Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition
Abstract
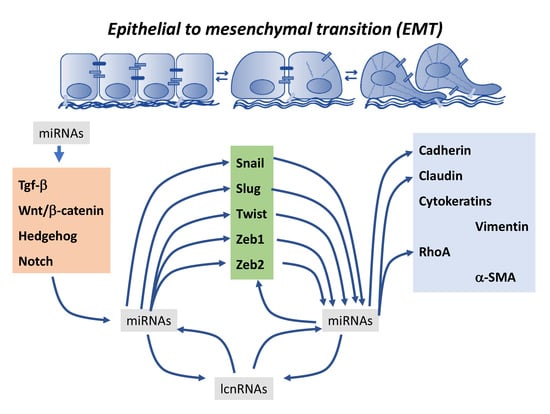
:1. Introduction
2. Contribution of microRNAs to Epithelial-To-Mesenchymal Transition Regulation
2.1. Regulation of Snail by microRNAs
2.2. Regulation of Slug by microRNAs
2.3. Co-Regulation of Snail and Slug by microRNAs
2.4. Regulation of Twist by microRNAs
2.5. Regulation of Zeb1 and Zeb2 by microRNAs
2.6. Co-Regulation of Epithelial-To-Mesenchymal Transition-Associated Transcription Factors by microRNAs
2.7. Regulation of Cell–Cell Contact and Cytoskeletal Proteins by microRNAs
3. Contribution of Long Non-Coding RNAs to Epithelial-To-Mesenchymal Transition Regulation
4. Cross-Talk between microRNAs and Long Non-Coding RNAs in Epithelial-To-Mesenchymal Transition
5. Conclusions and Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gonzalez, D.M.; Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014, 7, re8. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 78–96. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Garikapati, K.R.; Makani, V.K.K.; Nair, A.D.; Vangara, N.; Bhadra, U.; Pal Bhadra, M. Regulating BMI1 expression via miRNAs promote Mesenchymal to Epithelial Transition (MET) and sensitizes breast cancer cell to chemotherapeutic drug. PLoS ONE 2018, 13, e0190245. [Google Scholar] [CrossRef] [PubMed]
- Drago-García, D.; Espinal-Enríquez, J.; Hernández-Lemus, E. Network analysis of EMT and MET micro-RNA regulation in breast cancer. Sci. Rep. 2017, 7, 13534. [Google Scholar] [CrossRef] [PubMed]
- Ware, K.E.; Gilja, S.; Xu, S.; Shetler, S.; Jolly, M.K.; Wang, X.; Bartholf Dewitt, S.; Hish, A.J.; Jordan, S.; Eward, W.; et al. Induction of Mesenchymal-Epithelial Transitions in Sarcoma Cells. J. Vis. Exp. 2017, 122. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Chimenti, I.; Rosa, P.; Angelini, F.; Pagano, F.; Calogero, A.; Giacomello, A.; Messina, E. EMT/MET at the Crossroad of Stemness, Regeneration and Oncogenesis: The Ying-Yang Equilibrium Recapitulated in Cell Spheroids. Cancers 2017, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G.; Rath, B. Mesenchymal-Epithelial Transition and Circulating Tumor Cells in Small Cell Lung Cancer. Adv. Exp. Med. Biol. 2017, 994, 229–245. [Google Scholar] [PubMed]
- Barui, A.; Chowdhury, F.; Pandit, A.; Datta, P. Rerouting mesenchymal stem cell trajectory towards epithelial lineage by engineering cellular niche. Biomaterials 2018, 156, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Fritzenwanker, J.H.; Saina, M.; Technau, U. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev. Biol. 2004, 275, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Kraus, Y.; Technau, U. Gastrulation in the sea anemone Nematostella vectensis occurs by invagination and immigration: An ultrastructural study. Dev. Genes Evol. 2006, 216, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Smallhorn, M.; Murray, M.J.; Saint, R. The epithelial-mesenchymal transition of the Drosophila mesoderm requires the Rho GTP exchange factor Pebble. Development 2004, 131, 2641–2651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, S.; Miyagi, C.; Fukada, T.; Kagara, N.; Che, Y.S.; Hirano, T. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature 2004, 429, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Chuai, M.; Weijer, C.J. Regulation of cell migration during chick gastrulation. Curr. Opin. Genet. Dev. 2009, 19, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, Y.; Sukowati, E.W.; Alev, C.; Nakazawa, F.; Sheng, G. Involvement of dystroglycan in epithelial-mesenchymal transition during chick gastrulation. Cells Tissues Organs 2011, 193, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Patrakka, J.; Nukui, M.; Chi, L.; Niu, D.; Betsholtz, C.; Pikkarainen, T.; Vainio, S.; Tryggvason, K. Deficiency in Crumbs homolog 2 (Crb2) affects gastrulation and results in embryonic lethality in mice. Dev. Dyn. 2011, 240, 2646–2656. [Google Scholar] [CrossRef] [PubMed]
- Trask, M.C.; Tremblay, K.D.; Mager, J. Yin-Yang1 is required for epithelial-to-mesenchymal transition and regulation of Nodal signaling during mammalian gastrulation. Dev. Biol. 2012, 368, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Zohn, I.E.; Li, Y.; Skolnik, E.Y.; Anderson, K.V.; Han, J.; Niswander, L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 2006, 125, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Suzuki, K.; Tachibana, K.; Tanaka, H.; Yamada, G. Cessation of gastrulation is mediated by suppression of epithelial-mesenchymal transition at the ventral ectodermal ridge. Development 2007, 134, 4315–4324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakaya, Y.; Sukowati, E.W.; Wu, Y.; Sheng, G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 2008, 10, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Schoenwolf, G.C.; Yamada, G. The cessation of gastrulation: BMP signaling and EMT during and at the end of gastrulation. Cell Adhes. Migr. 2010, 4, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.; Deroo, T.; Fujita, Y.; Itasaki, N. A positive role of cadherin in Wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS ONE 2011, 6, e23899. [Google Scholar] [CrossRef] [PubMed]
- Martinovic, V.; Vukusic Pusic, T.; Restovic, I.; Bocina, I.; Filipovic, N.; Saraga-Babic, M.; Vukojevic, K. Expression of Epithelial and Mesenchymal Differentiation Markers in the Early Human Gonadal Development. Anat. Rec. 2017, 300, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Matoba, R.; Morizane, Y.; Shiode, Y.; Hirano, M.; Doi, S.; Toshima, S.; Araki, R.; Hosogi, M.; Yonezawa, T.; Shiraga, F. Suppressive effect of AMP-activated protein kinase on the epithelial-mesenchymal transition in retinal pigment epithelial cells. PLoS ONE 2017, 12, e0181481. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Wang, Y.; Zhang, H.; Li, L.; Bao, X. Dickkopf-1 inhibits Wnt3a-induced migration and epithelial-mesenchymal transition of human lens epithelial cells. Exp. Eye Res. 2017, 161, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zhu, Y.; Chen, C.; Chen, X.; Qin, Y.; Qu, B.; Luo, L.; Lin, H.; Wu, M.; Chen, W.; et al. Sprouty2 Suppresses Epithelial-Mesenchymal Transition of Human Lens Epithelial Cells through Blockade of Smad2 and ERK1/2 Pathways. PLoS ONE 2016, 11, e0159275. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.; Luo, C.; Zhang, X.; Ye, P.; Zhang, Y.; He, J.; Yao, K. Regulation of angiogenin expression and epithelial-mesenchymal transition by HIF-1α signaling in hypoxic retinal pigment epithelial cells. Biochim. Biophys. Acta 2016, 1862, 1594–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Kang, L.; Chen, J.; Xue, Y.; Yang, M.; Qin, B.; Yang, L.; Zhang, J.; Lu, H.; Guan, H. CtBP2 Regulates TGFβ2-Induced Epithelial-Mesenchymal Transition Through Notch Signaling Pathway in Lens Epithelial Cells. Curr. Eye Res. 2016, 41, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Petrash, J.M. Aldose Reductase Mediates Transforming Growth Factor β2 (TGF-β2)-Induced Migration and Epithelial-To-Mesenchymal Transition of Lens-Derived Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4198–4210. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiao, W.; Liu, X.; Zeng, M.; Luo, L.; Wu, M.; Ye, S.; Liu, Y. Blockade of Jagged/Notch pathway abrogates transforming growth factor β2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells. Curr. Mol. Med. 2014, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, M.; Xu, D.; Zhao, C.; Liu, G.; Wang, F. Overexpression of Snail in retinal pigment epithelial triggered epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2014, 446, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Bastiaans, J.; van Meurs, J.C.; van Holten-Neelen, C.; Nagtzaam, N.M.; van Hagen, P.M.; Chambers, R.C.; Hooijkaas, H.; Dik, W.A. Thrombin induces epithelial-mesenchymal transition and collagen production by retinal pigment epithelial cells via autocrine PDGF-receptor signaling. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8306–8314. [Google Scholar] [CrossRef] [PubMed]
- Pifer, P.M.; Farris, J.C.; Thomas, A.L.; Stoilov, P.; Denvir, J.; Smith, D.M.; Frisch, S.M. Grainyhead-like 2 inhibits the coactivator p300, suppressing tubulogenesis and the epithelial-mesenchymal transition. Mol. Biol. Cell 2016, 27, 2479–2492. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Picelli, S.; Torregrossa, R.; Colluto, L.; Ceol, M.; Del Prete, D.; D’Angelo, A.; Valle, G.; Anglani, F. Genes involved in TGFβ1-driven epithelial-mesenchymal transition of renal epithelial cells are topologically related in the human interactome map. BMC Genom. 2007, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Forino, M.; Torregrossa, R.; Ceol, M.; Murer, L.; Della Vella, M.; Del Prete, D.; D’Angelo, A.; Anglani, F. TGFβ1 induces epithelial-mesenchymal transition, but not myofibroblast transdifferentiation of human kidney tubular epithelial cells in primary culture. Int. J. Exp. Pathol. 2006, 87, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Chagraoui, J.; Lepage-Noll, A.; Anjo, A.; Uzan, G.; Charbord, P. Fetal liver stroma consists of cells in epithelial-to-mesenchymal transition. Blood 2003, 101, 2973–2982. [Google Scholar] [CrossRef] [PubMed]
- Rukstalis, J.M.; Habener, J.F. Snail2, a mediator of epithelial-mesenchymal transitions, expressed in progenitor cells of the developing endocrine pancreas. Gene Exp. Patterns 2007, 7, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.; Anderson, M.; Antin, P.B.; Limesand, S.W. One process for pancreatic β-cell coalescence into islets involves an epithelial-mesenchymal transition. J. Endocrinol. 2009, 203, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.J.; You, L.R.; Yu, C.T.; Hsu, W.H.; Tsai, M.J.; Tsai, S.Y. Endocardial cushion morphogenesis and coronary vessel development require chicken ovalbumin upstream promoter-transcription factor II. Arterioscler. Thromb. Vasc. Biol. 2012, 32, e135–e146. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, M.; Ikeda, K.; Nakatani, K.; Kawada, N.; Imanaka-Yoshida, K.; Yoshida, T.; Yamagishi, T.; Nakajima, Y. Rho kinases regulate endothelial invasion and migration during valvuloseptal endocardial cushion tissue formation. Dev. Dyn. 2006, 235, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Morabito, C.J.; Dettman, R.W.; Kattan, J.; Collier, J.M.; Bristow, J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev. Biol. 2001, 234, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, T.; Ando, K.; Nakamura, H. Roles of TGFβ and BMP during valvulo-septal endocardial cushion formation. Anat. Sci. Int. 2009, 84, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.R.; Robinson, J.Y.; Sanchez, N.S.; Townsend, T.A.; Arrieta, J.A.; Merryman, W.D.; Trykall, D.Z.; Olivey, H.E.; Hong, C.C.; Barnett, J.V. Common pathways regulate Type III TGFβ receptor-dependent cell invasion in epicardial and endocardial cells. Cell. Signal. 2016, 28, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Sridurongrit, S.; Larsson, J.; Schwartz, R.; Ruiz-Lozano, P.; Kaartinen, V. Signaling via the TGF-β type I receptor Alk5 in heart development. Dev. Biol. 2008, 322, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Niessen, K.; Fu, Y.; Chang, L.; Hoodless, P.A.; McFadden, D.; Karsan, A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008, 182, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Dyer, L.; Lockyer, P.; Wu, Y.; Saha, A.; Cyr, C.; Moser, M.; Pi, X.; Patterson, C. BMPER Promotes Epithelial-Mesenchymal Transition in the Developing Cardiac Cushions. PLoS ONE 2015, 10, e0139209. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, K.; Cooley, M.A.; Sugi, Y.; Barth, J.L.; Rasmussen, L.M.; Kern, C.B.; Argraves, K.M.; Argraves, W.S. Fibulin-1 suppresses endothelial to mesenchymal transition in the proximal outflow tract. Mech. Dev. 2015, 136, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.; Garside, V.C.; Fournier, M.; Smrz, J.; Vrljicak, P.; Umlandt, P.; Fuller, M.; Robertson, G.; Zhao, Y.; Tam, A.; et al. A Notch-dependent transcriptional hierarchy promotes mesenchymal transdifferentiation in the cardiac cushion. Dev. Dyn. 2014, 243, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Lee, S.; Bae, J.S.; Jee, J.G.; Cha, H.J.; Lee, Y.M. Thymosin β4 regulates cardiac valve formation via endothelial-mesenchymal transformation in zebrafish embryos. Mol. Cells 2014, 37, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, L.; Wadham, C.; Crocker, L.A.; Lardelli, M.; Khew-Goodall, Y. The protein tyrosine phosphatase Pez regulates TGFβ, epithelial-mesenchymal transition, and organ development. J. Cell Biol. 2007, 178, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Okagawa, H.; Markwald, R.R.; Sugi, Y. Functional BMP receptor in endocardial cells is required in atrioventricular cushion mesenchymal cell formation in chick. Dev. Biol. 2007, 306, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Fässler, R.; Mishina, Y.; Jiao, K.; Baldwin, H.S. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev. Biol. 2007, 301, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Liebner, S.; Cattelino, A.; Gallini, R.; Rudini, N.; Iurlaro, M.; Piccolo, S.; Dejana, E. β-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J. Cell Biol. 2004, 166, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Olivey, H.E.; Mundell, N.A.; Austin, A.F.; Barnett, J.V. Transforming growth factor-β stimulates epithelial-mesenchymal transformation in the proepicardium. Dev. Dyn. 2006, 235, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Compton, L.A.; Potash, D.A.; Mundell, N.A.; Barnett, J.V. Transforming growth factor-β induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev. Dyn. 2006, 235, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.R.; Sanchez, N.S.; Love, J.D.; Arrieta, J.A.; Hong, C.C.; Brown, C.B.; Austin, A.F.; Barnett, J.V. BMP2 signals loss of epithelial character in epicardial cells but requires the Type III TGFβ receptor to promote invasion. Cell. Signal. 2012, 24, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- Bax, N.A.; van Oorschot, A.A.; Maas, S.; Braun, J.; van Tuyn, J.; de Vries, A.A.; Groot, A.C.; Goumans, M.J. In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFβ-signaling and WT1. Basic Res. Cardiol. 2011, 106, 829–847. [Google Scholar] [CrossRef] [PubMed]
- Dokic, D.; Dettman, R.W. VCAM-1 inhibits TGFβ stimulated epithelial-mesenchymal transformation by modulating Rho activity and stabilizing intercellular adhesion in epicardial mesothelial cells. Dev. Biol. 2006, 299, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.C.; Travisano, S.; de la Pompa, J.L. Epithelial-to-mesenchymal transition in epicardium is independent of snail1. Genesis 2013, 51, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.L.; Baek, S.T.; Sung, C.Y.; Tallquist, M.D. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ. Res. 2011, 108, e15–e26. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M.; Nimura, K.; Mori, M.; Nakagami, H.; Kaneda, Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLoS ONE 2013, 8, e57829. [Google Scholar] [CrossRef] [PubMed]
- Von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 2011, 356, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ramírez, A.S.; Garay, E.; García-Carrancá, A.; Vázquez-Cuevas, F.G. The P2RY2 Receptor Induces Carcinoma Cell Migration and EMT Through Cross-Talk with Epidermal Growth Factor Receptor. J. Cell. Biochem. 2016, 117, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.R.; Kim, T.H.; Kim, Y.S.; Choi, K.C. Alteration of epithelial-mesenchymal transition markers in human normal ovaries and neoplastic ovarian cancers. Int. J. Oncol. 2015, 46, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Adham, S.A.; Al Harrasi, I.; Al Haddabi, I.; Al Rashdi, A.; Al Sinawi, S.; Al Maniri, A.; Ba-Omar, T.; Coomber, B.L. Immunohistological insight into the correlation between neuropilin-1 and epithelial-mesenchymal transition markers in epithelial ovarian cancer. J. Histochem. Cytochem. 2014, 62, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Thompson, E.W.; Quinn, M.A. Epithelial-mesenchymal interconversions in normal ovarian surface epithelium and ovarian carcinomas: An exception to the norm. J. Cell. Physiol. 2007, 213, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Rasti, A.; Madjd, Z.; Abolhasani, M.; Mehrazma, M.; Janani, L.; Saeednejad Zanjani, L.; Asgari, M. Cytoplasmic expression of Twist1, an EMT-related transcription factor, is associated with higher grades renal cell carcinomas and worse progression-free survival in clear cell renal cell carcinoma. Clin. Exp. Med. 2017, 18, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Landolt, L.; Eikrem, Ø.; Strauss, P.; Scherer, A.; Lovett, D.H.; Beisland, C.; Finne, K.; Osman, T.; Ibrahim, M.M.; Gausdal, G.; et al. Clear Cell Renal Cell Carcinoma is linked to Epithelial-to-Mesenchymal Transition and to Fibrosis. Physiol. Rep. 2017, 5, e13305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hong, Z.; Chai, Y.; Liu, Z.; Du, Y.; Li, Q.; Liu, Q. CSN5 promotes renal cell carcinoma metastasis and EMT by inhibiting ZEB1 degradation. Biochem. Biophys. Res. Commun. 2017, 488, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Feldkoren, B.; Hutchinson, R.; Rapoport, Y.; Mahajan, A.; Margulis, V. Integrin signaling potentiates transforming growth factor-β 1 (TGF-β1) dependent down-regulation of E-Cadherin expression—Important implications for epithelial to mesenchymal transition (EMT) in renal cell carcinoma. Exp. Cell Res. 2017, 355, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, T.; Hong, B.; Wang, F.; Zhou, C.; Du, X.; Chen, S.; Deng, X.; Duoerkun, S.; Li, Q.; et al. Vascular Endothelial Growth Inhibitor, a Cytokine of the Tumor Necrosis Factor Family, is Associated with Epithelial-Mesenchymal Transition in Renal Cell Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, C.; Daniele, S.; Romei, C.; Tavanti, L.; Neri, T.; Piano, I.; Celi, A.; Martini, C.; Trincavelli, M.L. The A(2B) Adenosine Receptor Modulates the Epithelial-Mesenchymal Transition through the Balance of cAMP/PKA and MAPK/ERK Pathway Activation in Human Epithelial Lung Cells. Front. Pharmacol. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Chang, S.; Ko, T.; Anker, J.; Agte, S.; Iams, W.; Choi, W.M.; Lee, K.; Cruz, M. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci. Rep. 2018, 8, 2918. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cheng, Y.; Gao, Y.; He, Z.; Zhou, W.; Chang, R.; Peng, Z.; Zheng, Y.; Duan, C.; Zhang, C. SH2B1 promotes epithelial-mesenchymal transition through the IRS1/β-catenin signaling axis in lung adenocarcinoma. Mol. Carcinog. 2018, 57, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Hu, S.; Tang, C.; Liu, G. Mesenchymal stem cells promote cell invasion and migration and autophagy-induced epithelial-mesenchymal transition in A549 lung adenocarcinoma cells. Cell Biochem. Funct. 2018, 36, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Ye, M.; Jing, P.; Xiong, J.; Han, Z.; Kong, J.; Li, M.; Lai, X.; Chang, N.; et al. FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snail protein. Cancer Lett. 2018, 419, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, C.; Wada, H.; Eguchi, H.; Hatano, H.; Gotoh, K.; Noda, T.; Yamada, D.; Asaoka, T.; Kawamoto, K.; Nagano, H.; et al. Clinical significance of CD13 and epithelial mesenchymal transition (EMT) markers in hepatocellular carcinoma. Jpn. J. Clin. Oncol. 2018, 48, 52–60. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Dong, W.; Hu, J.; Ren, X. AQP5 promotes hepatocellular carcinoma metastasis via NF-κB-regulated epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2017, 490, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Zhang, P.F.; Zhang, C.; Huang, X.Y.; Ding, Y.B.; Deng, B.; Bai, D.S.; Xu, Y.P. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med. 2018, 7, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, M.; Tsunedomi, R.; Yoshimura, K.; Hashimoto, N.; Matsukuma, S.; Ogihara, H.; Kanekiyo, S.; Iida, M.; Sakamoto, K.; Suzuki, N.; et al. Metastatic ability and the epithelial-mesenchyma transition in induced cancer stem-like hepatoma cells. Cancer Sci. 2018, 109, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- He, Y.M.; Xiao, Y.S.; Wei, L.; Zhang, J.Q.; Peng, C.H. CUL4B Promotes Metastasis and Proliferation in Pancreatic Cancer Cells by Inducing Epithelial-Mesenchymal Transition via the Wnt/β-catenin Signaling Pathway. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Puls, T.J.; Tan, X.; Whittington, C.F.; Voytik-Harbin, S.L. 3D collagen fibrillar microstructure guides pancreatic cancer cell phenotype and serves as a critical design parameter for phenotypic models of EMT. PLoS ONE 2017, 12, e0188870. [Google Scholar] [CrossRef] [PubMed]
- Nishino, H.; Takano, S.; Yoshitomi, H.; Suzuki, K.; Kagawa, S.; Shimazaki, R.; Shimizu, H.; Furukawa, K.; Miyazaki, M.; Ohtsuka, M. Grainyhead-like 2 (GRHL2) regulates epithelial plasticity in pancreatic cancer progression. Cancer Med. 2017, 6, 2686–2696. [Google Scholar] [CrossRef] [PubMed]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Natsuizaka, M.; Whelan, K.A.; Kagawa, S.; Tanaka, K.; Giroux, V.; Chandramouleeswaran, P.M.; Long, A.; Sahu, V.; Darling, D.S.; Que, J.; et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun. 2017, 8, 1758. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, X.; Bu, X.; Hou, N.; Chen, P. Alpha B-crystallin promotes the invasion and metastasis of colorectal cancer via epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2017, 489, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lang, T.; Jin, B.; Chen, F.; Zhang, Y.; Beuerman, R.W.; Zhou, L.; Zhang, Z. Luteolin inhibits colorectal cancer cell epithelial-to-mesenchymal transition by suppressing CREB1 expression revealed by comparative proteomics study. J. Proteom. 2017, 161, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.B.; Yan, J.S.; Yan, Y.Y.; Xie, T.C.; Xu, L.; Hu, G.H.; Xu, Y.F.; Liu, M. Downregulation of pigment epithelium-derived factor is associated with increased epithelial-mesenchymal transition in bladder cancer. Panminerva Med. 2017, 59, 9–14. [Google Scholar] [PubMed]
- Asnaghi, L.; Gezgin, G.; Tripathy, A.; Handa, J.T.; Merbs, S.L.; van der Velden, P.A.; Jager, M.J.; Harbour, J.W.; Eberhart, C.G. EMT-associated factors promote invasive properties of uveal melanoma cells. Mol. Vis. 2015, 21, 919–929. [Google Scholar] [PubMed]
- Lv, N.; Shan, Z.; Gao, Y.; Guan, H.; Fan, C.; Wang, H.; Teng, W. Twist1 regulates the epithelial-mesenchymal transition via the NF-κB pathway in papillary thyroid carcinoma. Endocrine 2016, 51, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, P. Suppression of DACH1 promotes migration and invasion of colorectal cancer via activating TGF-β-mediated epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2015, 460, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Keinan, O.; Kedan, A.; Gavert, N.; Selitrennik, M.; Kim, S.; Karn, T.; Becker, S.; Lev, S. The lipid-transfer protein Nir2 enhances epithelial-mesenchymal transition and facilitates breast cancer metastasis. J. Cell Sci. 2014, 127, 4740–4749. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bär, C.; Chatterjee, S.; Thum, T. Long Noncoding RNAs in Cardiovascular Pathology, Diagnosis, and Therapy. Circulation 2016, 134, 1484–1499. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Sandberg, M.B.; Eskildsen, T.; Nielsen, S.B.; Kalluri, R.; Sheikh, S.P. miR-21 promotes fibrogenic epithelial-to-mesenchymal transition of epicardial mesothelial cells involving Programmed Cell Death 4 and Sprouty-1. PLoS ONE 2013, 8, e56280. [Google Scholar] [CrossRef] [PubMed]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Nossent, A.Y.; Nielsen, S.B.; Sheikh, S.P. Islet-1 is a dual regulator of fibrogenic epithelial-to-mesenchymal transition in epicardial mesothelial cells. Exp. Cell Res. 2013, 319, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Lu, M.M.; Massera, D.; Epstein, J.A. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J. Biol. Chem. 2011, 286, 41036–41045. [Google Scholar] [CrossRef] [PubMed]
- Antonello, Z.A.; Reiff, T.; Ballesta-Illan, E.; Dominguez, M. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8-Escargot switch. EMBO J. 2015, 34, 2025–2041. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Zhao, Z.; Hong, S.H.; Bai, S.; He, Z.; Malik, F.; Xu, J.; Zhou, L.; Chen, W.; Martin-Trevino, R.; et al. Role of microRNA221 in regulating normal mammary epithelial hierarchy and breast cancer stem-like cells. Oncotarget 2015, 6, 3709–37021. [Google Scholar] [CrossRef] [PubMed]
- Salviano-Silva, A.; Lobo-Alves, S.C.; Almeida, R.C.; Malheiros, D.; Petzl-Erler, M.L. Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. Noncoding RNA 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.G.; Qiu, R.L.; Wu, Y.H.; Li, Z.X.; Xie, P.; Zhang, J.; Zhou, J.J.; Zeng, L.X.; Tang, J.; Maharjan, A.; et al. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver Int. 2014, 34, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikoff, N.; Attema, J.L.; Roslan, S.; Bert, A.G.; Schwarz, Q.P.; Gregory, P.A.; Goodall, G.J. Specificity protein 1 (Sp1) maintains basal epithelial expression of the miR-200 family: Implications for epithelial-mesenchymal transition. J. Biol. Chem. 2014, 289, 11194–11205. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Zhang, Y.H.; Li, R.B.; Zhou, L.Y.; An, T.; Zhang, R.C.; Zhai, M.; Huang, Y.; Yan, K.W.; Dong, Y.H.; et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 2018, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, L.; Bai, M.; Zhang, L.; Ma, F.; Yang, X.; Sun, S. Hypoxia-induced activation of Twist/miR-214/E-cadherin axis promotes renal tubular epithelial cell mesenchymal transition and renal fibrosis. Biochem. Biophys. Res. Commun. 2018, 495, 2324–2330. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.W.; Ng, S.W. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2017, 18, 1207. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhen, S.; Ye, Z.; Lu, J.; Wang, L.; Li, P.; Li, J.; Zheng, X.; Li, H.; Chen, W.; et al. A Feedback Loop Between miR-30a/c-5p and DNMT1 Mediates Cisplatin Resistance in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2017, 41, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ma, X.; Li, H.; Gao, Y.; Huang, Q.; Zhang, Y.; Bao, X.; Du, Q.; Luo, G.; Liu, K.; et al. miR-122 promotes metastasis of clear-cell renal cell carcinoma by downregulating Dicer. Int. J. Cancer 2018, 142, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chen, N.; Xiao, R.; Wang, W.; Pan, Z. miR-144-3p serves as a tumor suppressor for renal cell carcinoma and inhibits its invasion and metastasis by targeting MAP3K8. Biochem. Biophys. Res. Commun. 2016, 480, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Machackova, T.; Mlcochova, H.; Stanik, M.; Dolezel, J.; Fedorko, M.; Pacik, D.; Poprach, A.; Svoboda, M.; Slaby, O. MiR-429 is linked to metastasis and poor prognosis in renal cell carcinoma by affecting epithelial-mesenchymal transition. Tumour Biol. 2016, 37, 14653–14658. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, J.; Xu, R.; Zhu, X.; Liu, L.; Zhao, X. MicroRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in clear cell renal cells. Mol. Med. Rep. 2016, 13, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, X.; Liang, H.; Jin, Z. Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem. Biol. Interact. 2018, 286, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, Q.; Li, L.; Ma, R. The long non-coding RNA CRNDE promotes cervical cancer cell growth and metastasis. Biol. Chem. 2017, 399, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, P.; Kulkarni, P.; Majid, S.; Varahram, S.; Hashimoto, Y.; Bhat, N.S.; Shiina, M.; Deng, G.; Saini, S.; Tabatabai, Z.L.; et al. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol. Cancer Ther. 2018, 17, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.; Deng, X.; Luo, M.; Wang, X.; Hu, H.; Liu, C.; Zhong, M. Long noncoding RNA H19 promotes transforming growth factor-β-induced epithelial–mesenchymal transition by acting as a competing endogenous RNA of miR-370-3p in ovarian cancer cells. Onco Targets Ther. 2018, 11, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; He, H.; Sui, W.; Zhang, J.; Zhang, S.; Yang, D. Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR-138 and releasing EZH2 in oral squamous cell carcinoma. Int. J. Oncol. 2018, 52, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Shen, D.; Zhang, M.; Jiang, N.; Sun, F.; Yuan, S.; Wan, K. MiR-488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am. J. Cancer Res. 2017, 7, 2070–2080. [Google Scholar] [PubMed]
- Qiu, L.; Tang, Q.; Li, G.; Chen, K. Long non-coding RNAs as biomarkers and therapeutic targets: Recent insights into hepatocellular carcinoma. Life Sci. 2017, 191, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Tseng, J.H.; Bisogna, M.; Hoang, L.N.; Olvera, N.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Sood, A.K.; Levine, D.A.; Jelinic, P. miR-200c-driven Mesenchymal-To-Epithelial Transition is a Therapeutic Target in Uterine Carcinosarcomas. Sci. Rep. 2017, 7, 3614. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ye, X.L.; Xu, J.; Cao, M.G.; Fang, Z.Y.; Li, L.Y.; Guan, G.H.; Liu, Q.; Qian, Y.H.; Xie, D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 2017, 10, eaak9557. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, F.; Jia, L.T.; Yang, A.G. Missing Links in Epithelial-Mesenchymal Transition: Long Non-Coding RNAs Enter the Arena. Cell. Physiol. Biochem. 2017, 44, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, H.; Liu, X.; Xu, D.; Wang, F. MicroRNA-29b regulates TGF-β1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells by targeting AKT2. Exp. Cell Res. 2016, 345, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Turcatel, G.; Rubin, N.; El-Hashash, A.; Warburton, D. MIR-99a and MIR-99b modulate TGF-β induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS ONE 2012, 7, e31032. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, E.; Vasilaki, E.; Vorvis, C.; Iliopoulos, D.; Moustakas, A.; Kardassis, D.; Stournaras, C. Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-β and miR-24: Role in epithelial-to-mesenchymal transition. Oncogene 2012, 31, 2862–2875. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Joo, C.K. MicroRNA-124 Controls Transforming Growth Factor β1-Induced Epithelial-Mesenchymal Transition in the Retinal Pigment Epithelium by Targeting RHOG. Investig. Ophthalmol. Vis. Sci. 2016, 57, 12–22. [Google Scholar]
- Suzuki, T.; Mizutani, K.; Minami, A.; Nobutani, K.; Kurita, S.; Nagino, M.; Shimono, Y.; Takai, Y. Suppression of the TGF-β1-induced protein expression of SNAI1 and N-cadherin by miR-199a. Genes Cells. 2014, 19, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, J.; Lei, X.; Zhu, Z.; Pei, C.; Qin, L. MicroRNA-486-5p suppresses TGF-β2-induced proliferation, invasion and epithelial-mesenchymal transition of lens epithelial cells by targeting Smad2. J. Biosci. 2017, 42, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lv, Y.; Ouyang, Y.; Liu, S.; Li, D. The Role of miR-497/EIF3A Axis in TGFβ1-Induced Epithelial-Mesenchymal Transition and Extracellular Matrix in Rat Alveolar Epithelial Cells and Pulmonary Fibroblasts. J. Cell. Biochem. 2017, 118, 3401–3408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.W.; Wang, E.W.; Li, L.X.; Yi, S.H.; Li, L.C.; Xu, F.L.; Wang, D.L.; Wu, Y.Z.; Nian, W.Q. A regulatory loop involving miR-29c and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget 2016, 7, 85905–85916. [Google Scholar] [PubMed]
- Li, D.P.; Fan, J.; Wu, Y.J.; Xie, Y.F.; Zha, J.M.; Zhou, X.M. MiR-155 up-regulated by TGF-β promotes epithelial-mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am. J. Transl. Res. 2017, 9, 2956–2965. [Google Scholar] [PubMed]
- Xu, M.; Wang, G.; Zhou, H.; Cai, J.; Li, P.; Zhou, M.; Lu, Y.; Jiang, X.; Huang, H.; Zhang, Y.; et al. TGF-β1-miR-200a-PTEN induces epithelial-mesenchymal transition and fibrosis of pancreatic stellate cells. Mol. Cell. Biochem. 2017, 431, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; He, B.X.; Cai, J.H.; Tao, Q.; King-Yin Lam, A. MicroRNA-27a-3p Modulates the Wnt/β-Catenin Signaling Pathway to Promote Epithelial-Mesenchymal Transition in Oral Squamous Carcinoma Stem Cells by Targeting SFRP1. Sci. Rep. 2017, 7, 44688. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wang, J.; Han, J.; Luo, D.; Sun, Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing Wnt/β-cadherin signaling pathway. Exp. Cell Res. 2017, 360, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.W.; Xiao, J.Q.; Li, Z.Y.; Zheng, Y.C.; Zhang, N. Effects of microRNA-135a on the epithelial-mesenchymal transition, migration and invasion of bladder cancer cells by targeting GSK3β through the Wnt/β-catenin signaling pathway. Exp. Mol. Med. 2018, 50, e429. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Sun, W.; Xia, L.; Zhao, A.; Yu, Y.; Zhao, L.; Wang, H.; Huang, C.; Sun, S. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS ONE 2012, 7, e30771. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Katoh, M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int. J. Mol. Med. 2008, 22, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Khanbabaei, H.; Teimoori, A.; Mohammadi, M. The interplay between microRNAs and Twist1 transcription factor: A systematic review. Tumour Biol. 2016, 37, 7007–7019. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Veronese, A.; Pichiorri, F.; Lee, T.J.; Jeon, Y.J.; Volinia, S.; Pineau, P.; Marchio, A.; Palatini, J.; Suh, S.S.; et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011, 208, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Lagendijk, A.K.; Goumans, M.J.; Burkhard, S.B.; Bakkers, J. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 2011, 109, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Bonet, F.; Dueñas, Á.; López-Sánchez, C.; García-Martínez, V.; Aránega, A.E.; Franco, D. MiR-23b and miR-199a impair epithelial-to-mesenchymal transition during atrioventricular endocardial cushion formation. Dev. Dyn. 2015, 244, 1259–1275. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, K.; Ma, G.K.; Kuhnert, F.J.; Kuo, C.J.; Chang, C.P. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev. Biol. 2010, 347, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Mudduluru, G.; Ceppi, P.; Muppala, S.; Kozlowski, M.; Niklinski, J.; Papotti, M.; Allgayer, H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int. J. Cancer 2012, 130, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhao, L.; Li, J.; Chen, W.; Li, X. miR-30d Blocked Transforming Growth Factor β1-Induced Epithelial-Mesenchymal Transition by Targeting Snail in Ovarian Cancer Cells. Int. J. Gynecol. Cancer 2015, 25, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.P.; Zeng, C.; Xu, L.; Gong, J.; Fang, J.H.; Zhuang, S.M. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene 2014, 33, 4069–4076. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Ma, X.; Li, X.; Dong, H.; Yi, J.; Zeng, W.; Yang, Z. miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting Snail. Oncol. Rep. 2015, 34, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Long, J.; Du, R.; Ge, C.; Guo, K.; Xu, Y. miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 2016, 37, 8327–8335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, T.; Yang, K.; Zhang, M.; Wang, K. miR-486-5p suppresses prostate cancer metastasis by targeting Snail and regulating epithelial-mesenchymal transition. Onco Targets Ther. 2016, 9, 6909–6914. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, H.; Li, M.; Zhang, X.; Liu, Y.; Wang, Y. MicroRNA-374c-5p regulates the invasion and migration of cervical cancer by acting on the Foxc1/snail pathway. Biomed. Pharmacother. 2017, 94, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhu, Y.; Liang, Z.; Li, S.; Xu, X.; Wang, X.; Wu, J.; Hu, Z.; Meng, S.; Liu, B.; et al. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3β/Snail signaling. Cell Death Dis. 2016, 7, e2088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, M.; Chen, Q.; Zhang, Q. Downregulation of microRNA-145 promotes epithelial-mesenchymal transition via regulating Snail in osteosarcoma. Cancer Gene Ther. 2017, 24, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.Y.; Yoo, J.O.; An, H.J.; Bae, I.H.; Park, M.J.; Kim, J.; Han, Y.H. miR-5003-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization and direct targeting of E-cadherin. J. Mol. Cell Biol. 2016, 8, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Yu, J.C.; Hsieh, Y.H.; Yao, C.C.; Chao, J.I.; Chen, P.M.; Hsieh, H.Y.; Hsiung, C.N.; Chu, H.W.; Shen, C.Y.; et al. MicroRNA-30a increases tight junction protein expression to suppress the epithelial-mesenchymal transition and metastasis by targeting Slug in breast cancer. Oncotarget 2016, 7, 16462–16478. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, X.; Cai, X.; Huang, C.; Zheng, M. miR-497 inhibits epithelial mesenchymal transition in breast carcinoma by targeting Slug. Tumour Biol. 2016, 37, 7939–7950. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.H.; Wei, Y.P.; Shen, N.J.; Wang, Z.C.; Kan, T.; Yu, W.L.; Yi, B.; Zhang, Y.J. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell. Physiol. Biochem. 2013, 32, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Zhang, Z.G.; Ding, Z.Y.; Liang, H.F.; Song, J.; Tan, X.L.; Wu, J.J.; Li, G.Z.; Zeng, Z.; Zhang, B.X.; et al. MicroRNA-630 suppresses tumor metastasis through the TGF-β-miR-630-Slug signaling pathway and correlates inversely with poor prognosis in hepatocellular carcinoma. Oncotarget 2016, 7, 22674–22686. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Bai, Y.; Qiu, S.; Zheng, L.; Huang, L.; Liu, T.; Wang, X.; Liu, Y.; Xu, N.; Yan, X.; et al. MiR-203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial-mesenchymal transition via SNAI2. Oncotarget 2015, 6, 8914–8928. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiang, G.; Zhou, J.; Wang, H.; Gong, Z.; Zhang, Z.; Min, K.; Zhu, H.; Tan, Y. Down-regulation of miR-140 induces EMT and promotes invasion by targeting Slug in esophageal cancer. Cell. Physiol. Biochem. 2014, 34, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, J.; Wang, X.; Meng, S.; Shen, J.; Wang, S.; Xu, X.; Xie, B.; Liu, B.; Xie, L. MiR-22 suppresses epithelial-mesenchymal transition in bladder cancer by inhibiting Snail and MAPK1/Slug/vimentin feedback loop. Cell Death Dis. 2018, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Jiang, Y.P.; Chen, W.; Li, K.D.; Liu, X.; Gao, S.Y.; Feng, H.; Wang, S.S.; Jiang, J.; Ma, X.R.; et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget 2015, 6, 6797–6810. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Hünten, S.; Kaller, M.; Menssen, A.; Götz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hünten, S.; Hermeking, H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.H.; Tsai, M.F.; Gow, C.H.; Wu, S.G.; Liu, Y.N.; Chang, Y.L.; Yu, S.L.; Tsai, H.C.; Lin, S.W.; Chen, Y.W.; et al. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. 2017, 402, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jang, K.; Miller, P.; Picon-Ruiz, M.; Yeasky, T.M.; El-Ashry, D.; Slingerland, J.M. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene 2017, 36, 5199–5211. [Google Scholar] [CrossRef] [PubMed]
- Findlay, V.J.; Wang, C.; Nogueira, L.M.; Hurst, K.; Quirk, D.; Ethier, S.P.; Staveley O’Carroll, K.F.; Watson, D.K.; Camp, E.R. SNAI2 modulates colorectal cancer 5-fluorouracil sensitivity through miR145 repression. Mol. Cancer Ther. 2014, 13, 2713–2726. [Google Scholar] [CrossRef] [PubMed]
- Moes, M.; Le Béchec, A.; Crespo, I.; Laurini, C.; Halavatyi, A.; Vetter, G.; Del Sol, A.; Friederich, E. A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE 2012, 7, e35440. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Li, X.; Liang, X.; Li, L.; Cao, B.; Wang, B.; Ma, J.; Ding, F.; Wang, X.; Pang, D.; et al. MicroRNA-182 drives colonization and macroscopic metastasis via targeting its suppressor SNAI1 in breast cancer. Oncotarget 2017, 8, 4629–4641. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.N.; Yin, J.J.; Abou-Kheir, W.; Hynes, P.G.; Casey, O.M.; Fang, L.; Yi, M.; Stephens, R.M.; Seng, V.; Sheppard-Tillman, H.; et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013, 32, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Park, S.I.; McCauley, L.K.; Wang, C.Y. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013, 288, 10241–10253. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Huang, Z.; Chen, X.; Chen, S. miR-98 inhibits expression of TWIST to prevent progression of non-small cell lung cancers. Biomed. Pharmacother. 2017, 89, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hou, J.; Li, Z.; Zheng, Z.; Wei, J.; Song, D.; Hu, T.; Wu, Q.; Yang, J.Y.; Cai, J.C. miR-15a-3p and miR-16-1-3p Negatively Regulate Twist1 to Repress Gastric Cancer Cell Invasion and Metastasis. Int. J. Biol. Sci. 2017, 13, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, X.; Liu, F.; Fu, Y. microRNA-335 inhibits colorectal cancer HCT116 cells growth and epithelial-mesenchymal transition (EMT) process by targeting Twist1. Pharmazie 2017, 72, 475–481. [Google Scholar] [PubMed]
- Liu, S.; Cui, J.; Liao, G.; Zhang, Y.; Ye, K.; Lu, T.; Qi, J.; Wan, G. MiR-137 regulates epithelial-mesenchymal transition in gastrointestinal stromal tumor. Tumour Biol. 2014, 35, 9131–9138. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Sun, H.; Sun, B.; Zhu, D.; Zhao, X.; Wang, Y.; Gu, Q.; Dong, X.; Liu, F.; Zhang, Y.; et al. miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: An essential role for Twist-1 in HCC. Sci. Rep. 2016, 6, 23091. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Kaneuchi, M.; Watari, H.; Sudo, S.; Sakuragi, N. MicroRNA-106b modulates epithelial-mesenchymal transition by targeting TWIST1 in invasive endometrial cancer cell lines. Mol. Carcinog. 2014, 53, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, H.; Yin, X.; Long, L.; Xie, C.; Liu, Y.; Hui, L.; Lin, X.; Fang, Y.; Cao, Y.; et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene 2016, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Deng, R.; Zhang, H.; Dou, J.; Yuan, H.; Hou, G.; Du, Y.; Chen, Q.; Yu, J. miR-186 suppresses prostate cancer progression by targeting Twist1. Oncotarget 2016, 7, 33136–33151. [Google Scholar] [PubMed]
- Yu, J.; Xie, F.; Bao, X.; Chen, W.; Xu, Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol. Cancer 2014, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y.; Hou, Y.; Yang, Q.; Chen, S.; Wang, X.; Wang, Z.; Yang, Y.; Chen, C.; Wang, Z.; et al. The PDGF-D/miR-106a/Twist1 pathway orchestrates epithelial-mesenchymal transition in gemcitabine resistance hepatoma cells. Oncotarget 2015, 6, 7000–7010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gong, W.; Zuo, B.; Chu, B.; Tang, Z.; Zhang, Y.; Yang, Y.; Zhou, D.; Weng, M.; Qin, Y.; et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget 2016, 7, 78640–78652. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Dang, B.L.; Huang, J.Z.; Chen, M.; Wu, D.; Xu, M.L.; Li, R.; Yan, G.R. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cáncer. Oncotarget 2015, 6, 32701–32712. [Google Scholar] [PubMed]
- Yu, Y.; Zhao, Y.; Sun, X.H.; Ge, J.; Zhang, B.; Wang, X.; Cao, X.C. Down-regulation of miR-129-5p via the Twist1-Snail feedback loop stimulates the epithelial-mesenchymal transition and is associated with poor prognosis in breast cancer. Oncotarget 2015, 6, 34423–34436. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.F.; Zhou, C.F.; Wu, X.G.; He, L.N.; Wu, L.F.; Chen, X.J.; Yan, R.M.; Zhong, M.; Yu, Y.H.; Liang, L.; et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis. 2017, 8, 3220. [Google Scholar] [CrossRef] [PubMed]
- Drasin, D.J.; Guarnieri, A.L.; Neelakantan, D.; Kim, J.; Cabrera, J.H.; Wang, C.A.; Zaberezhnyy, V.; Gasparini, P.; Cascione, L.; Huebner, K.; et al. TWIST1-Induced miR-424 Reversibly Drives Mesenchymal Programming while Inhibiting Tumor Initiation. Cancer Res. 2015, 75, 1908–1921. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Karaayvaz, M.; Jia, N.; Kaneuchi, M.; Hamada, J.; Watari, H.; Sudo, S.; Ju, J.; Sakuragi, N. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene 2013, 32, 3286–3295. [Google Scholar] [CrossRef] [PubMed]
- Al-Khalaf, H.H.; Aboussekhra, A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. J. Biol. Chem. 2014, 289, 31433–31447. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Thiery, J.P. Loss of Git2 induces epithelial-mesenchymal transition by miR146a-Cnot6L-controlled expression of Zeb1. J. Cell Sci. 2013, 126, 2740–2746. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Yamaguchi, N.; Ohkawa, K.; Yoshida, K. Proteasome inhibitor-resistant cells cause EMT-induction via suppression of E-cadherin by miR-200 and ZEB1. Int. J. Oncol. 2015, 46, 2251–2260. [Google Scholar] [CrossRef] [PubMed]
- Filios, S.R.; Xu, G.; Chen, J.; Hong, K.; Jing, G.; Shalev, A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J. Biol. Chem. 2014, 289, 36275–36283. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, C.; Shi, M.; Wang, F.; Chen, X.; Diao, D.; Hu, M.; Yu, M.; Qian, L.; Guo, N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 2013, 32, 5272–5282. [Google Scholar] [CrossRef] [PubMed]
- Grassian, A.R.; Lin, F.; Barrett, R.; Liu, Y.; Jiang, W.; Korpal, M.; Astley, H.; Gitterman, D.; Henley, T.; Howes, R.; et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT). J. Biol. Chem. 2012, 287, 42180–42194. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Jiang, L.; Zhou, Y.; Qiu, W.; Fang, L.; Tan, R.; Wen, P.; Yang, J. The miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am. J. Physiol. Ren. Physiol. 2012, 302, F369–F379. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.B.; Zheng, L.; Yan, R.; Yang, J.; Ning, J.; Peng, L.; Zhou, Q.; Chen, L. miR302a-3p May Modulate Renal Epithelial-Mesenchymal Transition in Diabetic Kidney Disease by Targeting ZEB1. Nephron 2018, 138, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Peng, M.L.; Yeh, K.T.; Tsai, Y.Y.; Chiang, C.C.; Cheng, Y.W. Inactivation of p53 in pterygium influence miR-200a expression resulting in ZEB1/ZEB2 up-regulation and EMT processing. Exp. Eye Res. 2016, 146, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Díaz-López, A.; Díaz-Martín, J.; Moreno-Bueno, G.; Cuevas, E.P.; Santos, V.; Olmeda, D.; Portillo, F.; Palacios, J.; Cano, A. Zeb1 and Snail1 engage miR-200f transcriptional and epigenetic regulation during EMT. Int. J. Cancer 2015, 136, E62–E73. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Li, M.; An, J.; Zhao, B.; Zhong, W.; Gu, Q.; Cao, L.; Yang, H.; Hu, C. MicroRNA-33b inhibits lung adenocarcinoma cell growth, invasion, and epithelial-mesenchymal transition by suppressing Wnt/β-catenin/ZEB1 signaling. Int. J. Oncol. 2015, 47, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Zhang, C.; Liu, G.; Gu, R.; Wu, H. MicroRNA-126 Inhibits Proliferation, Migration, Invasion, and EMT in Osteosarcoma by Targeting ZEB1. J. Cell. Biochem. 2017, 118, 3765–3774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, R.; Xu, S.; Li, Y.; Zhao, P.; Dong, W.; Liu, Z.; Zhao, Q.; Tan, B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim. Biophys. Sin. 2018, 50, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Yang, Z.; Ye, W.; Xu, H.; Hua, X. MicroRNA-150 predicts a favorable prognosisin patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS ONE 2014, 9, e103965. [Google Scholar]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 6, 19943. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.D.; Ye, X.M.; Zhang, M.Y.; Zhu, H.Y.; Xi, W.J.; Huang, X.; Zhao, J.; Gu, B.; Zheng, G.X.; Yang, A.G.; et al. MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int. J. Cancer 2014, 135, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, H.; Han, X.; Ma, J.; Zhou, Y.; Chen, Z.; Zhou, H.; Xu, H.; Sun, Z.; Kong, B.; et al. ΔNp63α attenuates tumor aggressiveness by suppressing miR-205/ZEB1-mediated epithelial-mesenchymal transition in cervical squamous cell carcinoma. Tumour Biol. 2016, 37, 10621–10632. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, X.; Hu, B.; Wang, X.J.; Wang, Q.; Wang, W.L. The effects of Micro-429 on inhibition of cervical cancer cells through targeting ZEB1 and CRKL. Biomed. Pharmacother. 2016, 80, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, H.; Liu, Y.; Liu, W.; Liu, M.; Tang, H. miR-484 suppresses proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells. Cancer Cell Int. 2017, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Niu, Y.; Zhu, S.; Jin, Y.; Deng, S.; Chen, J.; Liu, Y.; He, C.; Yin, T.; et al. MiR-652 inhibits acidic microenvironment-induced epithelial-mesenchymal transition of pancreatic cancer cells by targeting ZEB1. Oncotarget 2015, 6, 39661–39675. [Google Scholar] [CrossRef] [PubMed]
- El Bezawy, R.; Cominetti, D.; Fenderico, N.; Zuco, V.; Beretta, G.L.; Dugo, M.; Arrighetti, N.; Stucchi, C.; Rancati, T.; Valdagni, R.; et al. miR-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR-ZEB1 axis. Cancer Lett. 2017, 395, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Majid, S.; Dar, A.A.; Saini, S.; Deng, G.; Chang, I.; Greene, K.; Tanaka, Y.; Dahiya, R.; Yamamura, S. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS ONE 2013, 8, e67686. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Kong, K.K.; Xu, X.K.; Chen, C.; Li, H.; Wang, F.Y.; Peng, X.F.; Zhang, Z.; Li, P.; Li, J.L.; et al. Downregulation of miR-205 is associated with glioblastoma cell migration, invasion, and the epithelial-mesenchymal transition, by targeting ZEB1 via the Akt/mTOR signaling pathway. Int. J. Oncol. 2018, 52, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Chandra Mangalhara, K.; Manvati, S.; Saini, S.K.; Ponnusamy, K.; Agarwal, G.; Abraham, S.K.; Bamezai, R.N.K. ERK2-ZEB1-miR-101-1 axis contributes to epithelial-mesenchymal transition and cell migration in cancer. Cancer Lett. 2017, 391, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Barbáchano, A.; Fernández-Barral, A.; Pereira, F.; Segura, M.F.; Ordóñez-Morán, P.; Carrillo-de Santa Pau, E.; González-Sancho, J.M.; Hanniford, D.; Martínez, N.; Costales-Carrera, A.; et al. SPROUTY-2 represses the epithelial phenotype of colon carcinoma cells via upregulation of ZEB1 mediated by ETS1 and miR-200/miR-150. Oncogene 2016, 35, 2991–3003. [Google Scholar] [CrossRef] [PubMed]
- Ungewiss, C.; Rizvi, Z.H.; Roybal, J.D.; Peng, D.H.; Gold, K.A.; Shin, D.H.; Creighton, C.J.; Gibbons, D.L. The microRNA-200/Zeb1 axis regulates ECM-dependent β1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci. Rep. 2016, 6, 18652. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Mei, Y.; Hawthorn, L.; Cowell, J.K. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene 2014, 33, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Das, R.; Townley, S.L.; Coutinho, I.; Hanson, A.R.; Centenera, M.M.; Stylianou, N.; Sweeney, K.; Soekmadji, C.; Jovanovic, L.; et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene 2017, 36, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.B.; Luo, H.P.; Shi, Q.; Hao, Z.N.; Ding, Y.; Wang, Q.S.; Li, S.B.; Xiao, G.C.; Tong, S.L. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J. Gastroenterol. 2014, 20, 6515–6522. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Li, Y.; Fang, N.; Liu, B.; Zu, L.; Chang, R.; Li, X.; Zhou, Q. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS ONE 2014, 9, e91827. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, M.; Shi, Y.; Luo, B.; Gong, G.; Li, J.; Wang, J.; Zhao, W.; Zi, Y.; Wu, X.; et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol. Rep. 2015, 34, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Yang, Z.; Zhang, P.; Liu, Y.; Shao, G. miR-154 inhibits migration and invasionof human non-small cell lung cancer by targeting ZEB2. Oncol. Lett. 2016, 12, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Fu, Z.; Gao, L.; Zhou, J.; Deng, X.; Luo, X.; Fang, W.; Luo, R. Direct interaction between miR-203 and ZEB2 suppresses epithelial-mesenchymal transition signaling and reduces lung adenocarcinoma chemoresistance. Acta Biochim. Biophys. Sin. 2016, 48, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Zhou, Y.; Yang, H.; Li, L.; Deng, X.; Cheng, C.; Xie, Y.; Luo, X.; Fang, W.; Liu, Z. Adirectly negative interaction of miR-203 and ZEB2 modulates tumor stemness and chemotherapy resistance in nasopharyngeal carcinoma. Oncotarget 2016, 7, 67288–67301. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Wu, Z.; Lin, L.; Zhou, M.; Wang, L.; Ma, H.; Xia, J.; Bin, J.; Liao, Y.; Liao, W. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget 2015, 6, 15222–15234. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, J.; Yuan, X.; Zhao, J.; Zhang, Z.; Weng, L.; Liu, J. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int. J. Oncol. 2016, 48, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Kurashige, J.; Kamohara, H.; Watanabe, M.; Hiyoshi, Y.; Iwatsuki, M.; Tanaka, Y.; Kinoshita, K.; Saito, S.; Baba, Y.; Baba, H. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann. Surg. Oncol. 2012, 19, S656–S664. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, L.; Li, C.; Zhao, L.; Ma, Y.; Zheng, H.; Li, Z.; Zhang, Y.; Wang, R.; Liu, Y.; et al. Ubiquitin ligase Cbl-b represses IGF-I-induced epithelial mesenchymal transition via ZEB2 and microRNA-200c regulation in gastric cancer cells. Mol. Cancer 2014, 13, 136. [Google Scholar] [CrossRef] [PubMed]
- Jiao, A.; Sui, M.; Zhang, L.; Sun, P.; Geng, D.; Zhang, W.; Wang, X.; Li, J. MicroRNA-200c inhibits the metastasis of non-small cell lung cancer cells by targeting ZEB2, anepithelial-mesenchymal transition regulator. Mol. Med. Rep. 2016, 13, 3349–3355. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sánchez-Tilló, E.; Lu, X.; Huang, L.; Clem, B.; Telang, S.; Jenson, A.B.; Cuatrecasas, M.; Chesney, J.; Postigo, A.; et al. The ZEB1 transcription factor acts in a negative feedback loop with miR200 downstream of Ras and Rb1 to regulate Bmi1 expression. J. Biol. Chem. 2014, 289, 4116–4125. [Google Scholar] [CrossRef] [PubMed]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Diepenbruck, M.; Tiede, S.; Saxena, M.; Ivanek, R.; Kalathur, R.K.R.; Lüönd, F.; Meyer-Schaller, N.; Christofori, G. miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nat. Commun. 2017, 8, 1168. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.K.; Yu, Y.; Xie, Y.G.; Wang, J.; Mao, J.F.; Zhang, B.; Wang, X.; Cao, X.C. miR-340 and ZEB1 negative feedback loop regulates TGF-β- mediated breast cancer progression. Oncotarget 2016, 7, 26016–26026. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Wang, M.; Guo, W.; Huang, S.; Wang, Z.; Zhao, X.; Du, H.; Song, L.; Peng, X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cáncer cells. Cell Tissue Res. 2014, 358, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Cogdell, D.; Hu, L.; Yang, D.; Sood, A.K.; Xue, F.; Zhang, W. MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol. Rep. 2014, 31, 2021–2028. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Cong, N.; Du, P.; Zhang, A.; Shen, F.; Su, J.; Pu, P.; Wang, T.; Zjang, J.; Kang, C.; Zhang, Q. Downregulated microRNA-200a promotes EMT and tumor growth through the Wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol. Rep. 2013, 29, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Eggers, J.C.; Martino, V.; Reinbold, R.; Schäfer, S.D.; Kiesel, L.; Starzinski-Powitz, A.; Schüring, A.N.; Kemper, B.; Greve, B.; Götte, M. microRNA miR-200b affects proliferation, invasiveness and stemness of endometriotic cells by targeting ZEB1, ZEB2 and KLF4. Reprod. Biomed. Online 2016, 32, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Lin, Y.; Zhang, H.; Wu, D. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem. Biophys. Res. Commun. 2015, 463, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Park, M.K.; Park, J.H.; Lee, H.J.; Shin, D.H.; Kang, Y.; Lee, C.H.; Kong, G. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene 2014, 33, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Ni, K.; Ke, J.; Zhang, W.; Feng, Y.; Mao, Q. miR-448 inhibits the epithelial-mesenchymal transition in breast cancer cells by directly targeting the E-cadherin repressor ZEB1/2. Exp. Biol. Med. 2018, 243, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Zheng, Y.; Zhao, Y.; Xiu, X.; Wang, J. miR-590-3p suppresses cancer cell migration, invasion and epithelial-mesenchymal transition in glioblastoma multiforme by targeting ZEB1 and ZEB2. Biochem. Biophys. Res. Commun. 2015, 468, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.; Liu, X.; Yu, T. miR-1271 inhibits migration, invasion and epithelial-mesenchymal transition by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2016, 472, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.M.; Wang, L.; Shen, H.; Jiang, C.F.; Ge, X.; Li, D.M.; Wen, Y.Y.; Sun, H.R.; Pan, M.H.; Li, W.; et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 2017, 36, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Zhang, L.; Bao, Y.; Li, B.; He, C.; Gao, M.; Feng, X.; Xu, W.; Zhang, X.; Wang, S. Epithelial-mesenchymal transition, a novel target of sulforaphane via COX-2/MMP2,9/Snail, ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. J. Nutr. Biochem. 2013, 24, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Lin, X.L.; Li, J.; Zhang, T.T.; Wang, H.Y.; Shi, J.W.; Yang, S.; Zhao, W.T.; Xie, R.Y.; Wei, F.; et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS ONE 2014, 9, e101330. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Duan, H.; Gan, L.; Liu, X.; Chen, F.; Shen, X.; Tang, Y.Q.; Wang, S. MicroRNA-1291 promotes endometrial fibrosis by regulating the ArhGAP29-RhoA/ROCK1 signaling pathway in a murine model. Mol. Med. Rep. 2017, 16, 4501–4510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, J.; Wang, B.; Ren, J.C.; Su, W.; Zhang, T. MicroRNA-10b Triggers the Epithelial-Mesenchymal Transition (EMT) of Laryngeal Carcinoma Hep-2 Cells by Directly Targeting the E-cadherin. Appl. Biochem. Biotechnol. 2015, 176, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Li, S.C.; Kao, Y.H.; Luo, H.L.; Cheng, Y.T.; Lin, P.R.; Tai, M.H.; Chiang, P.H. MiR-30a-5p Inhibits Epithelial-to-Mesenchymal Transition and Upregulates Expression of Tight Junction Protein Claudin-5 in Human Upper Tract Urothelial Carcinoma Cells. Int. J. Mol. Sci. 2017, 18, 1826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Xiao, H.X.; Tian, H.P.; Liu, Z.L.; Xia, S.S.; Zhou, T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int. J. Mol. Med. 2013, 31, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhou, X.; Liu, F.; Hu, C.; Zhu, X.; Luo, Y.; Wang, M.; Xu, X.; Yang, S.; Kanwar, Y.S.; et al. MicroRNA-129-5p modulates epithelial-to-mesenchymal transition by targeting SIP1 and SOX4 during peritoneal dialysis. Lab. Investig. 2015, 95, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, L.; Wang, A.; Xu, Y.; Luo, X.; Liu, X.; Hua, Y.; Zhang, D.; Wu, S.; Lin, T.; et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am. J. Transl. Res. 2017, 9, 3611–3622. [Google Scholar] [PubMed]
- Pang, L.; Li, B.; Zheng, B.; Niu, L.; Ge, L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol. Rep. 2017, 38, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wang, D.; Wei, A.; Lu, H.; Tan, C.; Li, A.; Tang, J.; Wang, Y.; He, S.; Liu, X.; et al. Deregulated expression of miR-107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin-1 and PTEN. Exp. Cell Res. 2017, 361, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.W.; Wen, W.; Jiang, C.; Fu, Q.B.; Gu, Y.J.; Lv, T.T.; Li, Z.D.; Xue, W. Inhibition of microRNA-214 promotes epithelial-mesenchymal transition process and induce interstitial cystitis in postmenopausal women by upregulating Mfn2. Exp. Mol. Med. 2017, 49, e357. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.X.; Zhou, K.C.; Cao, Y. MCRS1 overexpression, which is specifically inhibited by miR-129*, promotes the epithelial-mesenchymal transition and metastasis in non-small cell lung cancer. Mol. Cancer. 2014, 13, 245. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Han, Q.; Zhou, N.; Wang, S.; Lu, S.; Bai, C.; Zhao, R.C. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol. Oncol. 2013, 7, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xiao, L.; Liu, Y.; Wang, H.; Li, H.; Zhou, Q.; Pan, J.; Lei, B.; Huang, A.; Qi, S. MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation. Autophagy 2015, 11, 2213–2232. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Fan, Y.; Zhang, X.; Ma, J. miRNA-23 regulates high glucose induced epithelial to mesenchymal transition in human mesothelial peritoneal cells by targeting VDR. Exp. Cell Res. 2017, 360, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, J.; Zhang, X.; Zhou, X.; Lu, S.; Huang, X.; Shao, J.; Lou, G.; Yang, D.; Geng, Y.J. Role of microRNA-29b in angiotensin II-induced epithelial-mesenchymal transition in renal tubular epithelial cells. Int. J. Mol. Med. 2014, 34, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Miao, X.; Liu, L.; Zhang, Y.; Jin, X.; Luo, X.; Gao, H.; Deng, X. Methylation-mediated silencing of microRNA-211 promotes cell growth and epithelial to mesenchymal transition through activation of the Akt/β-catenin pathway in GBM. Oncotarget 2017, 8, 25167–25176. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, W. MiR-211 is epigenetically regulated by DNMT1 mediated methylation and inhibits EMT of melanoma cells by targeting RAB22A. Biochem. Biophys. Res. Commun. 2016, 476, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Zhang, K.; Li, C.; Chen, J.; Pan, Y.; Feng, B.; Lu, L.; Zhu, Z.; Wang, R.; Chen, L. Methylation-associated silencing of microRNA-129-3p promotes epithelial-mesenchymal transition, invasion and metastasis of hepatocellular cancer by targeting Aurora-A. Oncotarget 2016, 7, 78009–78028. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Ihira, K.; Xiong, Y.; Watari, H.; Hanley, S.J.; Yamada, T.; Hosaka, M.; Kudo, M.; Yue, J.; Sakuragi, N. Reactivation of epigenetically silenced miR-124 reverses the epithelial-to-mesenchymal transition and inhibits invasion in endometrial cancer cells via the direct repression of IQGAP1 expression. Oncotarget 2016, 7, 20260–20270. [Google Scholar] [CrossRef] [PubMed]
- Pieraccioli, M.; Imbastari, F.; Antonov, A.; Melino, G.; Raschellà, G. Activation of miR200 by c-Myb depends on ZEB1 expression and miR200 promoter methylation. Cell Cycle 2013, 12, 2309–2320. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.H.; Malouf, G.G.; Lu, E.; Sphyris, N.; Vijay, V.; Ramachandran, P.P.; Ueno, K.R.; Gaur, S.; Nicoloso, M.S.; Rossi, S.; et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci. Rep. 2013, 3, 2687. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Xiang, L.; Yin, L.; Liu, X.; Yang, D.; Zhou, J. Loss-of-function of miR-142 by hypermethylation promotes TGF-β-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017, 50, e12384. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lv, D.; Li, M.; Zhang, X.; Sun, G.; Bai, Y.; Chang, D. Hypermethylation of miRNA-589 promoter leads to upregulation of HDAC5 which promotes malignancy in non-small cell lung cancer. Int. J. Oncol. 2017, 50, 2079–2090. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lu, Z.; Chen, B.; Wu, X.; Dong, P.; Zheng, J. Salvianolic acid B-induced microRNA-152 inhibits liver fibrosis by attenuating DNMT1-mediated Patched1 methylation. J. Cell Mol. Med. 2015, 19, 2617–2632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.P.; Wang, X.L.; Zhao, W.; Qi, B.; Yang, Q.; Wan, H.Y.; Shuang, Z.Y.; Liu, M.; Li, X.; Li, S.; et al. DNA methylation-mediated repression of miR-941 enhances lysine (K)-specific demethylase 6B expression in hepatoma cells. J. Biol. Chem. 2014, 289, 24724–24735. [Google Scholar] [CrossRef] [PubMed]
- Zare, M.; Bastami, M.; Solali, S.; Alivand, M.R. Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: Diagnosis and therapeutic implications. J. Cell. Physiol. 2018, 233, 3729–3744. [Google Scholar] [CrossRef] [PubMed]
- Träger, M.M.; Dhayat, S.A. Epigenetics of epithelial-to-mesenchymal transition in pancreatic carcinoma. Int. J. Cancer 2017, 141, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fang, J. Epigenetic regulation of epithelial-mesenchymal transition. Cell. Mol. Life Sci. 2016, 73, 4493–4515. [Google Scholar] [CrossRef] [PubMed]
- Warns, J.A.; Davie, J.R.; Dhasarathy, A. Connecting the dots: Chromatin and alternative splicing in EMT. Biochem. Cell Biol. 2016, 94, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Nickel, A.; Stadler, S.C. Role of epigenetic mechanisms in epithelial-to-mesenchymal transition of breast cancer cells. Transl. Res. 2015, 165, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U. Aberrant DNA methylation of microRNA genes in human breast cancer—A critical appraisal. Cell Tissue Res. 2014, 356, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Cicchini, C.; de Nonno, V.; Battistelli, C.; Cozzolino, A.M.; De Santis Puzzonia, M.; Ciafrè, S.A.; Brocker, C.; Gonzalez, F.J.; Amicone, L.; Tripodi, M. Epigenetic control of EMT/MET dynamics: HNF4α impacts DNMT3s through miRs-29. Biochim. Biophys. Acta 2015, 1849, 919–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEv5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2017, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Ballarino, M. Long noncoding RNA regulation of pluripotency. Stem Cells Int. 2015, 2016, 1797692. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Shearwood, A.M.J.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Kurashige, J.; Nambara, S.; Komatsu, H.; Hirata, H.; Ueda, M.; Sakimura, S.; Uchi, R.; Takano, Y.; Shinden, Y.; et al. A Long Non-coding RNA Activated by Transforming Growth Factor-β is an Independent Prognostic Marker of Gastric Cancer. Ann. Surg. Oncol. 2015, 22, S915–S922. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Chen, L.Z.; Lu, Y.X.; Zhang, D.S.; Zeng, Z.L.; Pan, Z.Z.; Huang, P.; Wang, F.H.; Li, Y.H.; Ju, H.Q.; et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial–mesenchymal transition in colorectal cancer. Cell Death Dis. 2017, 8, e3011. [Google Scholar] [CrossRef] [PubMed]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.X.; Sun, J.X.; Zhao, J.H.; Yang, Y.C.; Shi, J.X.; Wu, Z.H.; Chen, X.W.; Gao, P.; Miao, Z.F.; Wang, Z.N. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat. Commun. 2017, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shi, H.; Ren, F.; Jia, Y.; Zhang, R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp. Cell Res. 2017, 359, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed. Pharmacother. 2017, 90, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Mehterov, N.; Ling, H.; Stanta, G.; Braicu, C.; Berindan-Neagoe, I. The “good-cop bad-cop” TGF-β role in breast cancer modulated by non-coding RNAs. Biochim. Biophys. Acta 2017, 1861, 1661–1675. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.X.; Jiang, Y.P.; Tang, Y.L.; Liang, X.H. The crosstalk between lncRNA and microRNA in cancer metastasis: Orchestrating the epithelial-mesenchymal plasticity. Oncotarget 2017, 8, 12472–12483. [Google Scholar] [CrossRef] [PubMed]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Raveh, E.; Abu-lail, R.; Mezan, S.; Gilon, M.; Gershtain, E.; Birman, T.; Gallula, J.; Schneider, T.; Barkali, M.; et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta 2014, 1843, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, Y.; Tang, Y.; Li, Q. MALAT1 Modulates TGF-β1-Induced Endothelial-to-Mesenchymal Transition through Downregulation of miR-145. Cell. Physiol. Biochem. 2017, 42, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated b TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Jiang, C.; Fang, J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 495, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.N.; Yan, T.H.; Yu, R.M.; Gao, Y.; Zeng, W.L.; Lu, S.W.; Que, H.X.; Liu, Z.P.; Jiang, J.H. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J. Cancer Res. Clin. Oncol. 2017, 143, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Cui, J.; Song, Y. Long noncoding RNA PVT1 promotes EMT via mediating microRNA-186 targeting of Twist1 in prostate cancer. Gene 2018, 654, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Han, T.; Zhang, T.; Ma, C.; Sun, C. LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway. Oncotarget 2017, 8, 36410–36422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J.; Song, H.; Chen, L.B. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget 2017, 9, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, T.; Wei, G.; Liu, L.; Chen, Q.; Xu, L.; Zhang, K.; Zeng, D.; Liao, R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016, 37, 11733–11741. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Xu, H.X.; Yu, Y.; He, J.D.; Wang, Z.; Xu, Y.J.; Wang, C.Y.; Zhang, H.M.; Zhang, R.X.; Zhang, J.J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zeng, Y.; Zhou, C.C.; Ye, W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch. Biochem. Biophys. 2018, 637, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Lu, Y.X.; Zhang, J.X.; Wei, X.L.; Wang, F.; Zeng, Z.L.; Pan, Z.Z.; Yuan, Y.F.; Wang, F.H.; Pelicano, H.; et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics 2017, 7, 4836–4849. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Qiu, K.; Li, M.; Liang, Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015, 589, 3175–3181. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, F.; Yin, C.; Zhuang, Y.; Yue, G.; Zhang, G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell. Physiol. Biochem. 2015, 36, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Qiu, M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem. Biophys. Res. Commun. 2018, 495, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xiao, J.; Chai, Y.; Du, Y.Y.; Liu, Z.; Huang, K.; Zhou, X.; Zhou, W. LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma Through Suppressing miR-152. Dig. Dis. Sci. 2017, 62, 3050–3058. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, H.; Kong, H.; Chen, Z.; Chen, B.; Zhou, M. The LncRNA-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging MiR-382. Cell. Physiol. Biochem. 2017, 42, 2145–2158. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhong, Z.; Huang, M.; Tian, Q.; Jiang, R.; Chen, J. LncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim. Biophys. Acta 2017, 1864, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Chen, R.; Xiong, H.; Qiu, F.; Liu, S.; Zhang, M.; Wang, F.; Wang, Y.; Zhou, X.; et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016, 383, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Fan, H.X.; Zhao, X.P.; Lv, P.; Fan, J.Y.; Zhang, Y.; Liu, M.; Tang, H. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016, 382, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Yao, B.; Dou, C.; Xu, M.; Xue, Y.; Ding, L.; Jia, Y.; Zhang, H.; Li, Q.; et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol. 2016, 37, 11429–11441. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.C.; Fu, W.M.; Wong, C.W.; Wang, Y.; Wang, W.M.; Hu, G.X.; Zhang, L.; Xiao, L.J.; Wan, D.C.; Zhang, J.F.; et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015, 6, 22513–22525. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Yao, B.; Li, Q.; Wang, L.; Wang, C.; Dou, C.; Xu, M.; Liu, Q.; Tu, K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer 2017, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Wang, S.; Zhang, H.; Han, H.; Ma, B.; Nan, W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial-Mesenchymal Transition in Osteosarcoma by Regulating the miR-221/ARHI Pathway. J. Cell. Biochem. 2017, 118, 4772–4781. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Li, O.; Zheng, W.; Xiao, W.Z.; Zhang, L.; Wu, D.; Cai, G.Y.; He, J.C.; Chen, X.M. LncRNA HOTAIR regulates HIF-1α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017, 8, e2772. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wang, S.H.; Cai, Q.; Jin, L.Y.; Zhou, D.; Ding, J.; Quan, Z.W. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomed. Pharmacother. 2017, 88, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, Z.; Wang, S.; Weng, M.; Zhou, D.; Li, C.; Wang, J.; Chen, E.; Quan, Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017, 7, 160247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Liang, F.; Zhang, J.W.; Wang, F.; Wang, L.; Kang, X.G. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother. Pharmacol. 2017, 79, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Luo, F.; Liu, Y.; Liu, X.; Shi, L.; Lu, X.; Liu, Q. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol. Appl. Pharmacol. 2015, 289, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.W.; Sun, M.; Xia, R.; Zhang, E.B.; Liu, X.H.; Zhang, Z.H.; Xu, T.P.; De, W.; Liu, B.R.; Wang, Z.X. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015, 6, e1802. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Wang, T.; Liu, W.H.; Li, X.C.; Tang, L.J.; Tian, F.Z. Enhancing HOTAIR/MiR-10b Drives Normal Liver Stem Cells Toward a Tendency to Malignant Transformation Through Inducing Epithelial-to-Mesenchymal Transition. Rejuvenation Res. 2015, 18, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Wang, L.F.; Zhao, Y.L.; Yang, T.; Li, W.; Zhao, J.; Yu, F.; Wang, L.; Meng, Y.L.; Liu, N.N.; et al. Nuclear factor of activated T cells 5 maintained by HOTAIR suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014, 16, 454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Nong, K.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014, 35, 9163–9169. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Expósito-Villén, A.; E. Aránega, A.; Franco, D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition. Non-Coding RNA 2018, 4, 14. https://doi.org/10.3390/ncrna4020014
Expósito-Villén A, E. Aránega A, Franco D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition. Non-Coding RNA. 2018; 4(2):14. https://doi.org/10.3390/ncrna4020014
Chicago/Turabian StyleExpósito-Villén, Almudena, Amelia E. Aránega, and Diego Franco. 2018. "Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition" Non-Coding RNA 4, no. 2: 14. https://doi.org/10.3390/ncrna4020014