Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity
Abstract
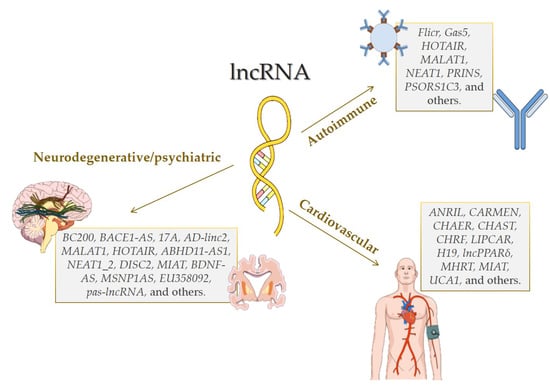
1. Introduction
2. Long Non-Coding RNAs in Complexes Diseases
2.1. Cardiovascular Diseases
2.2. Autoimmune Diseases
2.2.1. Rheumatoid Arthritis
2.2.2. Type 1 Diabetes
2.2.3. Systemic Lupus Erythematosus
2.2.4. Psoriasis
2.2.5. Other Autoimmune Diseases
2.3. Neurodegenerative Disorders
2.3.1. Alzheimer’s Disease
2.3.2. Parkinson’s Disease
2.3.3. Huntington’s Disease
2.3.4. Amyotrophic Lateral Sclerosis
2.4. Psychiatric Disorders
2.4.1. Schizophrenia
2.4.2. Depression and Anxiety Disorders
2.4.3. Autistic Spectrum Disorders
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hunter, D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Dieleman, J.L.; Baral, R.; Birger, M.; Bui, A.L.; Bulchis, A.; Chapin, A.; Hamavid, H.; Horst, C.; Johnson, E.K.; Joseph, J.; et al. US spending on personal health care and public health, 1996–2013. JAMA 2016, 316, 2627–2646. [Google Scholar] [CrossRef] [PubMed]
- Baird, P. The Human Genome Project, genetics and health. Community Genet. 2001, 4, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.H.G.S. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Irminger-Finger, I.; Kargul, J.; Laurent, G.J. Non-coding RNAs: A novel level of genome complexity. Int. J. Biochem. Cell Biol. 2014, 54, 286. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Liz, J.; Esteller, M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta 2016, 1859, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- Baira, E.; Greshock, J.; Coukos, G.; Zhang, L. Ultraconserved elements: Genomics, function and disease. RNA Biol. 2008, 5, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Aprile, M.; Esposito, R.; Ciccodicola, A. RNA-Seq and human complex diseases: Recent accomplishments and future perspectives. Eur. J. Hum. Genet. 2013, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, R.A.; Fraczek, M.G.; Parker, S.; Delneri, D.; O’Keefe, R.T. Non-coding RNAs and disease: The classical ncRNAs make a comeback. Biochem. Soc. Trans. 2016, 44, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Collins, F.S. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008, 24, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Ceman, S.; Saugstad, J. MicroRNAs: Meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease. Pharmacol. Ther. 2011, 130, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Welten, S.M.; Goossens, E.A.; Quax, P.H.; Nossent, A.Y. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc. Res. 2016, 110, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Mercer, T.R.; Bussotti, G.; Leonardi, T.; Haynes, K.R.; Crawford, J.; Brunck, M.E.; Cao, K.A.; Thomas, G.P.; Chen, W.Y.; et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat. Methods 2015, 12, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ning, Q. The emerging roles of long noncoding RNAs in common cardiovascular diseases. Hypertens. Res. 2015, 38, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Salviano-Silva, A.; Lobo-Alves, S.C.; Almeida, R.C.; Malheiros, D.; Petzl-Erler, M.L. Besides pathology: Long non-coding RNA in cell and tissue homeostasis. Non-Coding RNA 2018, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ma, W.; Huang, L.; Feng, D.; Cai, B. Long non-coding RNAs, a new important regulator of cardiovascular physiology and pathology. Int. J. Cardiol. 2015, 188, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Shah, A.; Shan, G. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [PubMed]
- D'Lima, N.G.; Ma, J.; Winkler, L.; Chu, Q.; Loh, K.H.; Corpuz, E.O.; Budnik, B.A.; Lykke-Andersen, J.; Saghatelian, A.; Slavoff, S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017, 13, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.D.; Andersen, R.E.; Liu, S.J.; Nowakowski, T.J.; Hong, S.J.; Gertz, C.C.; Salinas, R.D.; Zarabi, H.; Kriegstein, A.R.; Lim, D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015, 16, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Dallagiovanna, B.; Pereira, I.T.; Origa-Alves, A.C.; Shigunov, P.; Naya, H.; Spangenberg, L. lncRNAs are associated with polysomes during adipose-derived stem cell differentiation. Gene 2017, 610, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Harries, L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012, 40, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, R.; Zhang, K.; Chen, L.B. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J. Cell. Mol. Med. 2014, 18, 2425–2436. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.; Uranitsch, S.; Gerger, A.; Pichler, M.; Haybaeck, J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int. J. Mol. Sci. 2014, 15, 13993–14013. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.L.; Tuzova, A.V.; Bolton, E.M.; Lynch, T.H.; Perry, A.S. Long noncoding RNAs and prostate carcinogenesis: The missing ‘linc’? Trends Mol. Med. 2014, 20, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Kurochkin, I.V. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front. Genet. 2015, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601–8612. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, S.; Patel, T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief. Funct. Genom. 2016, 15, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Thakkar, N.; Chhatai, J.; Pal Bhadra, M.; Bhadra, U. Long non-coding RNA: Functional agent for disease traits. RNA Biol. 2017, 14, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Thum, T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Alvarez, A.; Hu, B.; Cheng, S.Y. Noncoding RNAs in cancer and cancer stem cells. Chin. J. Cancer 2013, 32, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Eades, G.; Zhang, Y.S.; Li, Q.L.; Xia, J.X.; Yao, Y.; Zhou, Q. Long non-coding RNAs in stem cells and cancer. World J. Clin. Oncol. 2014, 5, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta 2014, 1839, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Fatima, R.; Akhade, V.S.; Pal, D.; Rao, S.M. Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 2015, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xia, L.Q.; Lu, W.W.; Zhang, J.; Zhu, J.S. LncRNAs and cancer. Oncol. Lett. 2016, 12, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.K.D.M.; Rajkumar, T.; Mani, S. Perspectives of long non-coding RNAs in cancer. Mol. Biol. Rep. 2017, 44, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.V.; Choudhari, R.; Gadad, S.S. Long noncoding RNAs and cancer, an overview. Steroids 2018, 133, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Després, J.P.; Fullerton, H.J.; et al. Heart disease and stroke statistics—2016 update: A report from the American Heart Association. Circulation 2016, 133. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Statistical Classification of Diseases and Related Health Problems; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- American Heart Association. My Life Check—Life’s Simple 7. Available online: http://www.heart.org/HEARTORG/Conditions/My-Life-Check---Lifes-Simple-7_UCM_471453_Article.jsp#.WvFmGoiFPIV (accessed on 30 June 2016).
- Logue, J.; Murray, H.M.; Welsh, P.; Shepherd, J.; Packard, C.; Macfarlane, P.; Cobbe, S.; Ford, I.; Sattar, N. Obesity is associated with fatal coronary heart disease independently of traditional risk factors and deprivation. Heart 2011, 97, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Noels, H. Atherosclerosis: Current pathogenesis and therapeutic options. Nat. Med. 2011, 17, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K.; Witztum, J.L. Atherosclerosis. the road ahead. Cell 2001, 104, 503–516. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.S.; Cheng, J.; Cai, M.Y.; Yang, X.L.; Liu, X.G.; Zheng, B.Y.; Xiong, X.D. Association of lincRNA-p21 haplotype with coronary artery disease in a Chinese Han population. Dis. Mark. 2016, 2016, 9109743. [Google Scholar]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8, 326ra322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.J.; Ji, Y.X.; Zhang, P.; Deng, K.Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Rotllan, N.; Fernández-Hernando, C. Noncoding RNAs and atherosclerosis. Curr. Atheroscler. Rep. 2014, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Zhou, M.M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Katsuya, T.; Yasuda, O.; Oguro, R.; Yamamoto, K.; Ohishi, M.; Rakugi, H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys. Res. Commun. 2012, 419, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Willeit, J.; Kronenberg, F.; Xu, Q.; Kiechl, S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: A population-based, prospective study. J. Am. Coll. Cardiol. 2008, 52, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.D.; Biffi, A.; Rost, N.S.; Cortellini, L.; Furie, K.L.; Rosand, J. Chromosome 9p21 in ischemic stroke: Population structure and meta-analysis. Stroke 2010, 41, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Bilguvar, K.; Yasuno, K.; Niemelä, M.; Ruigrok, Y.M.; von Und Zu Fraunberg, M.; van Duijn, C.M.; van den Berg, L.H.; Mane, S.; Mason, C.E.; Choi, M.; et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat. Genet. 2008, 40, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Yasuno, K.; Bilguvar, K.; Bijlenga, P.; Low, S.K.; Krischek, B.; Auburger, G.; Simon, M.; Krex, D.; Arlier, Z.; Nayak, N.; et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat. Genet. 2010, 42, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; Masson, G.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Magnusson, K.P.; Grétarsdottir, S.; Steinthorsdottir, V.; Manolescu, A.; Jones, G.T.; Rinkel, G.J.; Blankensteijn, J.D.; Ronkainen, A.; et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat. Genet. 2008, 40, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Cluett, C.; McDermott, M.M.; Guralnik, J.; Ferrucci, L.; Bandinelli, S.; Miljkovic, I.; Zmuda, J.M.; Li, R.; Tranah, G.; Harris, T.; et al. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ. Cardiovasc. Genet. 2009, 2, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Palomaki, G.E.; Melillo, S.; Bradley, L.A. Association between 9p21 genomic markers and heart disease: A meta-analysis. JAMA 2010, 303, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Archer, K.; Broskova, Z.; Bayoumi, A.S.; Teoh, J.P.; Davila, A.; Tang, Y.; Su, H.; Kim, I.M. Long Non-coding RNAs as master regulators in cardiovascular diseases. Int. J. Mol. Sci. 2015, 16, 23651–23667. [Google Scholar] [CrossRef] [PubMed]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Arora, P. Long noncoding Mhrt RNA: Molecular crowbar unravel insights into heart failure treatment. Circ. Cardiovasc. Genet. 2015, 8, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Jiang, H.; Bei, Y.; Xiao, J.; Li, X. Long non-coding RNAs in cardiac remodeling. Cell. Physiol. Biochem. 2017, 41, 1830–1837. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Sun, L.; Zhang, Y.; Huang, Y.; Hou, Y.; Li, Q.; Guo, Y.; Feng, B.; Cui, L.; Wang, X.; et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J. Cell. Mol. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Coxon, F.P.; Miranda de Stegmann, D.; Bhogaraju, S.; Maddi, K.; et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Dawson, K.; Aflaki, M.; Nattel, S. Role of the Wnt-Frizzled system in cardiac pathophysiology: A rapidly developing, poorly understood area with enormous potential. J. Physiol. 2013, 591, 1409–1432. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Takahashi, N.; Xu, L.; Smithies, O.; Meissner, G. Early cardiac hypertrophy in mice with impaired calmodulin regulation of cardiac muscle Ca release channel. J. Clin. Investig. 2007, 117, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Zemljic-Harpf, A.E.; Miller, J.C.; Henderson, S.A.; Wright, A.T.; Manso, A.M.; Elsherif, L.; Dalton, N.D.; Thor, A.K.; Perkins, G.A.; McCulloch, A.D.; et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 2007, 27, 7522–7537. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.T.; Muchir, A.; Nagy, P.L.; Worman, H.J. LMNA cardiomyopathy: Cell biology and genetics meet clinical medicine. Dis. Model. Mech. 2011, 4, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Hullinger, T.G.; Montgomery, R.L.; Seto, A.G.; Dickinson, B.A.; Semus, H.M.; Lynch, J.M.; Dalby, C.M.; Robinson, K.; Stack, C.; Latimer, P.A.; et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012, 110, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.; Hua, F.; Li, Y.; Ma, J.; Gao, X.; Kelley, J.; Zhao, A.; Haddad, G.E.; Williams, D.L.; Browder, I.W.; et al. Blockade of MyD88 attenuates cardiac hypertrophy and decreases cardiac myocyte apoptosis in pressure overload-induced cardiac hypertrophy in vivo. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H985–H994. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Si, R.; Feng, Y.; Chen, H.H.; Zou, L.; Wang, E.; Zhang, M.; Warren, H.S.; Sosnovik, D.E.; Chao, W. Myocardial ischemia activates an injurious innate immune signaling via cardiac heat shock protein 60 and Toll-like receptor 4. J. Biol. Chem. 2011, 286, 31308–31319. [Google Scholar] [CrossRef] [PubMed]
- Dangwal, S.; Schimmel, K.; Foinquinos, A.; Xiao, K.; Thum, T. Noncoding RNAs in Heart Failure. Handb. Exp. Pharmacol. 2017, 243, 423–445. [Google Scholar] [PubMed]
- Zhou, X.; Zhang, W.; Jin, M.; Chen, J.; Xu, W.; Kong, X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017, 8, e2929. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, Y.; Zhang, Y.; Zhu, X.; Zhang, R.; Guan, L.; Tang, Q.; Jiang, H.; Huang, C.; Huang, H. MicroRNA-150 protects against pressure overload-induced cardiac hypertrophy. J. Cell. Biochem. 2015, 116, 2166–2176. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.H.; Yuan, Y.X.; Rao, S.L.; Wang, P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653–3660. [Google Scholar] [PubMed]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Frade, A.F.; Laugier, L.; Ferreira, L.R.; Baron, M.A.; Benvenuti, L.A.; Teixeira, P.C.; Navarro, I.C.; Cabantous, S.; Ferreira, F.M.; da Silva Cândido, D.; et al. Myocardial infarction-associated transcript, a long noncoding RNA, is overexpressed during dilated cardiomyopathy due to chronic Chagas disease. J. Infect. Dis. 2016, 214, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Zhang, L.; Dzau, V.J.; Pratt, R.E. H19, a developmentally regulated gene, is reexpressed in rat vascular smooth muscle cells after injury. J. Clin. Investig. 1994, 93, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Han, D.K.; Khaing, Z.Z.; Pollock, R.A.; Haudenschild, C.C.; Liau, G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J. Clin. Investig. 1996, 97, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Devlin, A.M.; Bottiglieri, T.; Domann, F.E.; Lentz, S.R. Tissue-specific changes in H19 methylation and expression in mice with hyperhomocysteinemia. J. Biol. Chem. 2005, 280, 25506–25511. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Gao, C.; Peng, G.; Greer, C.; Ren, S.; Wang, Y.; Xiao, X. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ. Res. 2011, 109, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.N.; Qian, L.; Hu, D.L.; Yu, Z.B.; Han, S.P.; Zhu, C.; Wang, X.; Hu, X. Effects of miR-19b overexpression on proliferation, differentiation, apoptosis and Wnt/β-catenin signaling pathway in P19 cell model of cardiac differentiation in vitro. Cell Biochem. Biophys. 2013, 66, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xu, H.; Cheng, J.; Zhang, Y.; Gao, C.; Fan, T.; Peng, B.; Li, B.; Liu, L.; Cheng, Z. Downregulation of long non-coding RNA H19 promotes P19CL6 cells proliferation and inhibits apoptosis during late-stage cardiac differentiation via miR-19b-modulated Sox6. Cell Biosci. 2016, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Cao, W.; Yang, J.J.; Shi, K.H.; Zhou, X.; Liu, L.P.; Li, J. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc. Pathol. 2016, 25, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.A.; Yeh, J.H.; Yan, D.; Xu, M.; Chan, A.C. Dusp5 negatively regulates IL-33-mediated eosinophil survival and function. EMBO J. 2015, 34, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.X. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 322–328. [Google Scholar] [PubMed]
- Cai, Y.; Yang, Y.; Chen, X.; He, D.; Zhang, X.; Wen, X.; Hu, J.; Fu, C.; Qiu, D.; Jose, P.A.; et al. Circulating “LncPPARδ” From Monocytes as a Novel Biomarker for Coronary Artery Diseases. Medicine 2016, 95, e2360. [Google Scholar] [CrossRef] [PubMed]
- Ehrenborg, E.; Skogsberg, J. Peroxisome proliferator-activated receptor delta and cardiovascular disease. Atherosclerosis 2013, 231, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, Y.N.; Yuan, H.H.; Zhang, T.T.; Sui, H.; Wei, X.L.; Liu, L.; Huang, P.; Zhang, W.J.; Bai, Y.X. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology 2014, 46, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Cai, W.; Ren, S.; Li, X.; Wang, Q.; Pan, H.; Zhao, M.; Li, J.; Zhang, Y.; Zhao, C.; et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015, 6, 23582–23593. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, F.; Cheng, L.; Yang, G.; Zhang, D.; Liu, J.; Chen, X.; Zhao, S. Prognostic and clinicopathological role of long non-coding RNA UCA1 in various carcinomas. Oncotarget 2017, 8, 28373. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, D.; Li, G.; Ming, X.; Tu, Y.; Tian, J.; Lu, H.; Yu, B. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell. Physiol. Biochem. 2015, 35, 1986–1998. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, B.; Liu, N.; Qi, C.; Xiao, Y.; Tian, X.; Li, T.; Liu, B. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed. Res. Int. 2016, 2016, 8079372. [Google Scholar] [CrossRef] [PubMed]
- Hrdlickova, B.; Kumar, V.; Kanduri, K.; Zhernakova, D.V.; Tripathi, S.; Karjalainen, J.; Lund, R.J.; Li, Y.; Ullah, U.; Modderman, R.; et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 2014, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Zemmour, D.; Pratama, A.; Loughhead, S.M.; Mathis, D.; Benoist, C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, E3472–E3480. [Google Scholar] [CrossRef] [PubMed]
- Mayama, T.; Marr, A.K.; Kino, T. Differential expression of glucocorticoid receptor noncoding RNA repressor Gas5 in autoimmune and inflammatory diseases. Horm. Metab. Res. 2016, 48, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Firestein, G.S. Evolving concepts of rheumatoid arthritis. Nature 2003, 423, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmüller, B.; Kunisch, E.; Franz, J.; Martinez-Gamboa, L.; Hernandez, M.M.; Pruss, A.; Ulbrich, N.; Erdmann, V.A.; Burmester, G.R.; Kinne, R.W. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003, 163, 901–911. [Google Scholar] [CrossRef]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Messemaker, T.C.; Frank-Bertoncelj, M.; Marques, R.B.; Adriaans, A.; Bakker, A.M.; Daha, N.; Gay, S.; Huizinga, T.W.; Toes, R.E.; Mikkers, H.M.; et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2016, 17, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Yu, H.C.; Yu, C.L.; Huang, H.B.; Koo, M.; Tung, C.H.; Lai, N.S. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol. Res. 2016, 64, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Zhu, L.; Lv, H.; Pei, C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int. J. Mol. Med. 2016, 38, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Motterle, A.; Gattesco, S.; Caille, D.; Meda, P.; Regazzi, R. Involvement of long non-coding RNAs in β cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 2015, 58, 1827–1835. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xue, L.; Zhu, Z.; Zhang, F.; Yang, R.; Yuan, X.; Jia, Z.; Liu, Q. Insights from lncRNAs profiling of MIN6 β cells undergoing inflammation. Mediat. Inflamm. 2016, 2016, 9275106. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Isenberg, D.A. Systemic lupus erythematosus. N. Engl. J. Med. 2008, 358, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, Z.; Yu, A.M.; Wang, W.; Wei, Z.; Akhter, E.; Maurer, K.; Costa Reis, P.; Song, L.; Petri, M.; et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS ONE 2014, 9, e93846. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, F.; Ma, J.; Zhang, X.; Wu, L.; Qu, B.; Xia, S.; Chen, S.; Tang, Y.; Shen, N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res. Ther. 2015, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, L.; Qian, J.; Qu, B.; Xia, S.; La, T.; Wu, Y.; Ma, J.; Zeng, J.; Guo, Q.; et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016, 75, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Sonkoly, E.; Nagy, N.; Németh, I.B.; Bata-Csörgo, Z.; Kemény, L.; Dobozy, A.; Széll, M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp. Dermatol. 2010, 19, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Bata-Csorgo, Z.; Pivarcsi, A.; Polyanka, H.; Kenderessy-Szabo, A.; Molnar, G.; Szentpali, K.; Bari, L.; Megyeri, K.; Mandi, Y.; et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005, 280, 24159–24167. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Göblös, A.; Bacsa, S.; Antal, M.; Németh, I.B.; Bata-Csörgő, Z.; Kemény, L.; Dobozy, A.; Széll, M. Expression and functional studies on the noncoding RNA, PRINS. Int. J. Mol. Sci. 2012, 14, 205–225. [Google Scholar] [CrossRef] [PubMed]
- Holm, S.J.; Sánchez, F.; Carlén, L.M.; Mallbris, L.; Ståhle, M.; O’Brien, K.P. HLA-Cw*0602 associates more strongly to psoriasis in the Swedish population than variants of the novel 6p21.3 gene PSORS1C3. Acta Derm. Venereol. 2005, 85, 2–8. [Google Scholar] [PubMed]
- Chang, Y.T.; Chou, C.T.; Shiao, Y.M.; Lin, M.W.; Yu, C.W.; Chen, C.C.; Huang, C.H.; Lee, D.D.; Liu, H.N.; Wang, W.J.; et al. Psoriasis vulgaris in Chinese individuals is associated with PSORS1C3 and CDSN genes. Br. J. Dermatol. 2006, 155, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tsoi, L.C.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Johnston, A.; Ding, J.; Stuart, P.E.; Xing, X.; Kochkodan, J.J.; et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J. Investig. Dermatol. 2014, 134, 1828–1838. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Iyer, M.K.; Stuart, P.E.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Sarkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Ahn, R.; Gupta, R.; Lai, K.; Chopra, N.; Arron, S.T.; Liao, W. Network analysis of psoriasis reveals biological pathways and roles for coding and long non-coding RNAs. BMC Genom. 2016, 17, 841. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ahn, R.; Lai, K.; Mullins, E.; Debbaneh, M.; Dimon, M.; Arron, S.; Liao, W. Landscape of long noncoding RNAs in psoriatic and healthy skin. J. Investig. Dermatol. 2016, 136, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Peng, H.; Tian, J.; Ma, J.; Tang, X.; Rui, K.; Tian, X.; Wang, Y.; Chen, J.; Lu, L.; et al. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjögren syndrome. Immunol. Res. 2016, 64, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Fang, X.; Jiang, Y.; Shen, F.; Hu, Z.; Li, X.; Huang, X. TNF-α induces vascular endothelial cells apoptosis through overexpressing pregnancy induced noncoding RNA in Kawasaki disease model. Int. J. Biochem. Cell Biol. 2016, 72, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNF-α expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, S.; Harada, H.; Furugaki, K.; Akamizu, T.; Ishikawa, N.; Ito, K.; Tamai, H.; Kuma, K.; Kubota, S.; Hiratani, H.; et al. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum. Mol. Genet. 2004, 13, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Fernandez-Jimenez, N.; Kratchmarov, R.; Luo, X.; Bhagat, G.; Green, P.H.; Schneider, R.; Kiledjian, M.; Bilbao, J.R.; Ghosh, S. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016, 352, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Doyle, K.M.; Kennedy, D.; Gorman, A.M.; Gupta, S.; Healy, S.J.; Samali, A. Unfolded proteins and endoplasmic reticulum stress in neurodegenerative disorders. J. Cell. Mol. Med. 2011, 15, 2025–2039. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Hardy, J.; Fischbeck, K.H. Toxic proteins in neurodegenerative disease. Science 2002, 296, 1991–1995. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein aggregation in the brain: The molecular basis for Alzheimer’s and Parkinson’s diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Y.; Chen, J.; Chi, H.; Yu, Z.; Wang, J.; Chen, C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1-AS expression. Mol. Med. Rep. 2014, 10, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, J. Identification of Alzheimer’s disease-associated long noncoding RNAs. Neurobiol. Aging 2015, 36, 2925–2931. [Google Scholar] [CrossRef] [PubMed]
- Magistri, M.; Velmeshev, D.; Makhmutova, M.; Faghihi, M.A. Transcriptomics profiling of Alzheimer’s disease reveal neurovascular defects, altered amyloid-β homeostasis, and deregulated expression of long noncoding RNAs. J. Alzheimers Dis. 2015, 48, 647–665. [Google Scholar] [CrossRef] [PubMed]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, K.R.; Healy, D.G.; Schapira, A.H.; Excellence, N.I.F.C. Non-motor symptoms of Parkinson’s disease: Diagnosis and management. Lancet Neurol. 2006, 5, 235–245. [Google Scholar] [CrossRef]
- Lesage, S.; Brice, A. Parkinson’s disease: From monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009, 18, R48–R59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.S.; Wang, Z.H.; Zhang, J.L.; Duan, Y.L.; Li, G.F.; Zheng, D.L. β-Asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression. Biomed. Pharmacother. 2016, 83, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Kraus, T.F.J.; Haider, M.; Spanner, J.; Steinmaurer, M.; Dietinger, V.; Kretzschmar, H.A. Altered long noncoding RNA expression precedes the course of Parkinson’s disease—A Preliminary Report. Mol. Neurobiol. 2017, 54, 2869–2877. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Huang, H.; Chen, Y.; Cao, M.; Zhou, H.; Zhang, Y. Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell. Mol. Neurobiol. 2017, 37, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Cookson, M.R. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat. Rev. Neurosci. 2010, 11, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Hossein-Nezhad, A.; Fatemi, R.P.; Ahmad, R.; Peskind, E.R.; Zabetian, C.P.; Hu, S.C.; Shi, M.; Wahlestedt, C.; Zhang, J.; Faghihi, M.A. Transcriptomic profiling of extracellular RNAs present in cerebrospinal fluid identifies differentially expressed transcripts in Parkinson’s disease. J. Parkinsons Dis. 2016, 6, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Guo, Y.; Rong, H.; Liu, T. The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 2017, 8, 24449–24456. [Google Scholar] [PubMed]
- Johnson, R.; Richter, N.; Jauch, R.; Gaughwin, P.M.; Zuccato, C.; Cattaneo, E.; Stanton, L.W. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genom. 2010, 41, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Francelle, L.; Galvan, L.; Gaillard, M.C.; Petit, F.; Bernay, B.; Guillermier, M.; Bonvento, G.; Dufour, N.; Elalouf, J.M.; Hantraye, P.; et al. Striatal long noncoding RNA Abhd11os is neuroprotective against an N-terminal fragment of mutant huntingtin in vivo. Neurobiol. Aging 2015, 36, 1601.e7–1601.e16. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.L.; Mao, Y.S.; Ji, J.L. The link between long noncoding RNAs and depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 73, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Chubb, J.E.; Bradshaw, N.J.; Soares, D.C.; Porteous, D.J.; Millar, J.K. The DISC locus in psychiatric illness. Mol. Psychiatry 2008, 13, 36–64. [Google Scholar] [CrossRef] [PubMed]
- Millar, J.K.; Wilson-Annan, J.C.; Anderson, S.; Christie, S.; Taylor, M.S.; Semple, C.A.; Devon, R.S.; St Clair, D.M.; Muir, W.J.; Blackwood, D.H.; et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000, 9, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; He, Q.; Li, M.; Chen, Y.; Liu, Y.; Wang, J. LncRNA MIAT: Myocardial infarction associated and more. Gene 2016, 578, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.Q.; Hu, H.L.; Ye, N.; Shen, Y.; Xu, Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015, 166, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.T.; Nikkel, S.M.; Fernandez, B.A.; Marshall, C.R.; Noor, A.; Lionel, A.C.; Prasad, A.; Pinto, D.; Joseph-George, A.M.; Noakes, C.; et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin. Genet. 2011, 80, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ionita-Laza, I.; Roos, J.L.; Boone, B.; Woodrick, S.; Sun, Y.; Levy, S.; Gogos, J.A.; Karayiorgou, M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012, 44, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Gianfrancesco, O.; Warburton, A.; Collier, D.A.; Bubb, V.J.; Quinn, J.P. Novel brain expressed RNA identified at the MIR137 schizophrenia-associated locus. Schizophr. Res. 2017, 184, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Steen, V.M.; Nepal, C.; Ersland, K.M.; Holdhus, R.; Nævdal, M.; Ratvik, S.M.; Skrede, S.; Håvik, B. Neuropsychological deficits in mice depleted of the schizophrenia susceptibility gene CSMD1. PLoS ONE 2013, 8, e79501. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Niu, W.; Kong, L.; He, M.; Jiang, K.; Chen, S.; Zhong, A.; Zhang, Q.; Li, W.; Lu, J.; et al. Long noncoding RNA as an indicator differentiating schizophrenia from major depressive disorder and generalized anxiety disorder in nonpsychiatric hospital. Biomark. Med. 2017, 11, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, X.; Niu, W.; Kong, L.; He, M.; Li, W.; Zhong, A.; Lu, J.; Zhang, L. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med. Sci. Monit. 2016, 22, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, P.A.; Flavell, C.R.; Widagdo, J.; Ratnu, V.S.; Troup, M.; Ragan, C.; Mattick, J.S.; Bredy, T.W. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatry 2015, 78, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.M.; Crawford, B.; Dempster, E.L.; Hannon, E.; Burrage, J.; Turecki, G.; Kaminsky, Z.; Mill, J. Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl. Psychiatry 2017, 7, e989. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.A.; Pur, D.R.; Vujcic, B.; Gupta, A.K. Suicidal behaviors in the dermatology patient. Clin. Dermatol. 2017, 35, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Rao, S.; Du, T.; Hu, H.; Liu, Z.; Shen, Y.; Xu, Q. Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene 2017, 601, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Bahi, A.; Chandrasekar, V.; Dreyer, J.L. Selective lentiviral-mediated suppression of microRNA124a in the hippocampus evokes antidepressants-like effects in rats. Psychoneuroendocrinology 2014, 46, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Kocerha, J.; Dwivedi, Y.; Brennand, K.J. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Mol. Psychiatry 2015, 20, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, X.; Sun, N.; Xu, Y.; Meng, Y.; Yang, C.; Wang, Y.; Zhang, K. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS ONE 2014, 9, e93388. [Google Scholar] [CrossRef] [PubMed]
- Mus, E.; Hof, P.R.; Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 10679–10684. [Google Scholar] [CrossRef] [PubMed]
- Kerin, T.; Ramanathan, A.; Rivas, K.; Grepo, N.; Coetzee, G.A.; Campbell, D.B. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci. Transl. Med. 2012, 4, 128ra140. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, J.J.; Grepo, N.; Wilkinson, B.; Evgrafov, O.V.; Knowles, J.A.; Campbell, D.B. Impact of the autism-associated long noncoding RNA MSNP1AS on neuronal architecture and gene expression in human neural progenitor cells. Genes 2016, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Beck, T.F.; Pearson, D.M.; Proud, M.B.; Cheung, S.W.; Scott, D.A. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am. J. Med. Genet. A 2009, 149A, 1758–1762. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.; Whibley, A.; Marshall, C.R.; Gianakopoulos, P.J.; Piton, A.; Carson, A.R.; Orlic-Milacic, M.; Lionel, A.C.; Sato, D.; Pinto, D.; et al. Disruption at the PTCHD1 locus on Xp22.11 in autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Ziats, M.N.; Rennert, O.M. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013, 49, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, X.; Ju, W.; Flory, M.; Zhong, J.; Jiang, S.; Wang, P.; Dong, X.; Tao, X.; Chen, Q.; et al. Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl. Psychiatry 2015, 5, e660. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Expression | Disease | Mechanism | Reference |
|---|---|---|---|---|
| ANRIL | Down | Coronary Artery Disease (CAD), Myocardial Infarction | Chromatin remodelling (interacts with PRC1 and PRC2); regulation of CDKN2A and CDKN2B expression. | [74,75,76,77,79] |
| MHRT | Up | Heart failure, Acute Myocardial Infarction, Hypertrophy, Cardiomyopathy | Chromatin remodelling (cardiac stress); interacts with BRG1 preventing its ligation to targets. | [65,88,89,90,91,92] |
| CHAST | Up | Heart failure, Aortic Stenosis, Cardiac Hypertrophy. | Cardiac remodelling; negatively regulates Plekhm1. | [71,93,94,95,96,97] |
| LIPCAR | Down | Initial Myocardial Infarction, Chronic Cardiovascular Disease | Cardiac remodeling; unknown mechanism. | [66,98] |
| CARMEN | Up | Cardiovascular Disease (Cardiac Hypertrophy) | Chromatin remodelling; interacts with SUZ12 and EZH2 (PRC2). | [68] |
| CHRF | Up | Cardiac Hypertrophy | Sponge for mir-489. | [67,101] |
| CHAER (mouse) | Up | Cardiac Hypertrophy | Epigenetic Regulator; inhibits H3L27 methylation. | [72] |
| MIAT | Up | Cardiac Hypertrophy, Myocardial Infarction | Sponge for mir-150. | [78,102,103,104,105,106] |
| H19 | Up | Atherosclerosis, Heart failure, Cardiac Hypertrophy | Epigenetic Regulator; suppresses miR-675 and miR-19b; repression of DUSP5/ERK1/2; regulating MAPK and NF-kB signalling pathway. | [69,107,108,109,110,111,112,113,114,115] |
| LncPPARδ | Up | Atherosclerosis, Cardiovascular Disease | Decreases cholesterol efflux and increases migration of leukocyte/monocytes into the arterial wall. | [116] |
| UCA1 | Down/Up (Biphasic) | Myocardial Infarction (after 12 h/72 h) | Unknown. | [122] |
| H19 | Up | Rheumatoid Arthritis | Unknown. | [127] |
| HOTAIR | Up | Rheumatoid Arthritis | Unknown. | [128] |
| C5T1lncRNA | Up | Rheumatoid Arthritis | Regulates C5 mRNA expression. | [129] |
| LOC10065295/LOC100506036 | Up | Rheumatoid Arthritis | Unknown. | [130] |
| MALAT 1 | Up | Rheumatoid Arthritis | Promotes the apoptosis of rheumatoid arthritis fibroblast-like synoviocytes and leads to the activation of the PI3K/AKT pathway. | [131] |
| NEAT1 | Up | Systemic Lupus Erythematosus (SLE) | Contributes to expression of a group of chemokines and cytokines, including IL-6 and CXCL10. | [137] |
| TMEVPG1 | Up | Sjogren’s Syndrome | Positively correlates expression with Th1 cell proportion among CD4+ T cells. | [147] |
| PINC | Up | Kawasaki disease | Promotes expression of apoptosis genes in human umbilical vascular endothelial cells (HUVEC). | [148] |
| PRINS | Up | Psoriasis | Interacts with NPM protein, which is associated with keratinocytes proliferation. | [139] |
| BACE1-AS | Up | Alzheimer’s Disease | Increases the stability of BACE1 mRNA. Aβ production. | [157,158] |
| 17A | Up | Alzheimer’s Disease | Alternative splicing of GPR51 mRNA, reducing canonical form of GABAB R2 and impairing GABAB signaling. Aβ production. | [159] |
| HAO2-AS, EBF3-AS, AD-linc1 and AD-linc2 | Up | Alzheimer’s Disease | Neurotoxicity. | [161] |
| n341006 | Down | Alzheimer’s Disease | Acting in the ubiquitin-proteasome system (UPS), affecting the turnover and degradation, is impaired in AD. | [160] |
| n336934 | Up | Alzheimer’s Disease | Cholesterol pathway. | [160] |
| MALAT1 | Up | Parkinson’s Disease | α-synuclein production. | [165] |
| HOTAIR | Up | Parkinson’s Disease | Increasing the stability and level of LRRK2 mRNA. | [170] |
| AL049437 | Up | Parkinson’s Disease | Apoptosis pathway. | [167] |
| AK021630 | Down | Parkinson’s Disease | Apoptosis pathway. | [167] |
| p21 | Up | Parkinson’s Disease | Targets p53 and H1F1 (apoptosis pathway). | [166] |
| SNHG1 | Up | Parkinson’s Disease | Affects the p53 stability (cellular proliferation). | [166] |
| HAR1F and HAR1R | Down | Huntington’s Disease | Unknown. | [171] |
| ABHD11-AS1 | Down | Huntington’s Disease | Affects the neuronal toxicity. | [172] |
| NEAT1_2 | Up | Amyotrophic Lateral Sclerosis (ALS) | Changes the TDP-43 and FUS/TLS functions, stress granules formation). | [173] |
| DISC2 | Down | Schizophrenia, Autistic Spectrum Disorders (ASD) | DISC2 regulates DISC1 expression. Translocation in both genes are involved with psychiatric disorders. | [174,175,176] |
| EU358092 | ? | Schizophrenia | Located in SZ- associated region 1p21.3 and co-expressed with mir-137; alters response to psychoactive drugs. | [182] |
| MIAT | Down | Schizophrenia, Fear-related anxiety | Increases pathogenic splice variants of DISC1 and ERBB4. Associated with risk SNPs rs1894720 and rs4274/Possibly interacts with BMI1, regulating Crybb1 expression. | [177,178,179] |
| BDNF-AS | ? | Depression | Repress BDNF expression, and neuronal outgrowth and differentiation. | [191,192] |
| MSNP1AS | Up | ASD | Decreases neurite number and length in neuronal progenitor cells; dysregulates the expression of genes involved in protein synthesis and chromatin remodelling. | [196] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cipolla, G.A.; De Oliveira, J.C.; Salviano-Silva, A.; Lobo-Alves, S.C.; Lemos, D.S.; Oliveira, L.C.; Jucoski, T.S.; Mathias, C.; Pedroso, G.A.; Zambalde, E.P.; et al. Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity. Non-Coding RNA 2018, 4, 13. https://doi.org/10.3390/ncrna4020013
Cipolla GA, De Oliveira JC, Salviano-Silva A, Lobo-Alves SC, Lemos DS, Oliveira LC, Jucoski TS, Mathias C, Pedroso GA, Zambalde EP, et al. Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity. Non-Coding RNA. 2018; 4(2):13. https://doi.org/10.3390/ncrna4020013
Chicago/Turabian StyleCipolla, Gabriel A., Jaqueline C. De Oliveira, Amanda Salviano-Silva, Sara C. Lobo-Alves, Debora S. Lemos, Luana C. Oliveira, Tayana S. Jucoski, Carolina Mathias, Gabrielle A. Pedroso, Erika P. Zambalde, and et al. 2018. "Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity" Non-Coding RNA 4, no. 2: 13. https://doi.org/10.3390/ncrna4020013
APA StyleCipolla, G. A., De Oliveira, J. C., Salviano-Silva, A., Lobo-Alves, S. C., Lemos, D. S., Oliveira, L. C., Jucoski, T. S., Mathias, C., Pedroso, G. A., Zambalde, E. P., & Gradia, D. F. (2018). Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity. Non-Coding RNA, 4(2), 13. https://doi.org/10.3390/ncrna4020013