka, N.; Mosley, A.L.; Gaudenz, K.; Bradford, W.D.; Glynn, E.; Li, H.; Sardiu, M.E.; Fleharty, B.; et al. Delayed Correlation of mRNA and Protein Expression in Rapamycin-Treated Cells and a Role for Ggc1 in Cellular Sensitivity to Rapamycin. Mol. Cell Proteom. 2010, 9, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Yi, C.; Tong, J.; Lu, P.; Wang, Y.; Zhang, J.; Sun, C.; Yuan, K.; Xue, R.; Zou, B.; Li, N.; et al. Formation of a Snf1-Mec1-Atg1 Module on Mitochondria Governs Energy Deprivation-Induced Autophagy by Regulating Mitochondrial Respiration. Dev. Cell 2017, 41, 59–71.e4. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.M.; Lyver, E.R.; Lesuisse, E.; Dancis, A.; Pain, D. GTP in the Mitochondrial Matrix Plays a Crucial Role in Organellar Iron Homoeostasis1. Biochem. J. 2006, 400, 163–168. [Google Scholar] [CrossRef] [Green Version]
- Lei, G.; Zhuang, L.; Gan, B. mTORC1 and Ferroptosis: Regulatory Mechanisms and Therapeutic Potential. BioEssays 2021, 43, 2100093. [Google Scholar] [CrossRef]
- Marobbio, C.M.T.; Pisano, I.; Porcelli, V.; Lasorsa, F.M.; Palmieri, L. Rapamycin Reduces Oxidative Stress in Frataxin-Deficient Yeast Cells. Mitochondrion 2012, 12, 156–161. [Google Scholar] [CrossRef]
- Santoro, A.; Anjomani Virmouni, S.; Paradies, E.; Villalobos Coa, V.L.; Al-Mahdawi, S.; Khoo, M.; Porcelli, V.; Vozza, A.; Perrone, M.; Denora, N.; et al. Effect of Diazoxide on Friedreich Ataxia Models. Hum. Mol. Genet. 2018, 27, 992–1001. [Google Scholar] [CrossRef] [Green Version]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-Directed Mutagenesis by Overlap Extension Using the Polymerase Chain Reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Kishita, Y.; Pajak, A.; Bolar, N.A.; Marobbio, C.M.T.; Maffezzini, C.; Miniero, D.V.; Monné, M.; Kohda, M.; Stranneheim, H.; Murayama, K.; et al. Intra-Mitochondrial Methylation Deficiency Due to Mutations in SLC25A26. Am. J. Hum. Genet. 2015, 97, 761–768. [Google Scholar] [CrossRef] [Green Version]
- Punzi, G.; Porcelli, V.; Ruggiu, M.; Hossain, M.F.; Menga, A.; Scarcia, P.; Castegna, A.; Gorgoglione, R.; Pierri, C.L.; Laera, L.; et al. SLC25A10 Biallelic Mutations in Intractable Epileptic Encephalopathy with Complex I Deficiency. Hum. Mol. Genet. 2018, 27, 499–504. [Google Scholar] [CrossRef]
- Fiermonte, G.; Parisi, G.; Martinelli, D.; De Leonardis, F.; Torre, G.; Pierri, C.L.; Saccari, A.; Lasorsa, F.M.; Vozza, A.; Palmieri, F.; et al. A New Caucasian Case of Neonatal Intrahepatic Cholestasis Caused by Citrin Deficiency (NICCD): A Clinical, Molecular, and Functional Study. Mol. Genet. Metab. 2011, 104, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Curcio, R.; Muto, L.; Pierri, C.L.; Montalto, A.; Lauria, G.; Onofrio, A.; Fiorillo, M.; Fiermonte, G.; Lunetti, P.; Vozza, A.; et al. New Insights about the Structural Rearrangements Required for Substrate Translocation in the Bovine Mitochondrial Oxoglutarate Carrier. Biochim. Biophys. Acta 2016, 1864, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Lunetti, P.; Gorgoglione, R.; Curcio, R.; Marra, F.; Pignataro, A.; Vozza, A.; Riley, C.L.; Capobianco, L.; Palmieri, L.; Dolce, V.; et al. Drosophila melanogaster Uncoupling Protein-4A (UCP4A) Catalyzes a Unidirectional Transport of Aspartate. Int. J. Mol. Sci. 2022, 23, 1020. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, G.; Lobefaro, N.; Paradies, E.; Vozza, A.; Punzi, G.; Marobbio, C.M.T. Overexpression in E. coli and Purification of the L. pneumophila Lpp2981 Protein. Mol. Biotechnol. 2014, 56, 157–165. [Google Scholar] [CrossRef]
- Torraco, A.; Stehling, O.; Stümpfig, C.; Rösser, R.; De Rasmo, D.; Fiermonte, G.; Verrigni, D.; Rizza, T.; Vozza, A.; Di Nottia, M.; et al. ISCA1 Mutation in a Patient with Infantile-Onset Leukodystrophy Causes Defects in Mitochondrial [4Fe–4S] Proteins. Hum. Mol. Genet. 2018, 27, 2739–2754. [Google Scholar] [CrossRef]
- Curcio, R.; Aiello, D.; Vozza, A.; Muto, L.; Martello, E.; Cappello, A.R.; Capobianco, L.; Fiermonte, G.; Siciliano, C.; Napoli, A.; et al. Cloning, Purification, and Characterization of the Catalytic C-Terminal Domain of the Human 3-Hydroxy-3-Methyl Glutaryl-CoA Reductase: An Effective, Fast, and Easy Method for Testing Hypocholesterolemic Compounds. Mol. Biotechnol. 2020, 62, 119–131. [Google Scholar] [CrossRef]
- Marobbio, C.M.T.; Punzi, G.; Pierri, C.L.; Palmieri, L.; Calvello, R.; Panaro, M.A.; Palmieri, F. Pathogenic Potential of SLC25A15 Mutations Assessed by Transport Assays and Complementation of Saccharomyces cerevisiae ORT1 Null Mutant. Mol. Genet. Metab. 2015, 115, 27–32. [Google Scholar] [CrossRef]
- Vozza, A.; Parisi, G.; De Leonardis, F.; Lasorsa, F.M.; Castegna, A.; Amorese, D.; Marmo, R.; Calcagnile, V.M.; Palmieri, L.; Ricquier, D.; et al. UCP2 Transports C4 Metabolites out of Mitochondria, Regulating Glucose and Glutamine Oxidation. Proc. Natl. Acad. Sci. USA 2014, 111, 960–965. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, V.; Fiermonte, G.; Longo, A.; Palmieri, F. The Human Gene SLC25A29, of Solute Carrier Family 25, Encodes a Mitochondrial Transporter of Basic Amino Acids. J. Biol. Chem. 2014, 289, 13374–13384. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, V.; Vozza, A.; Calcagnile, V.; Gorgoglione, R.; Arrigoni, R.; Fontanesi, F.; Marobbio, C.M.T.; Castegna, A.; Palmieri, F.; Palmieri, L. Molecular Identification and Functional Characterization of a Novel Glutamate Transporter in Yeast and Plant Mitochondria. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1859, 1249–1258. [Google Scholar] [CrossRef]
- Miniero, D.V.; Monné, M.; Di Noia, M.A.; Palmieri, L.; Palmieri, F. Evidence for non-essential salt bridges in the M-gates of mitochondrial carrier proteins. Int. J. Mol. Sci. 2022, 23, 5060. [Google Scholar] [CrossRef]
- Profilo, E.; Peña-Altamira, L.E.; Corricelli, M.; Castegna, A.; Danese, A.; Agrimi, G.; Petralla, S.; Giannuzzi, G.; Porcelli, V.; Sbano, L.; et al. Down-Regulation of the Mitochondrial Aspartate-Glutamate Carrier Isoform 1 AGC1 Inhibits Proliferation and N-Acetylaspartate Synthesis in Neuro2A Cells. Biochim. Biophys. Acta Mol. Basis. Dis. 2017, 1863, 1422–1435. [Google Scholar] [CrossRef]
- Palmieri, L.; Vozza, A.; Hönlinger, A.; Dietmeier, K.; Palmisano, A.; Zara, V.; Palmieri, F. The Mitochondrial Dicarboxylate Carrier Is Essential for the Growth of Saccharomyces Cerevisiae on Ethanol or Acetate as the Sole Carbon Source. Mol. Microbiol. 1999, 31, 569–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vozza, A.; De Leonardis, F.; Paradies, E.; De Grassi, A.; Pierri, C.L.; Parisi, G.; Marobbio, C.M.T.; Lasorsa, F.M.; Muto, L.; Capobianco, L.; et al. Biochemical Characterization of a New Mitochondrial Transporter of Dephosphocoenzyme A in Drosophila melanogaster. Biochim. Biophys. Acta (BBA)-Bioenerg. 2017, 1858, 137–146. [Google Scholar] [CrossRef]
- Ersoy Tunalı, N.; Marobbio, C.M.T.; Tiryakioğlu, N.O.; Punzi, G.; Saygılı, S.K.; Önal, H.; Palmieri, F. A Novel Mutation in the SLC25A15 Gene in a Turkish Patient with HHH Syndrome: Functional Analysis of the Mutant Protein. Mol. Genet. Metab. 2014, 112, 25–29. [Google Scholar] [CrossRef]
- Palmieri, F.; Klingenberg, M. Direct Methods for Measuring Metabolite Transport and Distribution in Mitochondria. In Methods in Enzymology; Biomembranes Part G: Bioenergetics: Biogenesis of Mitochondria, Organization, and Transport; Academic Press: Cambridge, MA, USA, 1979; Volume 56, pp. 279–301. [Google Scholar]
- Raho, S.; Capobianco, L.; Malivindi, R.; Vozza, A.; Piazzolla, C.; De Leonardis, F.; Gorgoglione, R.; Scarcia, P.; Pezzuto, F.; Agrimi, G.; et al. KRAS-Regulated Glutamine Metabolism Requires UCP2-Mediated Aspartate Transport to Support Pancreatic Cancer Growth. Nat. Metab. 2020, 2, 1373–1381. [Google Scholar] [CrossRef]
- Palmieri, F.; Indiveri, C.; Bisaccia, F.; Iacobazzi, V. Mitochondrial Metabolite Carrier Proteins: Purification, Reconstitution, and Transport Studies. In Methods in Enzymology; Mitochondrial Biogenesis and Genetics Part A; Academic Press: Cambridge, MA, USA, 1995; Volume 260, pp. 349–369. [Google Scholar]
- Gorgoglione, R.; Porcelli, V.; Santoro, A.; Daddabbo, L.; Vozza, A.; Monné, M.; Di Noia, M.A.; Palmieri, L.; Fiermonte, G.; Palmieri, F. The Human Uncoupling Proteins 5 and 6 (UCP5/SLC25A14 and UCP6/SLC25A30) Transport Sulfur Oxyanions, Phosphate and Dicarboxylates. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Pierri, C.L.; Palmieri, F.; De Grassi, A. Single-Nucleotide Evolution Quantifies the Importance of Each Site along the Structure of Mitochondrial Carriers. Cell Mol. Life Sci. 2014, 71, 349–364. [Google Scholar] [CrossRef]
- Pierri, C.L.; Parisi, G.; Porcelli, V. Computational Approaches for Protein Function Prediction: A Combined Strategy from Multiple Sequence Alignment to Molecular Docking-Based Virtual Screening. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1695–1712. [Google Scholar] [CrossRef]
- Lunetti, P.; Cappello, A.R.; Marsano, R.M.; Pierri, C.L.; Carrisi, C.; Martello, E.; Caggese, C.; Dolce, V.; Capobianco, L. Mitochondrial Glutamate Carriers from Drosophila melanogaster: Biochemical, Evolutionary and Modeling Studies. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1827, 1245–1255. [Google Scholar] [CrossRef] [Green Version]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G.J.-M.; Brandolin, G. Structure of Mitochondrial ADP/ATP Carrier in Complex with Carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The Mitochondrial Transporter Family (SLC25): Physiological and Pathological Implications. Pflug. Arch.-Eur. J. Physiol. 2004, 447, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Pierri, C.L. Structure and Function of Mitochondrial Carriers–Role of the Transmembrane Helix P and G Residues in the Gating and Transport Mechanism. FEBS Lett. 2010, 584, 1931–1939. [Google Scholar] [CrossRef] [Green Version]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Todisco, S.; Di Noia, M.A.; Onofrio, A.; Parisi, G.; Punzi, G.; Redavid, G.; De Grassi, A.; Pierri, C.L. Identification of New Highly Selective Inhibitors of the Human ADP/ATP Carriers by Molecular Docking and in Vitro Transport Assays. Biochem. Pharmacol. 2016, 100, 112–132. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L. Mitochondrial Metabolite Transport. Essays Biochem. 2010, 47, 37–52. [Google Scholar] [CrossRef]
- Palmieri, F.; Stipani, I.; Quagliariello, E.; Klingenberg, M. Kinetic Study of the Tricarboxylate Carrier in Rat Liver Mitochondria. Eur. J. Biochem. 1972, 26, 587–594. [Google Scholar] [CrossRef]
- Tragni, V.; Cotugno, P.; De Grassi, A.; Massari, F.; Di Ronzo, F.; Aresta, A.M.; Zambonin, C.; Sanzani, S.M.; Ippolito, A.; Pierri, C.L. Targeting Mitochondrial Metabolite Transporters in Penicillium Expansum for Reducing Patulin Production. Plant. Physiol. Biochem. 2021, 158, 158–181. [Google Scholar] [CrossRef]
- Palmieri, F.; Monné, M. Discoveries, Metabolic Roles and Diseases of Mitochondrial Carriers: A Review. Biochim. Biophys. Acta 2016, 1863, 2362–2378. [Google Scholar] [CrossRef]
- Palmieri, F. The Mitochondrial Transporter Family SLC25: Identification, Properties and Physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef]
- Monné, M.; Palmieri, F. Antiporters of the Mitochondrial Carrier Family. Curr. Top. Membr. 2014, 73, 289–320. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, J.J.; Kunji, E.R.S. The SLC25 Mitochondrial Carrier Family: Structure and Mechanism. Trends Biochem. Sci. 2020, 45, 244–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisaccia, F.; De Palma, A.; Palmieri, F. Identification and purification of the tricarboxylate carrier from rat liver mitochondria. Biochim. Biophys. Acta 1989, 977, 171–176. [Google Scholar] [CrossRef]
- Park, D.; Chiu, J.; Perrone, G.G.; Dilda, P.J.; Hogg, P.J. The Tumour Metabolism Inhibitors GSAO and PENAO React with Cysteines 57 and 257 of Mitochondrial Adenine Nucleotide Translocase. Cancer Cell Int. 2012, 12, 11. [Google Scholar] [CrossRef] [Green Version]
- Giangregorio, N.; Palmieri, F.; Indiveri, C. Glutathione controls the redox state of the mitochondrial carnitine/acylcarnitine carrier Cys residues by glutathionylation. Biochim. Biophys. Acta 2013, 1830, 5299–5304. [Google Scholar] [CrossRef]
- Giangregorio, N.; Tonazzi, A.; Console, L.; Lorusso, I.; De Palma, A.; Indiveri, C. The mitochondrial carnitine/acylcarnitine carrier is regulated by hydrogen sulfide via interaction with C136 and C155. Biochim. Biophys. Acta 2016, 1860, 20–27. [Google Scholar] [CrossRef]
- Capobianco, L.; Brandolin, G.; Palmieri, F. Transmembrane topography of the mitochondrial phosphate carrier explored by peptide-specific antibodies and enzymatic digestion. Biochemistry 1991, 30, 4963–4969. [Google Scholar] [CrossRef]
- Palmieri, F.; Bisaccia, F.; Capobianco, L.; Dolce, V.; Fiermonte, G.; Iacobazzi, V.; Zara, V. Transmembrane topology, genes and biogenesis of the mitochondrial phosphate and oxoglutarate carriers. J. Bioenerg. Biomembr. 1993, 25, 493–501. [Google Scholar] [CrossRef]
- Bisaccia, F.; Capobianco, L.; Brandolin, G.; Palmieri, F. Transmembrane topography of the mitochondrial oxoglutarate carrier assessed by peptide-specific antibodies and enzymatic cleavage. Biochemistry 1994, 33, 3705–3713. [Google Scholar] [CrossRef]
- Capobianco, L.; Bisaccia, F.; Michel, A.; Sluse, F.E.; Palmieri, F. The N- and C-termini of the tricarboxylate carrier are exposed to the cytoplasmic side of the inner mitochondrial membrane. FEBS Lett. 1995, 357, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Krämer, R.; Klingenberg, M. Reconstitution of adenine nucleotide transport from beef heart mitochondria. Biochemistry 1979, 18, 4209–4215. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, C.; Dierks, T.; Krämer, R.; Palmieri, F. Reaction mechanism of the reconstituted oxoglutarate carrier from bovine heart mitochondria. Eur. J. Biochem. 1991, 198, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.S.; Major, J.A.; Gremse, D.A.; Wood, D.O. High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 4108–4114. [Google Scholar] [CrossRef] [Green Version]
- Castegna, A.; Scarcia, P.; Agrimi, G.; Palmieri, L.; Rottensteiner, H.; Spera, I.; Germinario, L.; Palmieri, F. Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 17359–17370. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seccia, R.; De Santis, S.; Di Noia, M.A.; Palmieri, F.; Miniero, D.V.; Marmo, R.; Paradies, E.; Santoro, A.; Pierri, C.L.; Palmieri, L.; et al. Citrate Regulates the Saccharomyces cerevisiae Mitochondrial GDP/GTP Carrier (Ggc1p) by Triggering Unidirectional Transport of GTP. J. Fungi 2022, 8, 795. https://doi.org/10.3390/jof8080795
Seccia R, De Santis S, Di Noia MA, Palmieri F, Miniero DV, Marmo R, Paradies E, Santoro A, Pierri CL, Palmieri L, et al. Citrate Regulates the Saccharomyces cerevisiae Mitochondrial GDP/GTP Carrier (Ggc1p) by Triggering Unidirectional Transport of GTP. Journal of Fungi. 2022; 8(8):795. https://doi.org/10.3390/jof8080795
Chicago/Turabian StyleSeccia, Roberta, Silvia De Santis, Maria A. Di Noia, Ferdinando Palmieri, Daniela V. Miniero, Raffaele Marmo, Eleonora Paradies, Antonella Santoro, Ciro L. Pierri, Luigi Palmieri, and et al. 2022. "Citrate Regulates the Saccharomyces cerevisiae Mitochondrial GDP/GTP Carrier (Ggc1p) by Triggering Unidirectional Transport of GTP" Journal of Fungi 8, no. 8: 795. https://doi.org/10.3390/jof8080795
APA StyleSeccia, R., De Santis, S., Di Noia, M. A., Palmieri, F., Miniero, D. V., Marmo, R., Paradies, E., Santoro, A., Pierri, C. L., Palmieri, L., Marobbio, C. M. T., & Vozza, A. (2022). Citrate Regulates the Saccharomyces cerevisiae Mitochondrial GDP/GTP Carrier (Ggc1p) by Triggering Unidirectional Transport of GTP. Journal of Fungi, 8(8), 795. https://doi.org/10.3390/jof8080795







