Comparison of Primers for the Detection of Phytophthora (and Other Oomycetes) from Environmental Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mock Communities and Environmental Samples
| Publication | Gene Region 1 | Primer Set 2 | Study Location and Scale | Number of Samples | Sequencing Platform | % Oom | % Phyt | Species Detected |
|---|---|---|---|---|---|---|---|---|
| Coince et al. [15] | O-ITS | P14 | France: Beech forest | 20 root samples 20 soil samples | 454 | 0.8% | 2 Pythium 2 Phytophthora | |
| Vannini et al. [21] | O-ITS | P3 | Italy: Chestnut forests in the Latium region | 10 soil samples | 454 | 78% | 15 Phytophthora 18 oomycetes | |
| Català et al. [23] | P-ITS | P4 | Spain: Forests and plantations in northern Spain | 24 soil samples 15 water samples | 454 | >99% | 35 | |
| Sapkota and Nicolaisen [26] | O-ITS | P14 | Denmark: Agricultural field and carrots showing symptoms | 26 soil samples 11 carrot samples | 454 | 95% | 2 Phytophthora 65 oomycete | |
| Agler et al. [27] | O-ITS | P15 | Germany: Phyllosphere of wild Arabidopsis thaliana populations | 5 sites, two harvests | Illumina | na | Genus only | |
| Prigigallo et al. [28] | P-ITS | P4 | Italy: Soil and root samples from 8 potted nurseries | 8 pooled samples | 454 | >99% | 25 Phytophthora | |
| Riit et al. [29] | O-ITS | P1 3 | Estonia: Plant nurseries and surrounds | 20 soil samples | Illumina | 25% | Genus only | |
| Burgess et al. [19] Burgess et al. [30] | P-ITS | P4 | Australia: 5 states, soil samples from natural ecosystems | 640 soil samples | 454 | >99% | 68 | |
| Català et al. [17] | P-ITS | P4 | Spain: Two oak forests in eastern Spain | 23 soil samples 10 root samples | 454 | >99% | 13 Phytophthora | |
| Cerri et al. [31] | O-ITS | P14 | Italy: 5 freshwater ecosystems, some with reed dieback | 96 root, rhizosphere and soil samples | 454 | 88% | 523 OTUs 4 | |
| Bose et al. [32] | P-ITS | P4 | South Africa: Four sites from Eucalyptus and Acacia plantations and adjacent forests; soil samples | 120 soil samples | 454 | >99% | 32 Phytophthora | |
| Redondo et al. [20] | P-ITS | P16 | Sweden: 96 sites in 16 rivers over 2 years | 192 water samples (filtered) | PacBio | 74% | 36 Phytophthora | |
| Gómez et al. [33] | O-ITS | P3 | Spain: Declining oak | 52 soil samples | Illumina | 50% | 178 ASVs 5 | |
| Legeay et al. [22] | P-ITS | P11 | Mock Community: 24 species of Phytophthora and other fungi, eukaryotes, and bacteria | Mock communities | Illumina | >99% | 19 Phytophthora | |
| Legeay et al. [22] | P-ITS | P11 | France: Rhizosphere soil | 8 eDNA samples | Illumina | 95% | 7 Phytophthora | |
| Legeay et al. [22] | O-ITS | P17 | Mock Community: 24 species of Phytophthora and other fungi, eukaryotes, and bacteria | Mock communities | Illumina | 100% | 21 Phytophthora | |
| Legeay et al. [22] | O-ITS | P17 | France: Rhizosphere soil | 8 eDNA samples | Illumina | 97% | 1 Phytophthora | |
| Legeay et al. [22] | O-RAS 6 | P18 | Mock Community: 24 species of Phytophthora and other fungi, eukaryotes, and bacteria | Mock communities | Illumina | 100% | 22 Phytophthora | |
| Mora-Sala et al. [34] | P-ITS | P4 | Spain: 6 Quercus ilex stands in 3 regions | 150 soil samples 365 bait leaves | 454 | >99% | 37 Phytophthora | |
| Redekar et al. [35] | O-ITS | P3 | USA: Recycled irrigation water in a nursery across 12 months | 302 water filters | Illumina | 6% | 48 Phytophthora >50 oomycetes | |
| Redekar et al. [35] | O-ITS | P3 | USA: Recycled irrigation water in a nursery across 12 months | 82 bait leaves | Illumina | 55% | 26 Phytophthora 21 oomycetes | |
| Riddell et al. [18] | P-ITS | P4 | Britain: 14 gardens/amenity woodland sites | 140 soil samples | Illumina | >99% | 35 Phytophthora | |
| Sapp et al. [36] | O-cox2 | P13 | Spain: Andalusia, 22 trees in declining oak stands | 66 root samples | Illumina | n/a 7 | n/a 7 | |
| Foster et al. [37] | O-ITS | P14 | USA: Microbiome of roots of three cultivars of Rhododendron grown under different conditions in four nurseries | 120 root balls | Illumina | n/a | 3 Phytophthora 4 Pythium | |
| Green et al. [38] | P-ITS | P4 | Britain: 14 gardens/amenity woodland sites | 140 soil samples | Illumina | >99% | 23 Phytophthora | |
| Khdair et al. [25] | P-ITS | P4 | Australia: Parks and gardens in one city | 236 soil samples | 454 | >99% | 44 Phytophthora | |
| Legeay et al. [39] | P-ITS | P11 | French Guiana: Two sites in rainforest; 10 plots and up to 10 host families at each plot | 93 soil samples 264 bait leaves | Illumina | >99% | 6 Phytophthora | |
| Maciá-Vicente et al. [40] | O-cox2 | P19 | Germany: Naturally co-occurring Brassicaceae | 146 soil and root samples | Illumina | n/a | 951 ASVs | |
| Noel et al. [41] | O-ITS | P3 | USA: Soyabean rhizosphere communities (roots) 4 genotypes, 4 plots, and 6 replicates | 362 rhizosphere samples | Illumina | 20% of ASVs | 86% Pythium 3% Phytophthora | |
| Redekar et al. [42] | O-ITS | P3 | USA: Recycled irrigation water in a nursery across 12 months | 168 water ilters and leaf baits | Illumina | 50% | 32 Phytophthora >50 oomycetes | |
| Riddell et al. [43] | P-ITS | P4 | Britain:Phytophthora in water samples in juniper woodland (rain traps and rivers) over 12 months | 36 pooled water samples (filtered) | Illumina | >99% | 14 Phytophthora | |
| Bose et al. [44] | P-ITS | P4 | South Africa: Four sites from Eucalyptus and Acacia plantations and adjacent forests, root samples | 120 root samples | 454 | >99% | 27 Phytophthora | |
| Fiore-Donno and Bonkowski [45] | O-ITS | P20 | Germany: 3 established biodiversity sites; 50 grassland and 50 forest at each | 300 soil samples | Illumina | 96% | 31% known species | |
| Gyeltshen et al. [46] | P-ITS | P4 | Australia: Topsoil stockpiles (3) and adjacent forest | 42 bulk root samples from 20 plants species | Illumina | >99% | 23 Phytophthora | |
| Khaliq et al. [47] | P-ITS | P4 | Australia: Altitude survey, 3 roads, 20 sites per road, sample at disturbed edge and 50 m into natural vegetation | 120 pooled root samples | Illumina | >99% | 25 Phytophthora | |
| Landa et al. [48] | P-ITS | P4 | Britian: 14 sites—9 disturbed, 5 undisturbed | 132 soil samples | Illumina | 100% | 62 Phytophthora | |
| Landa et al. [48] | O-cox1 | P5 | Britian: 14 sites—9 disturbed, 5 undisturbed | 132 soil samples | Illumina | 71% | 11% | 52 Phytophthora |
| Marčiulynienė et al. [49] | O-ITS | P14 8 | Lithuania: 5 different tree species in 7 bare root forest nurseries | 350 root samples 350 soil samples | PacBio | 1.5% | 2 Phytophthora 33 oomycete | |
| Rossmann et al. [50] | O-ITS | P15 | Norway: Soil from internationally shipped plants | 73 soil samples (before and after enrichment) | Illumina | 72% | 5% | Genus only |
| Green et al. [51] | P-ITS | P4 | Britain: Water and root samples from nurseries | 400 water and root samples | Illumina | na | na | 63 Phytophthora |
2.2. Comparison of Primer Combination
2.3. Bioinformatic Analysis
2.4. Statistical Analysis
3. Results
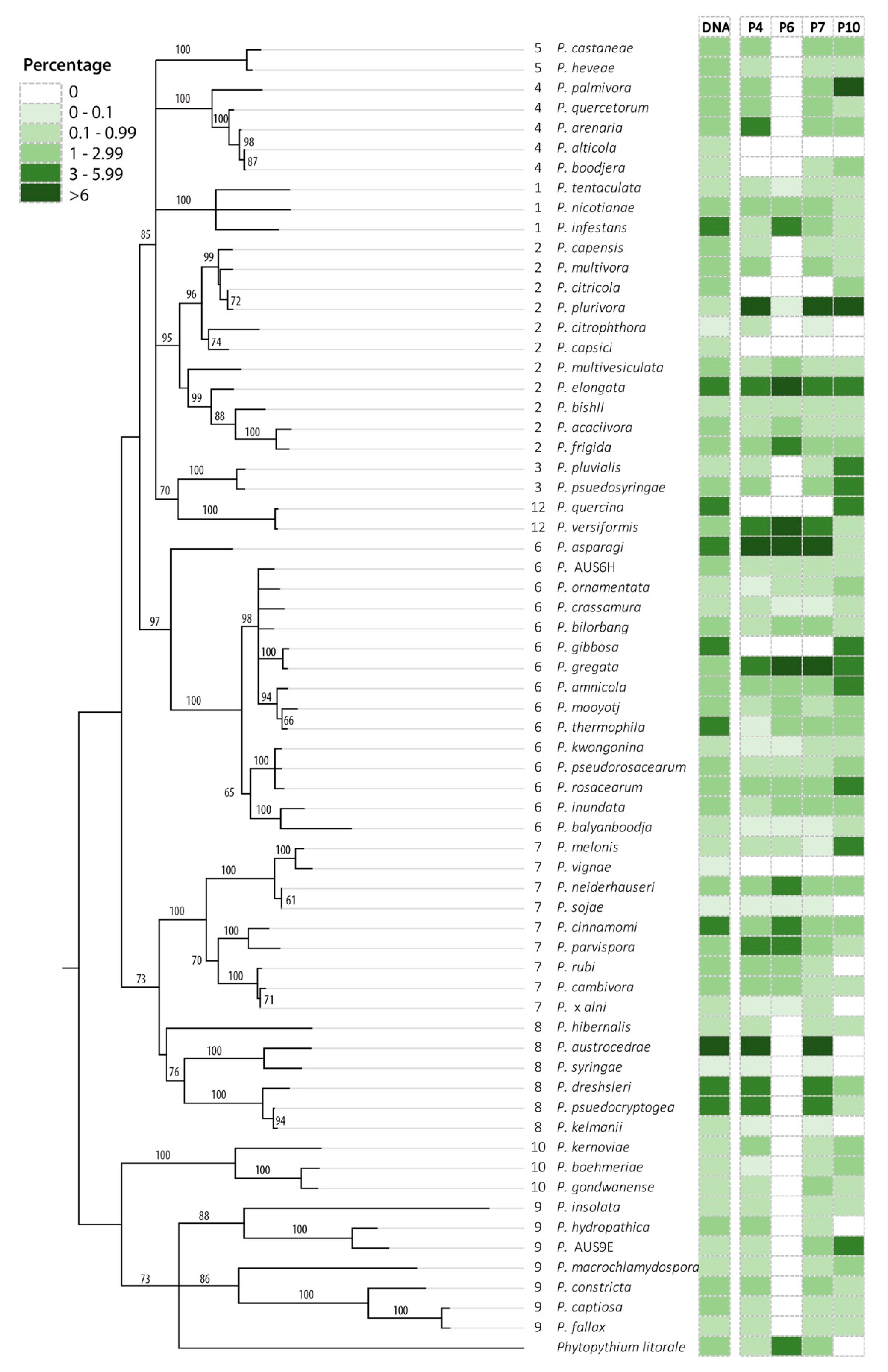
3.1. Phytophthora Detection in Mock Communities
3.2. Phytophthora Detection in eDNA Samples Spiked with the Mock Community
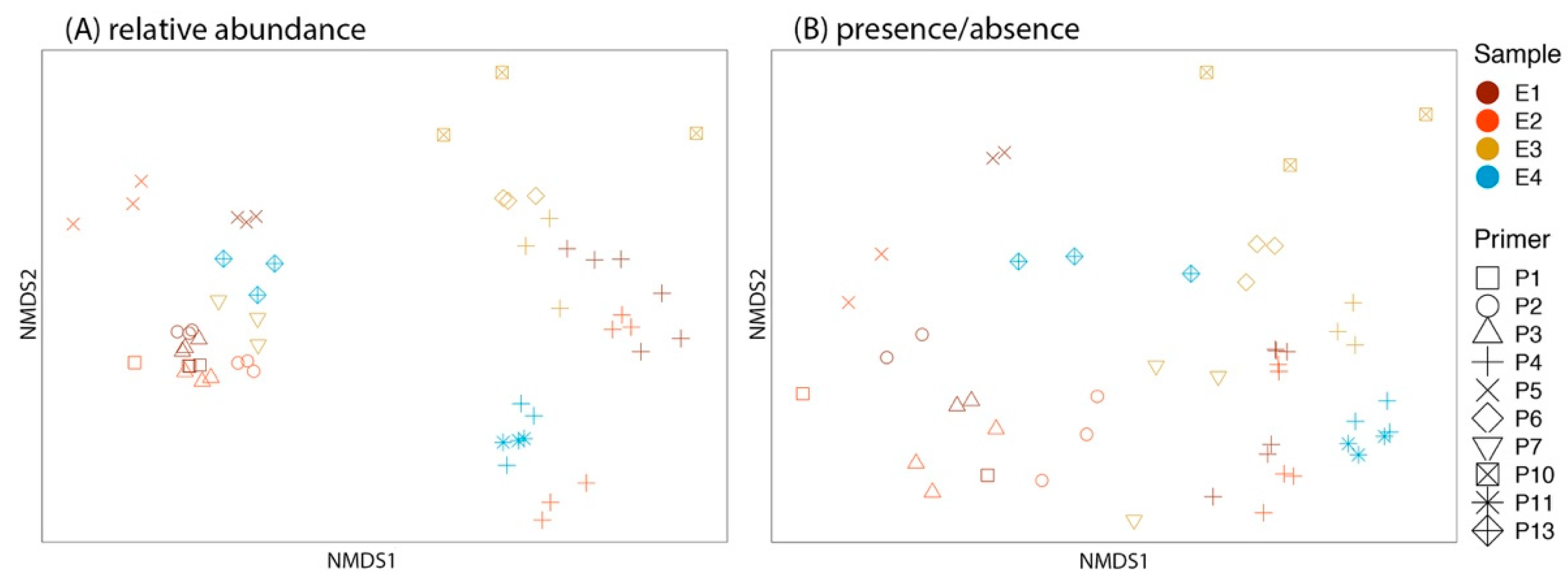
3.3. Phytophthora Detection in eDNA Samples
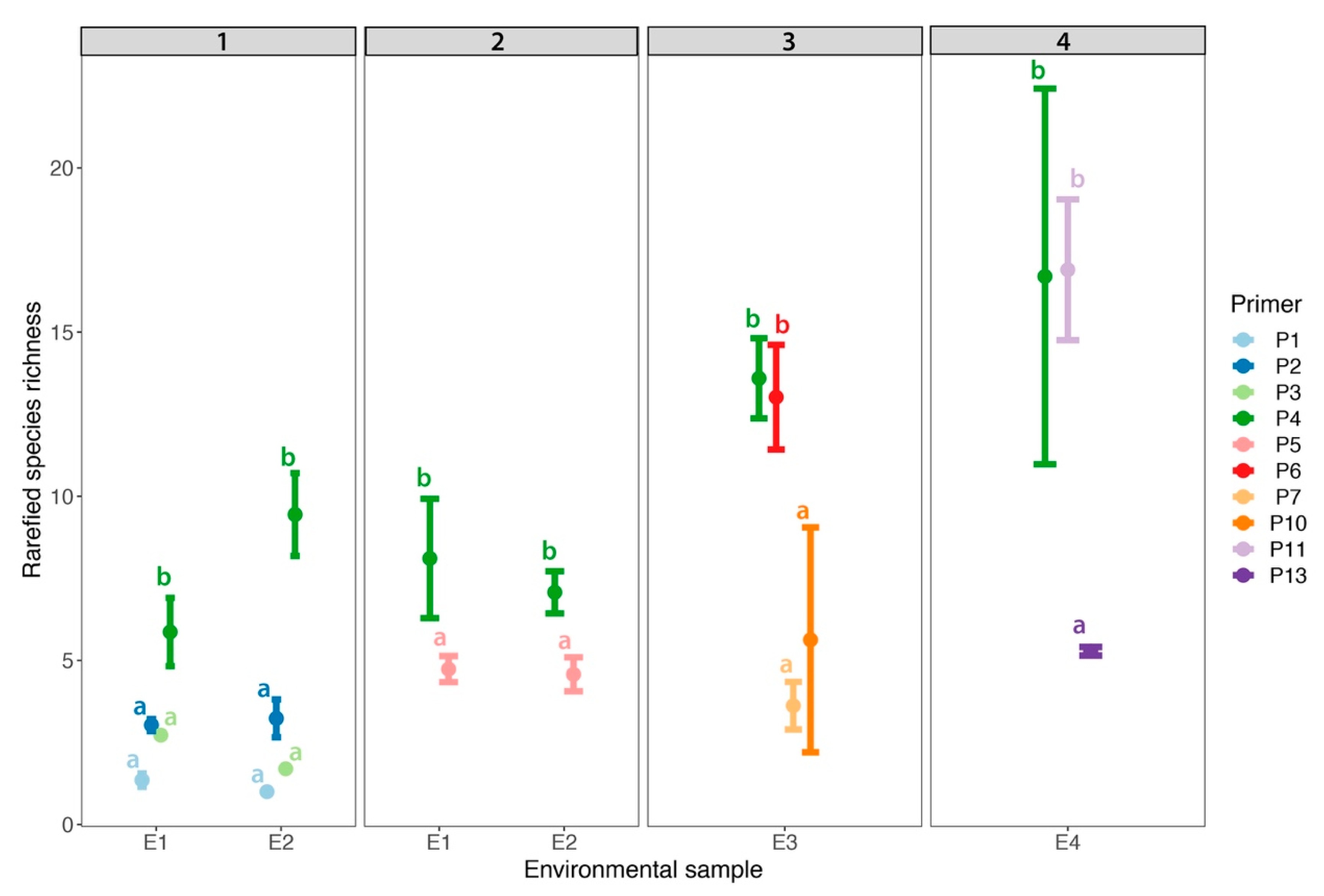
3.4. Technical Replicates
4. Discussion
4.1. Comparison of Primers
4.2. Technical Replicates
4.3. Hybrid Species
5. Conclusions
- Primers designed for oomycetes do not have the same sensitivity toward Phytophthora as the Phytophthora-specific primers. Studies that use oomycete-specific primers to study Phytophthora communities have probably underestimated Phytophthora diversity. The selection of primers is a trade-off between detecting Phytophthora or detecting oomycetes and will depend upon the study’s intent.
- Our results show that using multiple primer sets would reduce taxonomic biases and increase taxonomic coverage.
- While taking technical replicates separately through the process and assigning unique barcodes may be helpful, this could be an expensive option. We recommend conducting the PCR steps in triplicate and then combining them before adding barcodes.
- Use a phylogenetic approach to assign OTUs or ASVs to species rather than simple blast searches. By doing so, minor sequencing errors that do not influence phylogenetic placement will allow several OTUs to be assigned to the same species.
- Internal controls were not included in the current study but would be a valuable addition to any protocol. Green et al. [38] included four samples containing a mix of synthetic ‘Phytophthora’ sequences of known base composition on the plate as a check for sequence contamination. These can be synthetic reference sequences included in the initial PCR reactions as control samples to determine any cross-contamination during the amplification stage.
- Many Phytophthora species can hybridise, especially those commonly found in water. Care must be taken with metabarcoding studies as it is not possible to identify hybrids.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; APS Press: St. Paul, MN, USA, 1996; p. 562. [Google Scholar]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef]
- Hansen, E.M.; Reeser, P.W.; Sutton, W. Phytophthora beyond agriculture. Annu. Rev. Phytopathol. 2012, 50, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Cahill, D.M.; Rookes, J.E.; Wilson, B.A.; Gibson, L.; McDougall, K.L. Turner Review No. 17. Phytophthora cinnamomi and Australia’s biodiversity: Impacts, predictions and progress towards control. Aust. J. Bot. 2008, 56, 279–310. [Google Scholar] [CrossRef]
- Scott, P.; Bader, M.; Burgess, T.I.; Hardy, G.E.S.J.; Williams, N. Global biogeography and invasion risk of the plant destroyer genus Phytophthora. Environ. Sci. Policy 2019, 101, 175–182. [Google Scholar] [CrossRef]
- Brasier, C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008, 57, 792–808. [Google Scholar] [CrossRef]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguín Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavarriaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- Laliberté, E.; Lambers, H.; Burgess, T.I.; Wright, S.J. Tansley Review; Phosphorus limitation, soil-borne pathogens and the coexistence of plant species in hyperdiverse forests and shrublands. New Phytol. 2015, 206, 507–521. [Google Scholar] [CrossRef]
- Burgess, T.I.; López-Villamor, A.; Paap, T.; Williams, B.; Belhaj, R.; Crone, M.; Dunstan, W.A.; Howard, K.; Hardy, G.E.S. Towards a best practice methodology for the detection of Phytophthora species in soils. Plant Pathol. 2021, 70, 604–614. [Google Scholar] [CrossRef]
- Sarker, S.R.; McComb, J.A.; Burgess, T.I.; Hardy, G.E.S. Antimicrobials in Phytophthora isolation media and the growth of Phytophthora species. Plant Pathol. 2020, 69, 1426–1436. [Google Scholar] [CrossRef]
- Sarker, S.R.; McComb, J.; Burgess, T.I.; Hardy, G.E.S. Timing and abundance of sporangia production and zoospore release influences the recovery of different Phytophthora species by baiting. Fungal Biol. 2021, 125, 477–484. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Alberdi, A.; Aizpurua, O.; Gilbert, M.T.P.; Bohmann, K. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 2018, 9, 134–147. [Google Scholar] [CrossRef]
- Coince, A.; Caël, O.; Bach, C.; Lengellé, J.; Cruaud, C.; Gavory, F.; Morin, E.; Murat, C.; Marçais, B.; Buée, M. Below-ground fine-scale distribution and soil versus fine root detection of fungal and soil oomycete communities in a French beech forest. Fungal Ecol. 2013, 6, 223–235. [Google Scholar] [CrossRef]
- Scibetta, S.; Schena, L.; Chimento, A.; Cacciola, S.O.; Cooke, D.E.L. A molecular method to assess Phytophthora diversity in environmental samples. J. Microbiol. Methods 2012, 88, 356–368. [Google Scholar] [CrossRef]
- Català, S.; Berbegal, M.; Pérez-Sierra, A.; Abad-Campos, P. Metabarcoding and development of new real-time specific assays reveal Phytophthora species diversity in holm oak forests in eastern Spain. Plant Pathol. 2017, 66, 115–123. [Google Scholar] [CrossRef]
- Riddell, C.E.; Frederickson-Matika, D.; Armstrong, A.C.; Elliot, M.; Forster, J.; Hedley, P.E.; Morris, J.; Thorpe, P.; Cooke, D.E.L.; Pritchard, L.; et al. Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. PeerJ 2019, 7, e6931. [Google Scholar] [CrossRef]
- Burgess, T.I.; White, D.; McDougall, K.L.; Garnas, J.; Dunstan, W.A.; Català, S.; Carnegie, A.J.; Worboys, S.; Cahill, D.; Vettraino, A.-M.; et al. Distribution and diversity of Phytophthora across Australia. Pac. Conserv. Biol. 2017, 23, 150–162. [Google Scholar] [CrossRef]
- Redondo, M.A.; Boberg, J.; Stenlid, J.; Oliva, J. Functional traits associated with the establishment of introduced Phytophthora spp. in Swedish forests. J. Appl. Ecol. 2018, 55, 1538–1552. [Google Scholar] [CrossRef]
- Vannini, A.; Bruni, N.; Tomassini, A.; Franceschini, S.; Vettraino, A.M. Pyrosequencing of environmental soil samples reveals biodiversity of the Phytophthora resident community in chestnut forests. FEMS Microbiol. Ecol. 2013, 85, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Legeay, J.; Husson, C.; Cordier, T.; Vacher, C.; Marcais, B.; Buée, M. Comparison and validation of Oomycetes metabarcoding primers for Phytophthora high throughput sequencing. J. Plant Pathol. 2019, 101, 743–748. [Google Scholar] [CrossRef]
- Català, S.; Pérez-Sierra, A.; Abad-Campos, P. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS ONE 2015, 10, e0119311. [Google Scholar] [CrossRef] [PubMed]
- Abad, Z.G.; Burgess, T.I.; Redford, A.J.; Bienapfl, J.C.; Srivastava, S.; Mathew, R.; Jennings, K. IDphy: An International online resource for molecular and morphological identification of Phytophthora based on type specimens. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Khdair, M.Y.; Barber, P.A.; Hardy, G.E.S.J.; Shaw, C.; Steel, E.J.; McMains, C.; Burgess, T.I. Association of Phytophthora with declining vegetation in an urban forest environment. Microorganisms 2020, 8, 973. [Google Scholar] [CrossRef]
- Sapkota, R.; Nicolaisen, M. An improved high throughput sequencing method for studying oomycete communities. J. Microbiol. Methods 2015, 110, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Prigigallo, M.I.; Abdelfattah, A.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Cooke, D.E.; Schena, L. Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 2016, 106, 305–313. [Google Scholar] [CrossRef]
- Riit, T.; Tedersoo, L.; Drenkhan, R.; Runno-Paurson, E.; Kokko, H.; Anslan, S. Oomycete-specific ITS primers for identification and metabarcoding. MycoKeys 2016, 14, 17. [Google Scholar] [CrossRef]
- Burgess, T.I.; McDougall, K.L.; Scott, P.; Hardy, G.E.S.J.; Garnas, J. Predictors of Phytophthora diversity and distribution in natural areas across diverse Australian ecoregions. Ecography 2019, 42, 594–607. [Google Scholar] [CrossRef]
- Cerri, M.; Sapkota, R.; Coppi, A.; Ferri, V.; Foggi, B.; Gigante, D.; Lastrucci, L.; Selvaggi, R.; Venanzoni, R.; Nicolaisen, M.; et al. Oomycete communities associated with reed die-back syndrome. Front. Plant Sci. 2017, 8, 1550. [Google Scholar] [CrossRef]
- Bose, T.; Wingfield, M.J.; Roux, J.; Vivas, M.; Burgess, T.I. Community composition and distribution of Phytophthora species across adjacent native and non-native forests of South Africa. Fungal Ecol. 2018, 36, 17–25. [Google Scholar] [CrossRef]
- Gómez, F.J.R.; Navarro-Cerrillo, R.M.; Pérez-de-Luque, A.; Oβwald, W.; Vannini, A.; Morales-Rodríguez, C. Assessment of functional and structural changes of soil fungal and oomycete communities in holm oak declined dehesas through metabarcoding analysis. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mora-Sala, B.; Gramaje, D.; Abad-Campos, P.; Berbegal, M. Diversity of Phytophthora species associated with Quercus ilex L. in three spanish regions evaluated by NGS. Forests 2019, 10, 979. [Google Scholar] [CrossRef]
- Redekar, N.R.; Eberhart, J.L.; Parke, J.L. Diversity of Phytophthora, Pythium, and Phytopythium species in recycled irrigation water in a container nursery. Phytobiomes J. 2019, 3, 31–45. [Google Scholar] [CrossRef]
- Sapp, M.; Tyborski, N.; Linstädter, A.; López Sánchez, A.; Mansfeldt, T.; Waldhoff, G.; Bareth, G.; Bonkowski, M.; Rose, L.E. Site-specific distribution of oak rhizosphere-associated oomycetes revealed by cytochrome c oxidase subunit II metabarcoding. Ecol. Evol. 2019, 9, 10567–10581. [Google Scholar] [CrossRef]
- Foster, Z.S.L.; Weiland, J.E.; Scagel, C.F.; Grünwald, N.J. The composition of the fungal and oomycete microbiome of Rhododendron roots under varying growth conditions, nurseries, and cultivars. Phytobiomes J. 2020, 4, 156–164. [Google Scholar] [CrossRef]
- Green, S.; Riddell, C.E.; Frederickson-Matika, D.; Armstrong, A.; Elliot, M.; Forster, J.; Hedley, P.E.; Morris, J.; Thorpe, P.; Cooke, D.E.; et al. Diversity of woody-host infecting Phytophthora species in public parks and botanic gardens as revealed by metabarcoding, and opportunities for mitigation through best practice. Sibbaldia 2020, 18, 67–88. [Google Scholar] [CrossRef]
- Legeay, J.; Husson, C.; Boudier, B.; Louisanna, E.; Baraloto, C.; Schimann, H.; Marcais, B.; Buée, M. Surprising low diversity of the plant pathogen Phytophthora in Amazonian forests. Environ. Microbiol. 2020, 22, 5019–5032. [Google Scholar] [CrossRef]
- Maciá-Vicente, J.G.; Nam, B.; Thines, M. Root filtering, rather than host identity or age, determines the composition of root-associated fungi and oomycetes in three naturally co-occurring Brassicaceae. Soil Biol. Biochem. 2020, 146, 107806. [Google Scholar] [CrossRef]
- Noel, Z.A.; Chang, H.X.; Chilvers, M.I. Variation in soybean rhizosphere oomycete communities from Michigan fields with contrasting disease pressures. Appl. Soil Ecol. 2020, 150, 103435. [Google Scholar] [CrossRef]
- Redekar, N.R.; Bourret, T.B.; Eberhart, J.L.; Johnson, G.E.; Pitton, B.J.L.; Haver, D.L.; Oki, L.R.; Parke, J.L. The population of oomycetes in a recycled irrigation water system at a horticultural nursery in southern California. Water Res. 2020, 183, 116050. [Google Scholar] [CrossRef] [PubMed]
- Riddell, C.E.; Dun, H.F.; Elliot, M.; Armstrong, A.C.; Clark, M.; Forster, J.; Hedley, P.E.; Green, S. Detection and spread of Phytophthora austrocedri within infected Juniperus communis woodland and diversity of co-associated Phytophthoras as revealed by metabarcoding. For. Pathol. 2020, e12602. [Google Scholar] [CrossRef]
- Bose, T.; Wingfield, M.J.; Roux, J.; Vivas, M.; Burgess, T.I. Phytophthora species associated with roots of native and non-native trees in natural and managed forests. Microb. Ecol. 2021, 81, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Fiore-Donno, A.M.; Bonkowski, M. Different community compositions between obligate and facultative oomycete plant parasites in a landscape-scale metabarcoding survey. Biol. Fertil. Soils 2021, 57, 245–256. [Google Scholar] [CrossRef]
- Gyeltshen, J.; Dunstan, W.A.; Shaw, C.; Howard, K.; Grigg, A.H.; Hardy, G.E.S.J.; Burgess, T.I. Metabarcoding shows multiple Phytophthora species associated with individual plant species: Implications for restoration. Eur. J. Plant Pathol. 2021, 159, 359–369. [Google Scholar] [CrossRef]
- Khaliq, I.; Burgess, T.I.; Hardy, G.E.S.J.; White, D.; McDougall, K.L. Phytophthora and vascular plant species distributions along a steep elevation gradient. Biol. Invasions 2021, 23, 1443–1459. [Google Scholar] [CrossRef]
- Landa, B.B.; Arias-Giraldo, L.F.; Henricot, B.; Montes-Borrego, M.; Shuttleworth, L.A.; Pérez-Sierra, A. Diversity of Phytophthora species detected in disturbed and undisturbed british soils using high-throughput sequencing targeting ITS rRNA and COI mtDNA r egions. Forests 2021, 12, 229. [Google Scholar] [CrossRef]
- Marčiulynienė, D.; Marčiulynas, A.; Lynikienė, J.; Vaičiukynė, M.; Gedminas, A.; Menkis, A. DNA-Metabarcoding of belowground fungal communities in bare-root forest nurseries: Focus on different tree species. Microorganisms 2021, 9, 150. [Google Scholar] [CrossRef]
- Rossmann, S.; Lysøe, E.; Skogen, M.; Talgø, V.; Brurberg, M.B. DNA metabarcoding reveals broad presence of plant pathogenic oomycetes in soil from internationally traded plants. Front. Microbiol. 2021, 12, e637068. [Google Scholar] [CrossRef]
- Green, S.; Cooke, D.E.L.; Dunn, M.; Barwell, L.; Purse, B.V.; Chapman, D.S.; Valatin, G.; Schlenzig, A.; Barbrook, J.; Pettitt, T.; et al. PHYTO-THREATS: Addressing threats to UK forests and woodlands from Phytophthora; identifying risks of spread in trade and methods for mitigation. Forests 2021, 12, 1617. [Google Scholar] [CrossRef]
- Riit, T.; Tedersoo, L.; Drenkhan, R.; Runno-Paurson, E.; Kokko, H.; Anslan, S. Corrigendum for: “Oomycete-specific ITS primers for identification and metabarcoding” published in MycoKeys. MycoKeys 2018, 41, 119. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophthora and related Oomycetes. Fungal Genet. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Beakes, G.W.; Glockling, S.; Kruse, J.; Nam, B.; Nigrelli, L.; Ploch, S.; Shin, H.D.; Shivas, R.G.; Telle, S. Towards a universal barcode of oomycetes–a comparison of the cox1 and cox2 loci. Mol. Ecol. Resour. 2015, 15, 1275–1288. [Google Scholar] [CrossRef]
- Znajda, N.R.; Grooters, A.M.; Marsella, R. PCR-based detection of Pythium and Lagenidium DNA in frozen and ethanol-fixed animal tissues. Vet. Dermatol. 2002, 13, 187–194. [Google Scholar] [CrossRef]
- Lievens, B.; Hanssen, I.R.M.; Vanachter, A.C.R.C.; Cammue, B.P.A.; Thomma, B.P.H.J. Root and foot rot on tomato caused by Phytophthora infestans detected in Belgium. Plant Dis. 2004, 88, 86. [Google Scholar] [CrossRef]
- Foster, Z.S.L.; Albornoz, F.E.; Fieland, V.J.; Larsen, M.M.; Jones, F.A.; Tyler, B.M.; Nguyen, H.D.T.; Burgess, T.I.; Riddell, C.; Voglmayr, H.; et al. A new oomycete metabarcoding method using the rps10 gene. Phytobiomes 2022. [Google Scholar] [CrossRef]
- Martin, F.N.; Blair, J.E.; Coffey, M.D. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genet. Biol. 2014, 66, 19–32. [Google Scholar] [CrossRef]
- Hudspeth, D.S.S.; Nadler, S.A.; Hudspeth, M.E.S. A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia 2000, 92, 674–684. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innes, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Thines, M.; Zipper, R.; Schäuffele, D.; Spring, O. Characteristics of Pustula tragopogonis (syn. Albugo tragopogonis) newly occurring on cultivated sunflower in Germany. J. Phytopathol. 2006, 154, 88–92. [Google Scholar] [CrossRef]
- Drenth, A.; Wagels, G.; Smith, B.; Sendall, B.; O’Dwyer, C.; Irvine, G.; Irwin, J. Development of a DNA-based method for detection and identification of Phytophthora species. Australas. Plant Pathol. 2006, 35, 147–159. [Google Scholar] [CrossRef]
- Weir, B.S.; Paderes, E.P.; Anand, N.; Uchida, J.Y.; Pennycook, S.R.; Bellgard, S.E.; Beever, R.E. A taxonomic revision of Phytophthora Clade 5 including two new species, Phytophthora agathidicida and P. cocois. Phytotaxa 2015, 205, 21–38. [Google Scholar] [CrossRef]
- Khaliq, I.; Hardy, G.E.S.J.; White, D.; Burgess, T.I. eDNA from roots: A robust tool for determining Phytophthora communities in natural ecosystems. FEMS Microbiol. Ecol. 2018, 94, fiy048. [Google Scholar] [CrossRef]
- Edgar, R. Usearch; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2010.
- Zuur, A.F.; Leno, E.N. A protocol for conducting and presenting results of regression-type analyses. Methods Ecol. Evol. 2016, 7, 636–645. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. Version 2.5-6. 2016. Available online: http://CRAN.R-project.org/package=vegan (accessed on 1 July 2022).
- Lofgren, L.A.; Uehling, J.K.; Branco, S.; Bruns, T.D.; Martin, F.; Kennedy, P.G. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019, 28, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Dickie, I.A.; Wakelin, A.M.; Martínez-García, L.B.; Richardson, S.J.; Makiola, A.; Tylianakis, J.M. Oomycetes along a 120,000 year temperate rainforest ecosystem development chronosequence. Fungal Ecol. 2019, 39, 192–200. [Google Scholar] [CrossRef]
- Martin, F.N.; Albornoz, F.; Foster, Z.; Fieland, V.; Larson, M.; Jones, F.A.; Tyler, B.M.; Nguyen, H.; Voglmayr, H.; Burgess, T.I.; et al. The rps10 gene as a new barcode locus for oomycetes and its utility in metagenomics studies of environmental samples. Phytopathology 2019, 109, 101–102. [Google Scholar]
- Murray, D.C.; Coghlan, M.L.; Bunce, M. From benchtop to desktop: Important considerations when designing amplicon sequencing workflows. PLoS ONE 2015, 10, e0124671. [Google Scholar]
- Husson, C.; Aguayo, J.; Revellin, C.; Frey, P.; Ioos, R.; Marçais, B. Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genet. Biol. 2015, 77, 12–21. [Google Scholar] [CrossRef]
- Ioos, R.; Andrieux, A.; Marçais, B.; Frey, P. Genetic characterization of the natural hybrid species Phytophthora alni as inferred from nuclear and mitochondrial DNA analyses. Fungal Genet. Biol. 2006, 43, 511–529. [Google Scholar] [CrossRef]
- Reeser, P.W.; Sutton, W.; Hansen, E.M.; Remigi, P.; Adams, G.C. Phytophthora species in forest streams in Oregon and Alaska. Mycologia 2011, 103, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Hüberli, D.; Hardy, G.E.S.; White, D.; Williams, N.; Burgess, T.I. Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australas. Plant Pathol. 2013, 42, 251–260. [Google Scholar] [CrossRef]
- Nagel, J.H.; Gryzenhout, M.; Slippers, B.; Wingfield, M.J.; Hardy, G.E.S.; Stukely, M.J.C.; Burgess, T.I. Characterization of Phytophthora hybrids from ITS clade 6 associated with riparian ecosystems in South Africa and Australia. Fungal Biol. 2013, 117, 329–347. [Google Scholar] [CrossRef] [PubMed]
- Burgess, T.I. Molecular characterization of natural hybrids formed between five related indigenous clade 6 Phytophthora species. PLoS ONE 2015, 10, e0134225. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Richardson, P.A.; Hong, C. Phytophthora × stagnum nothosp. nov., a new hybrid from irrigation reservoirs at ornamental plant nurseries in Virginia. PLoS ONE 2014, 9, e103450. [Google Scholar] [CrossRef]
- Safaiefarahani, B.; Mostowfizadeh-Ghalamfarsa, R.; Hardy, G.E.S.; Burgess, T.I. Species from within the Phytophthora cryptogea complex and related species, P. erythroseptica and P. sansomeana, readily hybridize. Fungal Biol. 2016, 120, 975–987. [Google Scholar] [CrossRef]
- Man in’t Veld, W.A.; Rosendahl, K.C.H.M.; Hong, C. Phytophthora × serendipita sp. nov. and P. × pelgrandis, two destructive pathogens generated by natural hybridization. Mycologia 2012, 104, 1390–1396. [Google Scholar] [CrossRef] [Green Version]

| Code | D | Primer | Sequence | Size | AT | Run1 | Run2 | Run3 | Run4 | Reference for Primer | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 1 (O) | PCR1 | F | ITS1oo | #F-GGA AGG ATC ATT ACC ACA | 900 | 60 | Y | Riit et al. [29] | |||
| R | ITS4ngs | #R-GTC CTS CGC TTA TTG ATA TGC | Tedersoo et al. [53] | ||||||||
| P2 (P) | PCR1 | F | ITS1oo | #F-GGA AGG ATC ATT ACC ACA | 350 | 60 | Y | Riit et al. [29] | |||
| R | ITS7 | #R-GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| P3 (O) | PCR1 | F | ITS6 | #F-GAA GGT GAA GTC GTA ACA AGG | 500 | 60 | Y | Cooke et al. [54] | |||
| R | ITS7 | #R-GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| P4 (P) | PRC1 | F | 18Ph2F | GGA TAG ACT GTT GCA ATT TTC AGT | 400 | 60 | Y | Y | Y | Y | Scibetta et al. [16] |
| R | 5.8S1R | GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| PCR2 | F | ITS6 | #F-GAA GGT GAA GTC GTA ACA AGG | 350 | 60 | Cooke et al. [54] | |||||
| R | 5.8S1R | #R-GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| P5 1 (O) | PCR1 | F | COX1levup-F | TCA WCW MGA TGG CTT TTT TCA AC | nd | 52 | Y | Choi et al. [55] | |||
| R | COX1levlo-R | CYT CHG GRT GWC CRA AAA ACC AAA | Choi et al. [55] | ||||||||
| PCR2 | F | Hvshort-F | #F-GNA TGA AYA AYA THA GYT TYT GG | 500 | 52 | Landa et al. [48] | |||||
| R | COX1levlo-R | #R-CYT CHG GRT GWC CRA AAA ACC AAA | Choi et al. [55] | ||||||||
| P6 (O) | PCR1 | F | ITS6 | GAA GGT GAA GTC GTA ACA AGG | nd | 60 | Y | Cooke et al. [54] | |||
| R | ITS2P | GCA GCG TTC TTC ATC GAT GT | Znajda et al. [56] | ||||||||
| PCR2 | F | OOMUP18Sc | #F-TGC GGA AGG ATC ATT ACC ACA C | 350 | 60 | Lievens et al. [57] | |||||
| R | 5.8S1R | #R-GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| P7 (O) | PCR1 | F | OOMUP18Sc | TGC GGA AGG ATC ATT ACC ACA C | 400 | 60 | Y | Lievens et al. [57] | |||
| R | ITS2P | GCA GCG TTC TTC ATC GAT GT | Znajda et al. [56] | ||||||||
| P8 2 (O) | PCR1 | F | ITS6 | GAA GGT GAA GTC GTA ACA AGG | na | 60 | Y | Cooke et al. [54] | |||
| R | 5.8S1R | GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| PCR2 | F | OOMUP18Sc | #F-TGC GGA AGG ATC ATT ACC ACA C | na | 60 | Lievens et al. [57] | |||||
| R | 5.8S1R | #R-GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| P9 2 (O) | PCR1 | F | rps10-F | #F-GTT GGT TAG AGY ARA AGA CT | 550 | 59 | Y | Foster et al. [58] | |||
| R | rps10-R | #R-ATR YYT AGA AAG AYT YGA ACT | Foster et al. [58] | ||||||||
| P10 (O) | PCR1 | F | PRV9-F | GTT GGT TAG AGT AAA AGA CT | na | 59 | Y | Martin et al. [59] | |||
| R | PRV9-R | GTA TAC TCT AAC CAA CTG AGT | Martin et al. [59] | ||||||||
| PCR2 | F | rps10-F | #F-GTT GGT TAG AGY ARA AGA CT | 550 | 59 | Foster et al. [58] | |||||
| R | rps10-R | #R-ATR YYT AGA AAG AYT YGA ACT | Foster et al. [58] | ||||||||
| P11 (P) | PCR1 | F | Oom18S | GCG CAT CGT GCT AGG GAT AG | nd | Y | Legeay et al. [22] | ||||
| R | ITS7 | GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| PCR2 | F | 18Ph2F | #F-GAA GGT GAA GTC GTA ACA AGG | 400 | Scibetta et al. [16] | ||||||
| R | 5.8S1R | #R-GCA RRG ACT TTC GTC CCY RC | Scibetta et al. [16] | ||||||||
| P12 3 (O) | PCR1 | F | ITS1oo(c) | #F-GGA AGG ATC ATT ACC ACAC | Y | Riit et al. [52] | |||||
| R | ITS7 | #R-GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| P13 (O) | PCR1 | F | Cox2hud-F | #F-GGC AAA TGG GTT TTC AAG ATC C | Y | Hudspeth et al. [60] | |||||
| R | Cox233D8r | #R-GAA TAT TCA TAR STC CAR TAC C | Sapp et al. [36] | ||||||||
| P14 (O) | PCR1 | F | ITS6 | GAA GGT GAA GTC GTA ACA AGG | Cooke et al. [54] | ||||||
| R | ITS4 | TCC TCC GCT TAT TGA TAT GC | White et al. [61] | ||||||||
| PCR2 | F | ITS6 | #F-GAA GGT GAA GTC GTA ACA AGG | Cooke et al. [54] | |||||||
| R | ITS7 | #R-GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| P15 (O) | PCR1 | F | ITS1O | #F-CGGAAGGATCATTACCAC | Thines et al. [62] | ||||||
| R | 5.8S-O-Rev | #R-AGCCTAGACATCCACTGCTG | Agler et al. [27] | ||||||||
| P16 4 (P) | PCR1 | F | A2 | ACT TTC CAC GTG AAC CGT TTC AA | Drenth et al. [63] | ||||||
| R | I2 | GAT ATC AGG TCC AAT TGA GAT GC | Drenth et al. [63] | ||||||||
| P17 (O) | PCR1 | F | DC6 | GAG GGA CTT TTG GGT AAT CA | Cooke et al. [54] | ||||||
| R | ITS7 | GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| PCR2 | F | Oom18S | #F-GCG CAT CGT GCT AGG GAT AG | Legeay et al. [22] | |||||||
| R | ITS7 | #R-GAG CGT TCT TCA TCG ATG TGC | Cooke et al. [54] | ||||||||
| P18 (O) | PCR1 | F | Yph1F | #F-CGA CCA TKG GTG TGG ACT TT | Weir et al. [64] | ||||||
| R | Yph2R | #R-ACG TTC TCM CAG GCG TAT CT | Weir et al. [64] | ||||||||
| P19 (O) | PCR1 | F | Cox2hud-F | #F-GGC AAA TGG GTT TTC AAG ATC C | Hudspeth et al. [60] | ||||||
| R | Cox2-RC4 | #R-TGA TTW AYN CCA CAA ATT TCR CTA CAT TG | Choi et al. [55] | ||||||||
| P20 (O) | PCR1 | F | S1777F | GGT GAA CCT GCG GAA GGA | Fiore-Donno and Bonkowski [45] | ||||||
| R | 5.8SOomR | TCT TCA TCG DTG TGC GAG C | Fiore-Donno and Bonkowski [45] | ||||||||
| PCR2 | F | S1786StraF | #F-GCG GAA GGA TCA TTA CCA C | Fiore-Donno and Bonkowski [45] | |||||||
| R | 5.8SOomR | #R-TCT TCA TCG DTG TGC GAG C | Fiore-Donno and Bonkowski [45] | ||||||||
| P21 (O) | PCR1 | F | ITS3oo | #F-AGT ATG YYT GTA TCA GTG TC | Riit et al. [52] | ||||||
| R | ITS4 | #R-TCC TCC GCT TAT TGA TAT GC | White et al. [61] |
| Metabarcoding Run | Run1 | Run1 | Run1 | Run1 | Run2 | Run2 | Run3 | Run3 | Run3 | Run3 | Run4 | Run4 | Run4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primer Combination | P1 | P2 | P3 | P4 | P4 | P5 | P4 | P6 | P7 | P10 | P4 | P11 | P13 |
| Gene Region Amplified | ITS | ITS | ITS | ITS | ITS | COX1 | ITS | ITS | ITS | RPS | ITS | ITS | COX2 |
| Sample | MIX1 | MIX1 | MIX1 | MIX1 | MIX1 | MIX1 | MIX2 | MIX2 | MIX2 | MIX2 | MIX3 | MIX3 | MIX3 |
| Average number of reads | 6 684 | 11,196 | 23,109 | 37,755 | 14,511 | 4105 | 18,942 | 8756 | 24,607 | 10,045 | 13,003 | 13,594 | 13,179 |
| Phytophthoraspecies detected | 37 | 46 | 46 | 46 | 45 | 25 | 59 | 33 | 60 | 55 | 47 | 34 | 47 |
| Species missed from MIX | 13 | 4 | 4 | 4 | 5 | 25 | 7 | 33 | 6 | 11 | 14 | 25 | 14 |
| Relationship between reads and DNA concentration | 0.613 | 0.665 | 0.613 | 0.75 | 0.732 | 0.095 | 0.650 | 0.261 | 0.526 | 0.037 | 0.385 | 0.063 | 0.143 |
| Sample | E1 | E1 | E1 | E1 | E1 | E1 | E3 | E3 | E3 | E3 | E4 | E4 | E4 |
| Average number of reads | 21,546 | 6002 | 28,349 | 12,084 | 27,587 | 6440 | 25,009 | 22,083 | 15,698 | 18,203 | 23,155 | 16,158 | 7879 |
| % Phytophthora reads | 0.02 | 0.14 | 0.06 | 100 | 99.93 | 7.49 | 100 | 99.72 | 0.17 | 41.55 | 99.73 | 87.16 | 3.25 |
| % Oomycete reads | 0.02 | 1.03 | 0.30 | 100 | 100 | 40 | 100 | 99.72 | 0.17 | 100 | 99.96 | 99.94 | 3.25 |
| Phytophthoraspecies detected | 2 | 2 | 3 | 11 | 15 | 4 | 23 | 16 | 11 | 10 | 29 | 23 | 13 |
| Species missed from eDNA sample | 14 | 14 | 15 | 5 | 1 | 11 | 3 | 10 | 15 | 16 | 3 | 9 | 19 |
| Sample | E2 | E2 | E2 | E2 | E2 | E2 | |||||||
| Average number of reads | 11,326 | 8844 | 9142 | 65,551 | 19,638 | 3957 | |||||||
| % Phytophthora reads | 0 | 0.15 | 0.04 | 77.83 | 100 | 44.21 | |||||||
| % Oomycete reads | 0 | 0.16 | 0.05 | 100 | 100 | 60.37 | |||||||
| Phytophthoraspecies detected | 0 | 7 | 2 | 13 | 11 | 3 | |||||||
| Species missed from eDNA sample | 16 | 9 | 14 | 3 | 5 | 13 | |||||||
| Sample | E1+ MIX1 | E1+ MIX1 | E3+ MIX2 | E3+ MIX2 | E3+ MIX2 | E3+ MIX2 | E4+ MIX3 | E4+ MIX3 | E4+ MIX3 | ||||
| Average number of reads | 36,097 | 3247 | 21,696 | 11,490 | 10,001 | 16,514 | 16,136 | 14,872 | 15,740 | ||||
| % Phytophthora reads | 100 | 76.44 | 99.87 | 97.56 | 2.41 | 88.05 | 99.61 | 99.41 | 99.48 | ||||
| % Oomycete reads | 100 | 83 | 100 | 99.98 | 2.42 | 100 | 99.96 | 99.90 | 99.48 | ||||
| Phytophthoraspecies detected | 46 | 25 | 57 | 30 | 41 | 51 | 38 | 36 | 48 | ||||
| Species missed from MIX | 6 | 25 | 10 | 37 | 26 | 16 | 22 | 24 | 12 | ||||
| Species missed from eDNA sample | 1 | 5 | 2 | 10 | 3 | 2 | 9 | 10 | 7 |
| Phytophthora Species | Clade | P4 | P4 | P4 | P4 | P6 | P6 | P6 | P6 | P10 | P10 | P10 | P10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | AV | 1 | 2 | 3 | AV | 1 | 2 | 3 | AV | ||
| P. nicotianae | 1 | 20.5 | 10.4 | 7.32 | 12.3 | 7.52 | 10.2 | 0.01 | 3.03 | ||||
| P. acaciivora | 2 | 0.01 | 11.1 | 0.01 | 2.73 | 4.88 | 4.65 | 0.01 | 1.82 | ||||
| P. capensis | 2 | 1.61 | 0.70 | 0.89 | 0.25 | ||||||||
| P. elongata | 2 | 10.9 | 0.04 | 0.02 | 3.49 | 0.03 | 0.06 | 0.01 | 0.02 | ||||
| P. multivora | 2 | 0.01 | 9.61 | 19.3 | 10.8 | 0.02 | 0.01 | ||||||
| P. plurivora | 2 | 0.03 | 0.02 | 0.25 | 0.13 | 63.9 | 0.10 | 17.9 | |||||
| P. arenaria | 4 | 5.92 | 12.9 | 8.70 | 8.84 | ||||||||
| P. boodjera | 4 | 1.39 | 0.34 | ||||||||||
| P. palmivora | 4 | 4.45 | 10.5 | 5.99 | 0.05 | 29.9 | 13.1 | ||||||
| P. amnicola | 6 | 8.38 | 6.73 | 2.29 | 5.31 | 2.25 | 8.17 | 14.6 | 10.4 | ||||
| P. asparagi | 6 | 16.5 | 7.19 | 0.04 | 7.03 | 4.68 | 13.5 | 4.37 | 5.16 | ||||
| P. bilorbang | 6 | 0.02 | 6.91 | 0.07 | 0.57 | 2.05 | 0.57 | ||||||
| P. gibbosa 1 | 6 | 23.0 | 0.03 | 6.44 | |||||||||
| P. gregata | 6 | 0.15 | 0.09 | 0.01 | 0.07 | 7.02 | 0.20 | 4.22 | 4.76 | 0.12 | 0.03 | ||
| P. inundata | 6 | 0.01 | 0.02 | 0.01 | 0.02 | 0.04 | 0.01 | ||||||
| P. moyootj | 6 | 0.01 | 0.02 | 0.01 | 0.01 | ||||||||
| P. rosacearum | 6 | 2.26 | 0.01 | 0.55 | 0.01 | 0.01 | |||||||
| P. thermophila | 6 | 0.03 | 0.06 | 3.12 | 1.96 | 0.03 | 0.01 | ||||||
| P. cambivora | 7 | 0.03 | 5.18 | 1.28 | 4.65 | 7.67 | 6.64 | 6.12 | |||||
| P. cinnamomi | 7 | 9.02 | 0.04 | 0.01 | 2.88 | 12.1 | 6.70 | 0.01 | 4.16 | 6.85 | 2.98 | ||
| P. niederhauserii | 7 | 8.07 | 7.06 | 13.2 | 10.1 | 8.52 | 15.8 | 18.8 | 15.5 | 28.2 | 12.3 | ||
| P. drechsleri | 8 | 0.01 | 0.01 | 0.01 | 0.01 | 0.05 | 0.03 | 0.03 | |||||
| P. pseudocryptogea | 8 | 0.01 | 0.02 | 7.02 | 3.08 | ||||||||
| P. syringae | 8 | 0.01 | 2.77 | 2.26 | 1.67 | ||||||||
| P. AUS12A | 12 | 15.8 | 23.1 | 27.5 | 22.7 | 44.3 | 12.7 | 42.6 | 40.9 | 99.9 | 9.87 | 34.8 | 46.4 |
| P. versiformis | 12 | 0.06 | 0.01 | 0.02 | 3.92 | 13.3 | 5.35 | 5.52 | |||||
| No. species detected | 20 | 19 | 21 | 23 | 15 | 14 | 14 | 16 | 2 | 7 | 8 | 11 | |
| No. species not detected | 6 | 7 | 5 | 3 | 11 | 12 | 12 | 10 | 24 | 17 | 16 | 15 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burgess, T.I.; White, D.; Sapsford, S.J. Comparison of Primers for the Detection of Phytophthora (and Other Oomycetes) from Environmental Samples. J. Fungi 2022, 8, 980. https://doi.org/10.3390/jof8090980
Burgess TI, White D, Sapsford SJ. Comparison of Primers for the Detection of Phytophthora (and Other Oomycetes) from Environmental Samples. Journal of Fungi. 2022; 8(9):980. https://doi.org/10.3390/jof8090980
Chicago/Turabian StyleBurgess, Treena I., Diane White, and Sarah J. Sapsford. 2022. "Comparison of Primers for the Detection of Phytophthora (and Other Oomycetes) from Environmental Samples" Journal of Fungi 8, no. 9: 980. https://doi.org/10.3390/jof8090980





