Interactions between Bacteria and Aspergillus fumigatus in Airways: From the Mycobiome to Molecular Interactions
Abstract
:1. Introduction
2. Pulmonary Mycobiota and Chronic Respiratory Infections
3. Limitations of Studies and Laboratory Methodologies
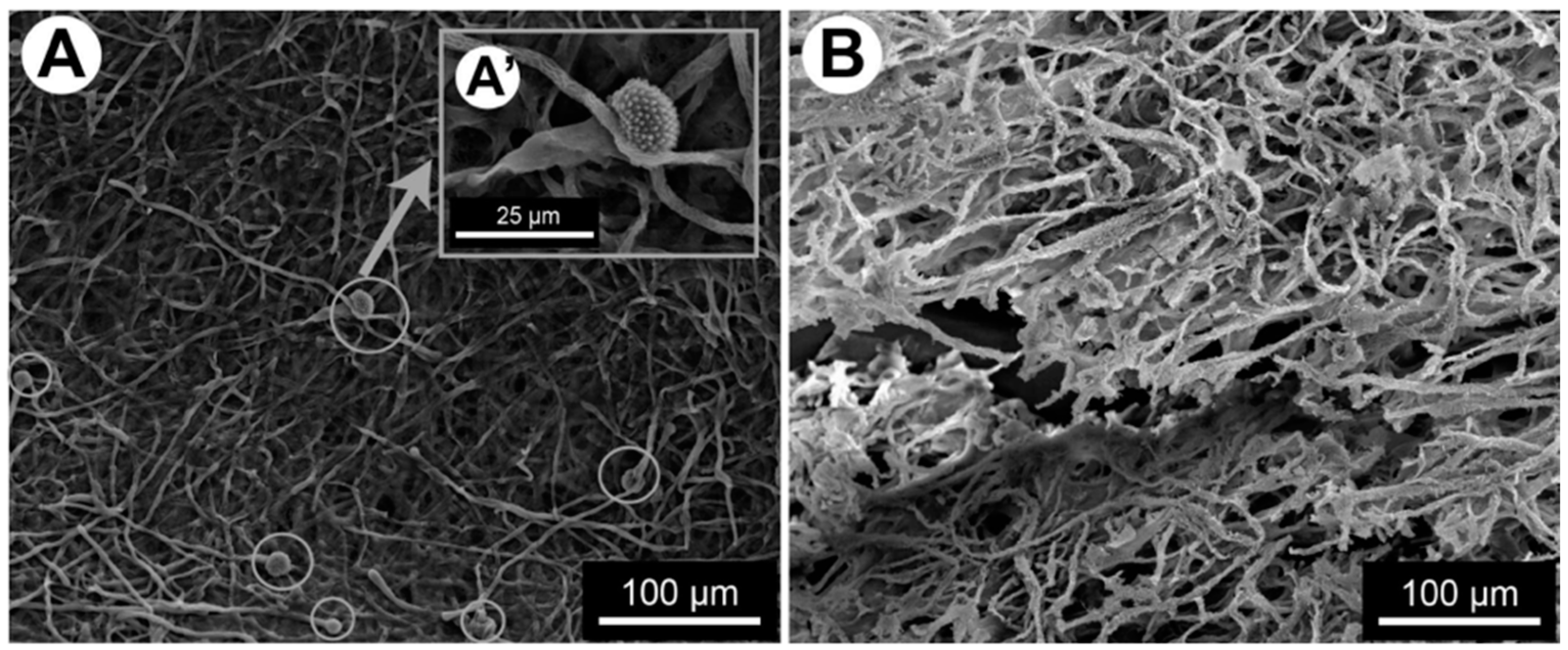
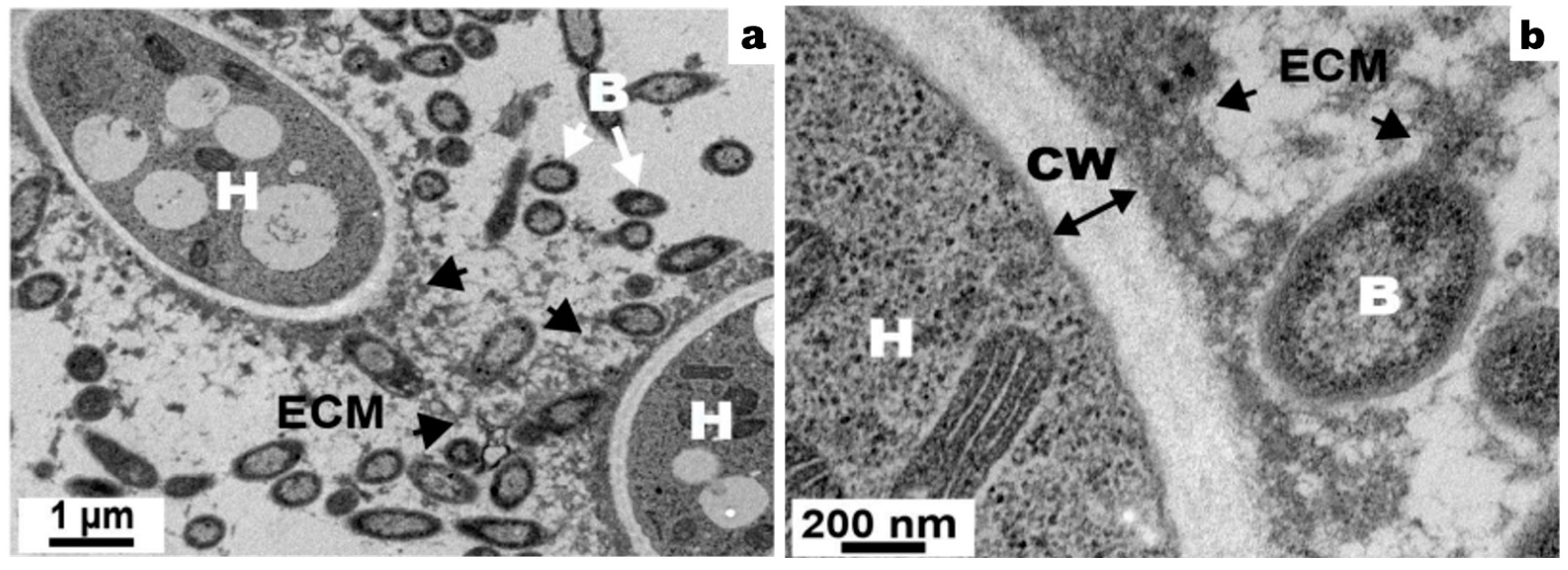
4. Formation of Mixed Filamentous Fungi–Bacteria Biofilm
5. Extracellular Matrix Production
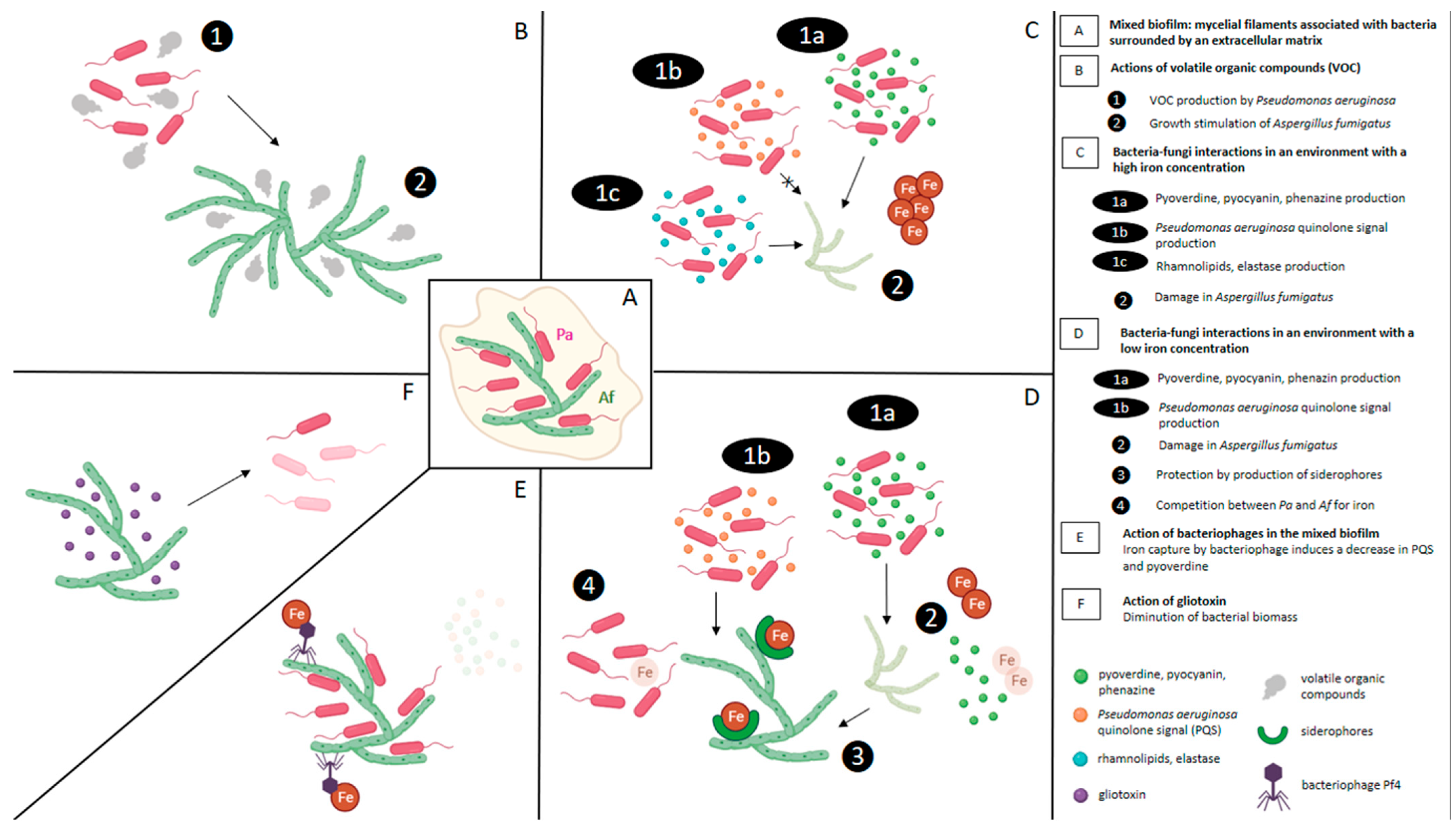
6. Social Interactions between Gram-Negative Bacteria and A. fumigatus in Mixed Biofilm
6.1. Antibiosis: The Most Common Social Interaction
6.2. Effect of Antibiosis Induced by the Bacteria on the Development of the Fungi
6.3. Strain-Dependent Interaction Effect
7. Effect of Extracellular Soluble Molecules of Bacteria on A. fumigatus Development
7.1. Role of P. aeruginosa Pyoverdine and Phenazines
7.2. Role of P. aeruginosa Quinolone Signal (PQS)
7.3. Role of Rhamnolipids and Elastase
7.4. Role of Volatile Organic Compounds
7.5. Role of Bacteriophages
8. Effect of Extracellular Soluble Molecules of Fungi on Bacterial Development (Figure 3)
8.1. Role of Gliotoxin
8.2. Role of A. fumigatus Siderophores
9. Role of Iron in Biofilm Interactions
10. Other Mechanisms
10.1. Apoptosis and ROS Production
10.2. Role of Oxygen and pH Gradients
11. Susceptibility of Polymicrobial Biofilms
12. Polymicrobial Biofilms Evasion of Immune System
13. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically Important Bacterial–Fungal Interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The Polymicrobial Nature of Biofilm Infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef]
- Boisvert, A.-A.; Cheng, M.P.; Sheppard, D.C.; Nguyen, D. Microbial Biofilms in Pulmonary and Critical Care Diseases. Ann. Am. Thorac. Soc. 2016, 13, 1615–1623. [Google Scholar] [CrossRef]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in Multispecies Biofilms: Do They Actually Matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival Strategies of Infectious Biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Parsek, M.R.; Singh, P.K. Bacterial Biofilms: An Emerging Link to Disease Pathogenesis. Annu. Rev. Microbiol. 2003, 57, 677–701. [Google Scholar] [CrossRef]
- Nguyen, L.D.N.; Viscogliosi, E.; Delhaes, L. The Lung Mycobiome: An Emerging Field of the Human Respiratory Microbiome. Front. Microbiol. 2015, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Portillo, E.; Orts, D.; Llorca, E.; Fernández, C.; Antón, J.; Ferrer, C.; Gálvez, B.; Esteban, V.; Revelles, E.; Pérez-Martín, C.; et al. The Domestic Environment and the Lung Mycobiome. Microorganisms 2020, 8, 1717. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- van Tilburg Bernardes, E.; Gutierrez, M.W.; Arrieta, M.-C. The Fungal Microbiome and Asthma. Front. Cell. Infect. Microbiol. 2020, 10, 583418. [Google Scholar] [CrossRef]
- Rick, E.; Woolnough, K.F.; Seear, P.J.; Fairs, A.; Satchwell, J.; Richardson, M.; Monteiro, W.R.; Craner, M.; Bourne, M.; Wardlaw, A.J.; et al. The Airway Fungal Microbiome in Asthma. Clin. Exp. Allergy 2020, 50, 1325–1341. [Google Scholar] [CrossRef]
- The Mucofong Investigation Group; Soret, P.; Vandenborght, L.-E.; Francis, F.; Coron, N.; Enaud, R.; Avalos, M.; Schaeverbeke, T.; Berger, P.; Fayon, M.; et al. Respiratory Mycobiome and Suggestion of Inter-Kingdom Network during Acute Pulmonary Exacerbation in Cystic Fibrosis. Sci. Rep. 2020, 10, 3589. [Google Scholar] [CrossRef]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The Airway Microbiota in Cystic Fibrosis: A Complex Fungal and Bacterial Community—Implications for Therapeutic Management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Felton, I.; James, P.; Cox, M.J.; Bilton, D.; Schelenz, S.; Loebinger, M.R.; Cookson, W.O.C.; Simmonds, N.J.; Moffatt, M.F. The Fungal Airway Microbiome in Cystic Fibrosis and Non-Cystic Fibrosis Bronchiectasis. J. Cyst. Fibros. 2021, 20, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Francis, F.; Enaud, R.; Soret, P.; Lussac-Sorton, F.; Avalos-Fernandez, M.; MucoFong Investigation Group; Bui, S.; Fayon, M.; Thiébaut, R.; Delhaes, L. New Insights in Microbial Species Predicting Lung Function Decline in CF: Lessons from the MucoFong Project. J. Clin. Med. 2021, 10, 3725. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Dicker, A.J.; Keir, H.R.; Poh, M.E.; Pang, S.L.; Mac Aogáin, M.; Chua, B.Q.Y.; Tan, J.L.; Xu, H.; Koh, M.S.; et al. A High-Risk Airway Mycobiome Is Associated with Frequent Exacerbation and Mortality in COPD. Eur. Respir. J. 2021, 57, 2002050. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, E.M.H.; Eagan, T.M.L.; Wiker, H.G.; Leiten, E.O.; Husebø, G.R.; Knudsen, K.S.; Tangedal, S.; Sanseverino, W.; Paytuví-Gallart, A.; Nielsen, R. A Longitudinal Study of the Pulmonary Mycobiome in Subjects with and without Chronic Obstructive Pulmonary Disease. PLoS ONE 2022, 17, e0267195. [Google Scholar] [CrossRef]
- Charlson, E.S.; Bittinger, K.; Chen, J.; Diamond, J.M.; Li, H.; Collman, R.G.; Bushman, F.D. Assessing Bacterial Populations in the Lung by Replicate Analysis of Samples from the Upper and Lower Respiratory Tracts. PLoS ONE 2012, 7, e42786. [Google Scholar] [CrossRef]
- Angebault, C.; Payen, M.; Woerther, P.-L.; Rodriguez, C.; Botterel, F. Combined Bacterial and Fungal Targeted Amplicon Sequencing of Respiratory Samples: Does the DNA Extraction Method Matter? PLoS ONE 2020, 15, e0232215. [Google Scholar] [CrossRef]
- Enaud, R.; Vandenborght, L.-E.; Coron, N.; Bazin, T.; Prevel, R.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Lamireau, T.; Delhaes, L. The Mycobiome: A Neglected Component in the Microbiota-Gut-Brain Axis. Microorganisms 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Manavathu, E.K.; Vager, D.L.; Vazquez, J.A. Development and Antimicrobial Susceptibility Studies of in Vitro Monomicrobial and Polymicrobial Biofilm Models with Aspergillus Fumigatus and Pseudomonas Aeruginosa. BMC Microbiol. 2014, 14, 53. [Google Scholar] [CrossRef]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas Aeruginosa and Their Small Diffusible Extracellular Molecules Inhibit Aspergillus Fumigatus Biofilm Formation: Pseudomonas Inhibits Aspergillus Biofilms. FEMS Microbiol. Lett. 2010, 313, 96–102. [Google Scholar] [CrossRef]
- Melloul, E.; Luiggi, S.; Anaïs, L.; Arné, P.; Costa, J.-M.; Fihman, V.; Briard, B.; Dannaoui, E.; Guillot, J.; Decousser, J.-W.; et al. Characteristics of Aspergillus Fumigatus in Association with Stenotrophomonas Maltophilia in an In Vitro Model of Mixed Biofilm. PLoS ONE 2016, 11, e0166325. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, J.; Kamei, K.; Watanabe, H. Disruption of Aspergillus Fumigatus Biofilm by Streptococcus Pneumoniae: Mycelial Fragmentation by Hydrogen Peroxide. J. Infect. Chemother. 2020, 26, 831–837. [Google Scholar] [CrossRef]
- Kaur, J.; Pethani, B.P.; Kumar, S.; Kim, M.; Sunna, A.; Kautto, L.; Penesyan, A.; Paulsen, I.T.; Nevalainen, H. Pseudomonas Aeruginosa Inhibits the Growth of Scedosporium Aurantiacum, an Opportunistic Fungal Pathogen Isolated from the Lungs of Cystic Fibrosis Patients. Front. Microbiol. 2015, 6, 866. [Google Scholar] [CrossRef]
- Bautista-Hernández, L.A.; Gómez-Olivares, J.L.; Buentello-Volante, B.; Dominguez-Lopez, A.; Garfias, Y.; Acosta-García, M.C.; Calvillo-Medina, R.P.; Bautista-de Lucio, V.M. Negative Interaction of Staphylococcus Aureus on Fusarium Falciforme Growth Ocular Isolates in an in Vitro Mixed Biofilm. Microb. Pathog. 2019, 135, 103644. [Google Scholar] [CrossRef] [PubMed]
- Ramírez Granillo, A.; Canales, M.G.M.; Espíndola, M.E.S.; Martínez Rivera, M.A.; de Lucio, V.M.B.; Tovar, A.V.R. Antibiosis Interaction of Staphylococccus Aureus on Aspergillus Fumigatus Assessed in Vitro by Mixed Biofilm Formation. BMC Microbiol. 2015, 15, 33. [Google Scholar] [CrossRef]
- Loussert, C.; Schmitt, C.; Prevost, M.-C.; Balloy, V.; Fadel, E.; Philippe, B.; Kauffmann-Lacroix, C.; Latgé, J.P.; Beauvais, A. In Vivo Biofilm Composition of Aspergillus Fumigatus. Cell. Microbiol. 2010, 12, 405–410. [Google Scholar] [CrossRef]
- Reichhardt, C.; Ferreira, J.A.G.; Joubert, L.-M.; Clemons, K.V.; Stevens, D.A.; Cegelski, L. Analysis of the Aspergillus Fumigatus Biofilm Extracellular Matrix by Solid-State Nuclear Magnetic Resonance Spectroscopy. Eukaryot. Cell 2015, 14, 1064–1072. [Google Scholar] [CrossRef]
- Briard, B.; Rasoldier, V.; Bomme, P.; ElAouad, N.; Guerreiro, C.; Chassagne, P.; Muszkieta, L.; Latgé, J.-P.; Mulard, L.; Beauvais, A. Dirhamnolipids Secreted from Pseudomonas Aeruginosa Modify Anjpegungal Susceptibility of Aspergillus Fumigatus by Inhibiting Β1,3 Glucan Synthase Activity. ISME J. 2017, 11, 1578–1591. [Google Scholar] [CrossRef]
- Lee, M.J.; Geller, A.M.; Bamford, N.C.; Liu, H.; Gravelat, F.N.; Snarr, B.D.; Le Mauff, F.; Chabot, J.; Ralph, B.; Ostapska, H.; et al. Deacetylation of Fungal Exopolysaccharide Mediates Adhesion and Biofilm Formation. mBio 2016, 7, e00252-00216. [Google Scholar] [CrossRef]
- Briard, B.; Fontaine, T.; Samir, P.; Place, D.E.; Muszkieta, L.; Malireddi, R.K.S.; Karki, R.; Christgen, S.; Bomme, P.; Vogel, P.; et al. Galactosaminogalactan Activates the Inflammasome to Provide Host Protection. Nature 2020, 588, 688–692. [Google Scholar] [CrossRef]
- Lee, M.J.; Liu, H.; Barker, B.M.; Snarr, B.D.; Gravelat, F.N.; Al Abdallah, Q.; Gavino, C.; Baistrocchi, S.R.; Ostapska, H.; Xiao, T.; et al. The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps. PLoS Pathog. 2015, 11, e1005187. [Google Scholar] [CrossRef]
- Chen, S.C.-A.; Patel, S.; Meyer, W.; Chapman, B.; Yu, H.; Byth, K.; Middleton, P.G.; Nevalainen, H.; Sorrell, T.C. Pseudomonas Aeruginosa Inhibits the Growth of Scedosporium and Lomentospora In Vitro. Mycopathologia 2018, 183, 251–261. [Google Scholar] [CrossRef]
- Homa, M.; Sándor, A.; Tóth, E.; Szebenyi, C.; Nagy, G.; Vágvölgyi, C.; Papp, T. In Vitro Interactions of Pseudomonas Aeruginosa With Scedosporium Species Frequently Associated With Cystic Fibrosis. Front. Microbiol. 2019, 10, 441. [Google Scholar] [CrossRef]
- Smith, K.; Rajendran, R.; Kerr, S.; Lappin, D.F.; Mackay, W.G.; Williams, C.; Ramage, G. Aspergillus Fumigatus Enhances Elastase Production in Pseudomonas aeruginosa Co-Cultures. Med. Mycol. 2015, 53, 645–655. [Google Scholar] [CrossRef]
- Melloul, E.; Roisin, L.; Durieux, M.-F.; Woerther, P.-L.; Jenot, D.; Risco, V.; Guillot, J.; Dannaoui, E.; Decousser, J.-W.; Botterel, F. Interactions of Aspergillus Fumigatus and Stenotrophomonas Maltophilia in an in Vitro Mixed Biofilm Model: Does the Strain Matter? Front. Microbiol. 2018, 9, 2850. [Google Scholar] [CrossRef]
- Nogueira, M.F.; Pereira, L.; Jenull, S.; Kuchler, K.; Lion, T. Klebsiella Pneumoniae Prevents Spore Germination and Hyphal Development of Aspergillus Species. Sci. Rep. 2019, 9, 218. [Google Scholar] [CrossRef]
- Nazik, H.; Moss, R.B.; Karna, V.; Clemons, K.V.; Banaei, N.; Cohen, K.; Choudhary, V.; Stevens, D.A. Are Cystic Fibrosis Aspergillus Fumigatus Isolates Different? Intermicrobial Interactions with Pseudomonas. Mycopathologia 2017, 182, 315–318. [Google Scholar] [CrossRef]
- Ferreira, J.A.G.; Penner, J.C.; Moss, R.B.; Haagensen, J.A.J.; Clemons, K.V.; Spormann, A.M.; Nazik, H.; Cohen, K.; Banaei, N.; Carolino, E.; et al. Inhibition of Aspergillus Fumigatus and Its Biofilm by Pseudomonas Aeruginosa Is Dependent on the Source, Phenotype and Growth Conditions of the Bacterium. PLoS ONE 2015, 10, e0134692. [Google Scholar] [CrossRef]
- Shirazi, F.; Ferreira, J.A.G.; Stevens, D.A.; Clemons, K.V.; Kontoyiannis, D.P. Biofilm Filtrates of Pseudomonas Aeruginosa Strains Isolated from Cystic Fibrosis Patients Inhibit Preformed Aspergillus Fumigatus Biofilms via Apoptosis. PLoS ONE 2016, 11, e0150155. [Google Scholar] [CrossRef]
- Anand, R.; Moss, R.B.; Sass, G.; Banaei, N.; Clemons, K.V.; Martinez, M.; Stevens, D.A. Small Colony Variants of Pseudomonas Aeruginosa Display Heterogeneity in Inhibiting Aspergillus Fumigatus Biofilm. Mycopathologia 2018, 183, 263–272. [Google Scholar] [CrossRef]
- Wurster, S.; Sass, G.; Albert, N.D.; Nazik, H.; Déziel, E.; Stevens, D.A.; Kontoyiannis, D.P. Live Imaging and Quantitative Analysis of Aspergillus fumigatus Growth and Morphology during Inter-Microbial Interaction with Pseudomonas aeruginosa. Virulence 2020, 11, 1329–1336. [Google Scholar] [CrossRef]
- Sass, G.; Shrestha, P.; Stevens, D.A. Pseudomonas Aeruginosa Virulence Factors Support Voriconazole Effects on Aspergillus Fumigatus. Pathogens 2021, 10, 519. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.-C.; Dietl, A.-M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas Aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus Fumigatus Biofilm. J. Bacteriol. 2018, 200, 24. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.-C.; Dietl, A.-M.; Visca, P.; Haas, H.; et al. Aspergillus-Pseudomonas Interaction, Relevant to Competition in Airways. Med. Mycol. 2019, 57, S228–S232. [Google Scholar] [CrossRef]
- Nazik, H.; Sass, G.; Ansari, S.R.; Ertekin, R.; Haas, H.; Déziel, E.; Stevens, D.A. Novel Intermicrobial Molecular Interaction: Pseudomonas Aeruginosa Quinolone Signal (PQS) Modulates Aspergillus Fumigatus Response to Iron. Microbiology 2020, 166, 44–55. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Chatterjee, P.; Stevens, D.A. Under Nonlimiting Iron Conditions Pyocyanin Is a Major Antifungal Molecule, and Differences between Prototypic Pseudomonas aeruginosa Strains. Med. Mycol. 2021, 59, 453–464. [Google Scholar] [CrossRef]
- Briard, B.; Heddergott, C.; Latgé, J.-P. Volatile Compounds Emitted by Pseudomonas Aeruginosa Stimulate Growth of the Fungal Pathogen Aspergillus Fumigatus. mBio 2016, 7, e00219-16. [Google Scholar] [CrossRef] [PubMed]
- Briard, B.; Mislin, G.L.A.; Latgé, J.-P.; Beauvais, A. Interactions between Aspergillus Fumigatus and Pulmonary Bacteria: Current State of the Field, New Data, and Future Perspective. J. Fungi 2019, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Penner, J.C.; Ferreira, J.A.G.; Secor, P.R.; Sweere, J.M.; Birukova, M.K.; Joubert, L.-M.; Haagensen, J.A.J.; Garcia, O.; Malkovskiy, A.V.; Kaber, G.; et al. Pf4 Bacteriophage Produced by Pseudomonas Aeruginosa Inhibits Aspergillus Fumigatus Metabolism via Iron Sequestration. Microbiology 2016, 162, 1583–1594. [Google Scholar] [CrossRef]
- Secor, P.R.; Sass, G.; Nazik, H.; Stevens, D.A. Effect of Acute Predation with Bacteriophage on Intermicrobial Aggression by Pseudomonas Aeruginosa. PLoS ONE 2017, 12, e0179659. [Google Scholar] [CrossRef]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus Fumigatus Inhibits Pseudomonas Aeruginosa in Co-Culture: Implications of a Mutually Antagonistic Relationship on Virulence and Inflammation in the CF Airway. Front. Microbiol. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Sass, G.; Ansari, S.R.; Dietl, A.-M.; Déziel, E.; Haas, H.; Stevens, D.A. Intermicrobial Interaction: Aspergillus Fumigatus Siderophores Protect against Competition by Pseudomonas Aeruginosa. PLoS ONE 2019, 14, e0216085. [Google Scholar] [CrossRef]
- Zheng, H.; Kim, J.; Liew, M.; Yan, J.K.; Herrera, O.; Bok, J.W.; Kelleher, N.L.; Keller, N.P.; Wang, Y. Redox Metabolites Signal Polymicrobial Biofilm Development via the NapA Oxidative Stress Cascade in Aspergillus. Curr. Biol. 2015, 25, 29–37. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Anand, R.; Clemons, K.V.; Stevens, D.A. Effect of Anaerobiasis or Hypoxia on Pseudomonas Aeruginosa Inhibition of Aspergillus Fumigatus Biofilm. Arch. Microbiol. 2017, 199, 881–890. [Google Scholar] [CrossRef]
- Roisin, L.; Melloul, E.; Woerther, P.-L.; Royer, G.; Decousser, J.-W.; Guillot, J.; Dannaoui, E.; Botterel, F. Modulated Response of Aspergillus Fumigatus and Stenotrophomonas Maltophilia to Antimicrobial Agents in Polymicrobial Biofilm. Front. Cell. Infect. Microbiol. 2020, 10, 574028. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Hengge, R. Stress Responses Go Three Dimensional—The Spatial Order of Physiological Differentiation in Bacterial Macrocolony Biofilms. Environ. Microbiol. 2014, 16, 1455–1471. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus Aureus Biofilms Prevent Macrophage Phagocytosis and Attenuate Inflammation In Vivo. J. Immunol. Baltim. Md 1950 2011, 186, 6585–6596. [Google Scholar] [CrossRef]
- Jesaitis, A.J.; Franklin, M.J.; Berglund, D.; Sasaki, M.; Lord, C.I.; Bleazard, J.B.; Duffy, J.E.; Beyenal, H.; Lewandowski, Z. Compromised Host Defense on Pseudomonas Aeruginosa Biofilms: Characterization of Neutrophil and Biofilm Interactions. J. Immunol. Baltim. Md 1950 2003, 171, 4329–4339. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida Albicans Biofilms Do Not Trigger Reactive Oxygen Species and Evade Neutrophil Killing. J. Infect. Dis. 2012, 206, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Günther, F.; Wabnitz, G.H.; Stroh, P.; Prior, B.; Obst, U.; Samstag, Y.; Wagner, C.; Hänsch, G.M. Host Defence against Staphylococcus Aureus Biofilms Infection: Phagocytosis of Biofilms by Polymorphonuclear Neutrophils (PMN). Mol. Immunol. 2009, 46, 1805–1813. [Google Scholar] [CrossRef]
- Staudinger, B.J.; Muller, J.F.; Halldórsson, S.; Boles, B.; Angermeyer, A.; Nguyen, D.; Rosen, H.; Baldursson, O.; Gottfreðsson, M.; Guðmundsson, G.H.; et al. Conditions Associated with the Cystic Fibrosis Defect Promote Chronic Pseudomonas Aeruginosa Infection. Am. J. Respir. Crit. Care Med. 2014, 189, 812–824. [Google Scholar] [CrossRef]
- Mishra, M.; Byrd, M.S.; Sergeant, S.; Azad, A.K.; Parsek, M.R.; McPhail, L.; Schlesinger, L.S.; Wozniak, D.J. Pseudomonas Aeruginosa Psl Polysaccharide Reduces Neutrophil Phagocytosis and the Oxidative Response by Limiting Complement-Mediated Opsonization. Cell. Microbiol. 2012, 14, 95–106. [Google Scholar] [CrossRef]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The Exopolysaccharide Alginate Protects Pseudomonas Aeruginosa Biofilm Bacteria from IFN-Gamma-Mediated Macrophage Killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef]
- Gravelat, F.N.; Beauvais, A.; Liu, H.; Lee, M.J.; Snarr, B.D.; Chen, D.; Xu, W.; Kravtsov, I.; Hoareau, C.M.Q.; Vanier, G.; et al. Aspergillus Galactosaminogalactan Mediates Adherence to Host Constituents and Conceals Hyphal β-Glucan from the Immune System. PLoS Pathog. 2013, 9, e1003575. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Debourgogne, A.; Monpierre, L.; Sy, K.A.; Valsecchi, I.; Decousser, J.-W.; Botterel, F. Interactions between Bacteria and Aspergillus fumigatus in Airways: From the Mycobiome to Molecular Interactions. J. Fungi 2023, 9, 900. https://doi.org/10.3390/jof9090900
Debourgogne A, Monpierre L, Sy KA, Valsecchi I, Decousser J-W, Botterel F. Interactions between Bacteria and Aspergillus fumigatus in Airways: From the Mycobiome to Molecular Interactions. Journal of Fungi. 2023; 9(9):900. https://doi.org/10.3390/jof9090900
Chicago/Turabian StyleDebourgogne, Anne, Lorra Monpierre, Khadeeja Adam Sy, Isabel Valsecchi, Jean-Winoc Decousser, and Françoise Botterel. 2023. "Interactions between Bacteria and Aspergillus fumigatus in Airways: From the Mycobiome to Molecular Interactions" Journal of Fungi 9, no. 9: 900. https://doi.org/10.3390/jof9090900





