Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders
Abstract
:1. Introduction
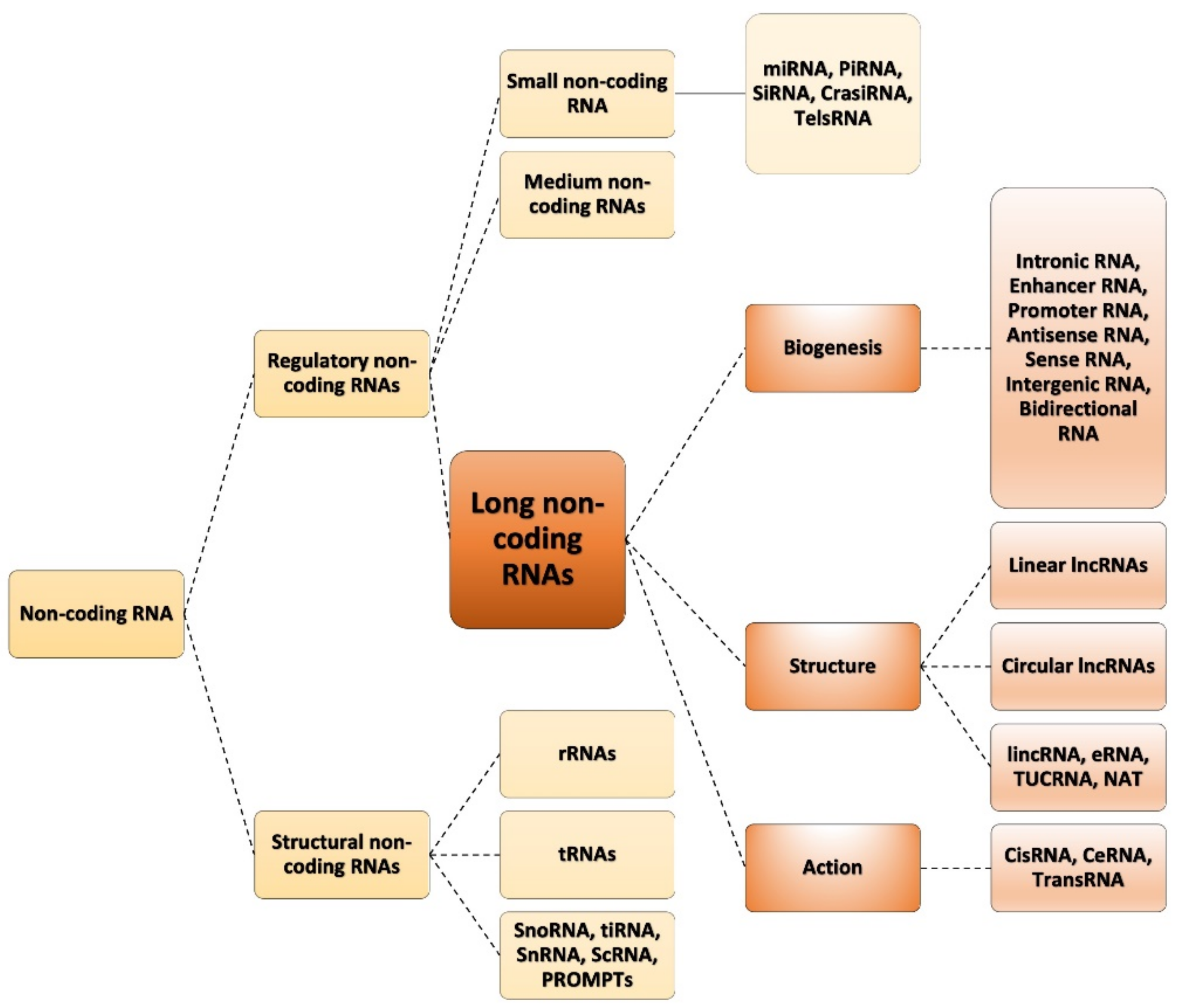
1.1. Classification of ncRNAs
1.2. Biogenesis of lncRNAs
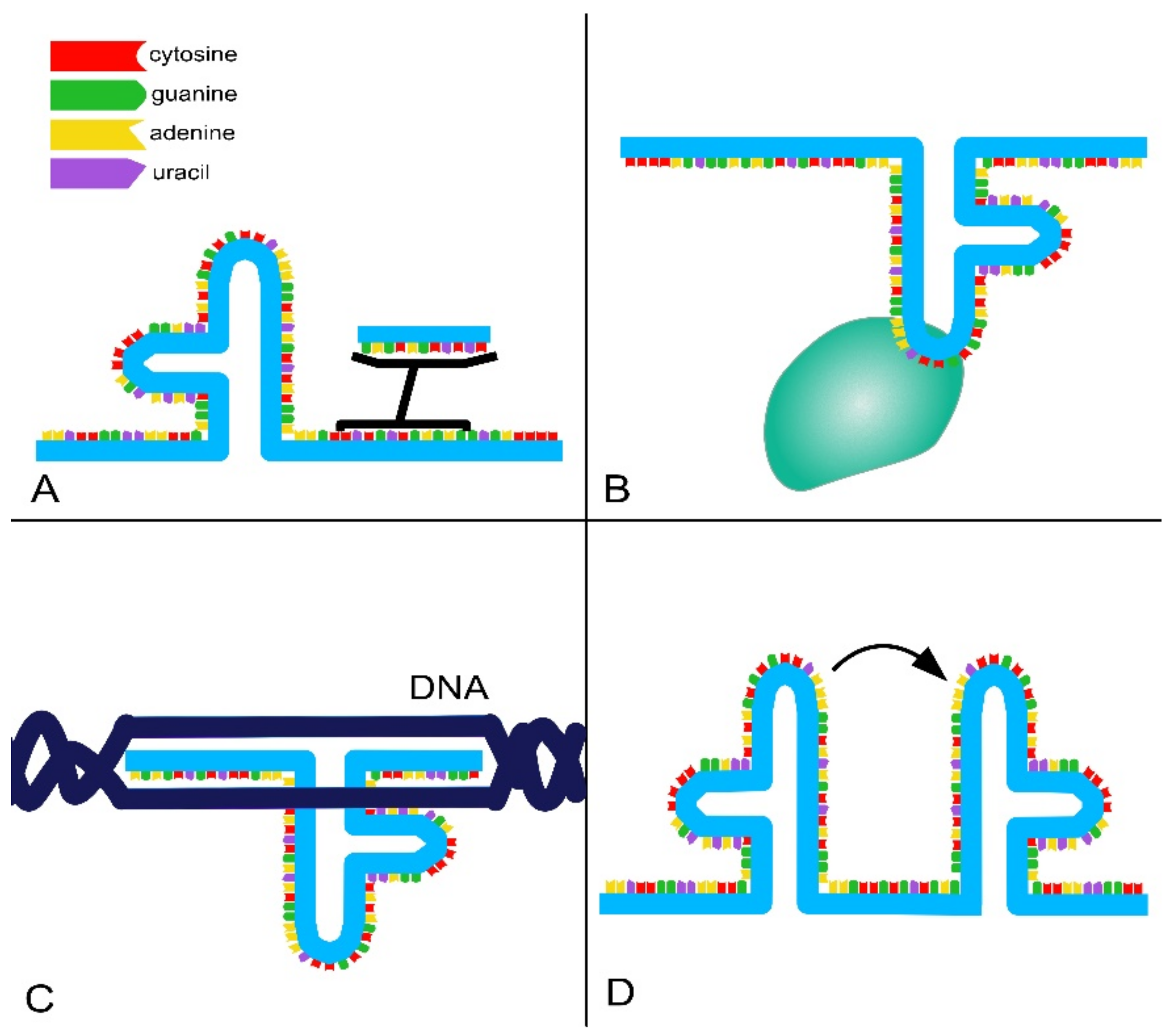
- RNA-binding domains. Thanks to their ability to base pair with other RNAs, lncRNAs can recognize and bind mRNAs, miRNAs and other lncRNAs, modulating target levels and function;
- Protein-binding domains. Proteins are a major partner of lncRNAs, forming ribonucleoprotein complexes (RNPs) that act as chaperones, transport aids or effectors (including phase-separation seeding). This type of interaction involves conformational changes in the protein, RNA or both;
- DNA-binding domains. Currently, there is a lack of extensive evidence for direct and functional interaction between lncRNAs and DNA as well as a lack of a consensus regarding the role and function of these interactions. However, it is known that RNA–DNA hybrids or triplex structures can allow single strands of RNA to interact with DNA duplexes through pair–base interactions. These direct interactions can efficiently and selectively direct RNA signals to genomic loci through base-pairing interactions. However, such interactions can also expose the genome to deamination and damage;
- Conformational switch. LncRNAs can act as regulatory devices by allosterically coupling binding domains with the switching of structural conformations and thereby activating or suppressing linked functional domains.
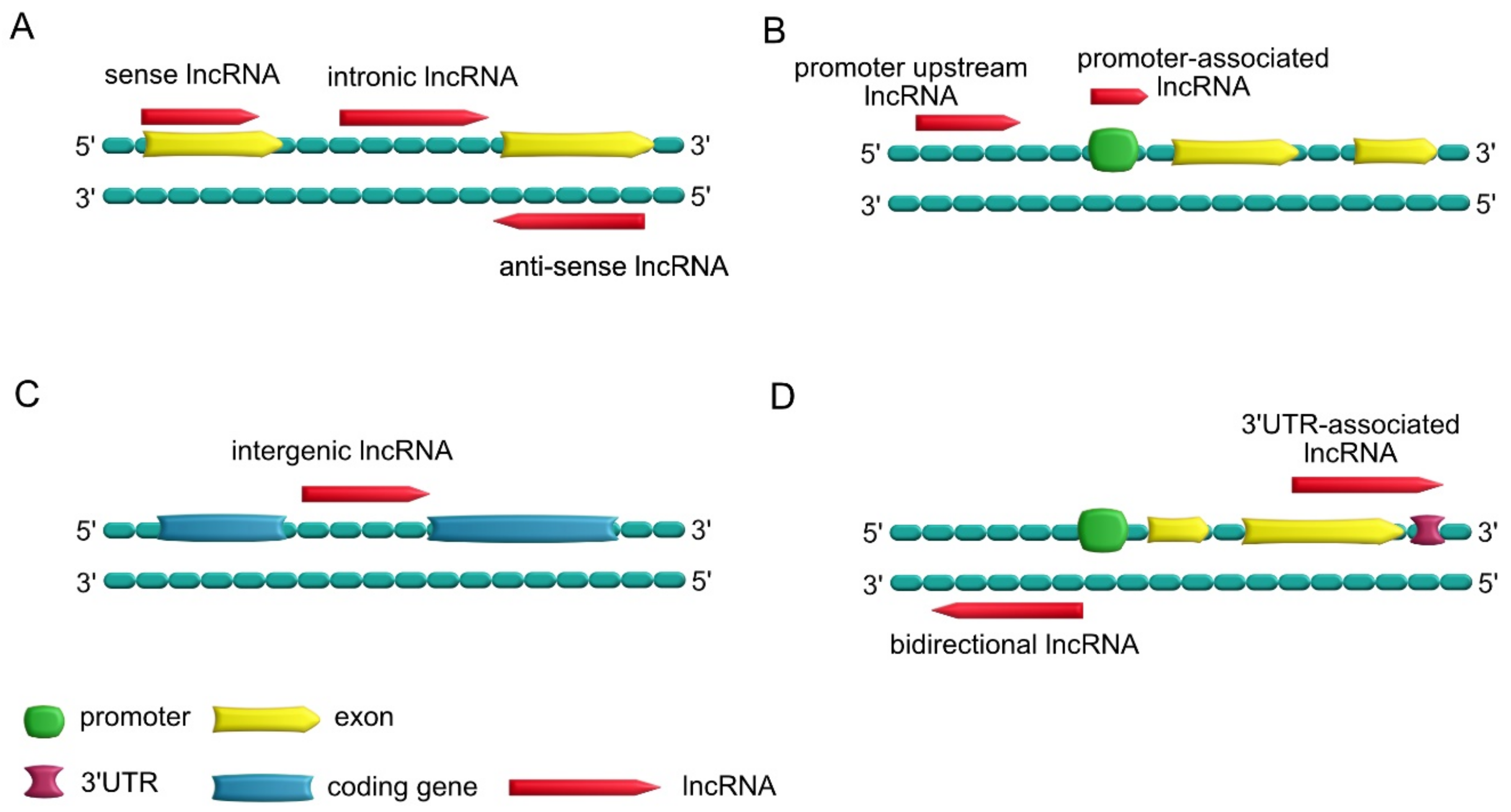
1.3. Types of lncRNAs
- Sense lncRNAs, which overlap one or more exons of neighboring mRNAs on the same strand; antisense lncRNAs, which overlap one or more exons of neighboring mRNAs on the opposite strand; intronic lncRNAs, which are transcribed from introns of a second transcript (sometimes may represent pre-mRNA sequences);
- Promoter upstream lncRNAs, which are located upstream of a promoter; promoter-associated lncRNAs, which are close to the promoter;
- Intergenic lncRNAs, which lie within the genomic interval between two genes;
- Bidirectional lncRNAs, which have promoters in common with protein-encoding genes but are transcribed in the opposite direction; 3′UTR-associated lncRNAs, which are transcribed from a protein-coding gene’s 3′UTR region.
1.4. Evolution, Conservation and Stability of lncRNAs
2. Functional Roles of lncRNA in Cellular Processes
2.1. Mechanisms of Action
2.2. LncRNAs as Chromatin Regulators
2.3. Transcriptional, Post-Transcriptional and Post-Translational Regulation
3. LncRNAs in Neurological and Neurodegenerative Disorders
3.1. Alzheimer’s Disease
3.2. Amyotrophic Lateral Sclerosis (ALS)
4. LncRNAs in Neurodevelopmental and Neuropsychiatric Disorders
4.1. Autism Spectrum Disorder
4.2. Schizophrenia
5. Final Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-Coding RNAs: Multi-Tasking Molecules in the Cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.C.; Morris, K.V.; Wood, M.J.A. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lekka, E.; Hall, J. Noncoding RNA s in disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar] [CrossRef]
- Chen, K.-W.; Chen, J.-A. Functional Roles of Long Non-coding RNAs in Motor Neuron Development and Disease. J. Biomed. Sci. 2020, 27, 38. [Google Scholar] [CrossRef]
- Clark, M.B.; Amaral, P.D.P.; Schlesinger, F.J.; Dinger, M.; Taft, R.J.; Rinn, J.L.; Ponting, C.P.; Stadler, P.F.; Morris, K.V.; Morillon, A.; et al. The Reality of Pervasive Transcription. PLoS Biol. 2011, 9, e1000625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.-H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2016, 18, 558–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Distefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tang, Y.; Sun, H.; Lin, X.; Jiang, B. The roles of long noncoding RNAs in myocardial pathophysiology. Biosci. Rep. 2019, 39, 20190966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Ao, L.; Yang, J. Long non-coding RNAs in diseases related to inflammation and immunity. Ann. Transl. Med. 2019, 7, 494. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mongelli, A.; Martelli, F.; Farsetti, A.; Gaetano, C. The Dark That Matters: Long Non-coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 2019, 10, 369. [Google Scholar] [CrossRef] [Green Version]
- Sparber, P.; Filatova, A.; Khantemirova, M.; Skoblov, M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med. Genom. 2019, 12 (Suppl. S2), 42. [Google Scholar] [CrossRef] [Green Version]
- Jaé, N.; Dimmeler, S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef]
- Sun, B.; Liu, C.; Zhang, L.; Luo, G.; Liang, S. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer (Review). Oncol. Lett. 2019, 19, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijesinghe, S.N.; Nicholson, T.; Tsintzas, K.; Jones, S.W. Involvements of long noncoding RNAs in obesity-associated inflammatory diseases. Obes. Rev. 2021, 22, 13156. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Wu, H.-J.; Hsu, J.-M.; Chang, S.-S.; Labaff, A.M.; Li, C.-W.; Wang, Y.; Hsu, J.L.; Hung, M.-C. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150. [Google Scholar]
- Tuck, A.C.; Tollervey, D. RNA in pieces. Trends Genet. 2011, 27, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.; Lim, D.A. Forging our understanding of lncRNAs in the brain. Cell Tissue Res. 2018, 371, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Carninci, P. Long non-coding RNA modifies chromatin: Epigenetic silencing by long non-coding RNAs. BioEssays 2011, 33, 830–839. [Google Scholar] [CrossRef] [Green Version]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zuo, X.; Deng, H.; Liu, X.; Liu, L.; Ji, A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013, 97, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Tsagakis, I.; Douka, K.; Birds, I.; Aspden, J.L. Long non-coding RNAs in development and disease: Conservation to mechanisms. J. Pathol. 2020, 250, 480–495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Dong, Y.; Wang, Y.; Gao, J.; Lv, J.; Sun, J.; Li, M.; Wang, M.; Zhao, Z.; Wang, J.; et al. Long non-codingRNAs in ocular diseases: New and potential therapeutic targets. FEBS J. 2019, 286, 2261–2272. [Google Scholar] [CrossRef] [Green Version]
- Wan, P.; Su, W.; Zhuo, Y. The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 54, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesterova, T.B.; Slobodyanyuk, S.Y.; Elisaphenko, E.A.; Shevchenko, A.I.; Johnston, C.; Pavlova, M.E.; Rogozin, I.B.; Kolesnikov, N.N.; Brockdorff, N.; Zakian, S.M. Characterization of the Genomic Xist Locus in Rodents Reveals Conservation of Overall Gene Structure and Tandem Repeats but Rapid Evolution of Unique Sequence. Genome Res. 2001, 11, 833–849. [Google Scholar] [CrossRef] [Green Version]
- Chodroff, R.A.; Goodstadt, L.; Sirey, T.M.; Oliver, P.L.; Davies, K.E.; Green, E.D.; Molnár, Z.; Ponting, C.P. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010, 11, R72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, J.M.; Kim, S.O.; Inoue, K.; Smola, M.J.; Lee, D.M.; Schertzer, M.; Wooten, J.S.; Baker, A.R.; Sprague, D.; Collins, D.W.; et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 2018, 50, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Sprague, D.; Waters, S.; Kirk, J.M.; Wang, J.R.; Samollow, P.B.; Waters, P.D.; Calabrese, J.M. Nonlinear sequence similarity between the Xist and Rsx long noncoding RNAs suggests shared functions of tandem repeat domains. RNA 2019, 25, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.N.; Miller, S.C.; Varani, G.; Calabrese, J.M.; Magnuson, T. Multimodal Long Noncoding RNA Interaction Networks: Control Panels for Cell Fate Specification. Genet. 2019, 213, 1093–1110. [Google Scholar] [CrossRef] [PubMed]
- Chillón, I.; Marcia, M. The molecular structure of long non-coding RNAs: Emerging patterns and functional implications. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 662–690. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Rasmussen, T.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Moss, W.N.; Rutenberg-Schoenberg, M.; Simon, M.D. Probing Xist RNA Structure in Cells Using Targeted Structure-Seq. PLoS Genet. 2015, 11, e1005668. [Google Scholar] [CrossRef] [Green Version]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Somarowthu, S.; Pyle, A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017, 13, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR Forms an Intricate and Modular Secondary Structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chillón, I.; Pyle, A.M. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 2016, 44, 9462–9471. [Google Scholar] [CrossRef] [Green Version]
- Pintacuda, G.; Young, A.N.; Cerase, A. Function by Structure: Spotlights on Xist Long Non-coding RNA. Front. Mol. Biosci. 2017, 4, 90. [Google Scholar] [CrossRef] [Green Version]
- Cirillo, D.; Blanco, M.; Armaos, A.; Buness, A.; Avner, P.; Guttman, M.; Cerase, A.; Tartaglia, G.G. Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods 2017, 14, 5–6. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.R.; Dinger, M.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.; Rossetti, G.; Panzeri, I.; De Candia, P.; Bonnal, R.J.P.; Rossi, R.L.; Geginat, J.; Abrignani, S. Role of microRNAs and long-non-coding RNAs in CD4+T-cell differentiation. Immunol. Rev. 2013, 253, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, B.; Eblackshaw, S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front. Genet. 2014, 5, 164. [Google Scholar] [CrossRef] [Green Version]
- Park, J.Y.; Lee, J.E.; Park, J.B.; Yoo, H.; Lee, S.-H.; Kim, J.H. Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development. Brain Tumor Res. Treat. 2014, 2, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Vondervoort, I.I.G.M.; Gordebeke, P.M.; Ekhoshab, N.; Tiesinga, P.H.E.; Buitelaar, J.K.; Ekozicz, T.; Easchrafi, A.; Glennon, J.C. Long non-coding RNAs in neurodevelopmental disorders. Front. Mol. Neurosci. 2013, 6, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2009, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Petazzi, P.; Sandoval, J.; Szczesna, K.; Jorge, O.C.; Roa, L.; Sayols, S.; Gomez, A.; Huertas, D.; Esteller, M. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013, 10, 1197–1203. [Google Scholar] [CrossRef] [Green Version]
- Ziats, M.N.; Rennert, O.M. Aberrant Expression of Long Noncoding RNAs in Autistic Brain. J. Mol. Neurosci. 2012, 49, 589–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastori, C.; Peschansky, V.; Barbouth, D.; Mehta, A.; Silva, J.P.; Wahlestedt, C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 2014, 133, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-F.; Gao, J.; Liu, C.-M. The Role of Non-Coding RNAs in Neurodevelopmental Disorders. Front. Genet. 2019, 10, 1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bánfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E.; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012, 22, 1646–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Johnstone, T.; Christiano, R.; Mackowiak, S.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef] [Green Version]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N. Long non-coding RNAs: Mechanism of action and functional utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhuang, Y.; Zhao, X.; Li, X. Long Non-coding RNA in Neuronal Development and Neurological Disorders. Front. Genet. 2019, 9, 744. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van Der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St-Laurent, G.; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef] [Green Version]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Gunawardane, L.; Niazi, F.; Jahanbani, F.; Chen, X.; Valadkhan, S. A Novel RNA Motif Mediates the Strict Nuclear Localization of a Long Noncoding RNA. Mol. Cell. Biol. 2014, 34, 2318–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, A.; Lin, H.; Shatabda, S. Locate-R: Subcellular localization of long non-coding RNAs using nucleotide compositions. Genomics 2020, 112, 2583–2589. [Google Scholar] [CrossRef]
- Cerase, A.; Tartaglia, G.G. Long non-coding RNA-polycomb intimate rendezvous. Open Biol. 2020, 10, 200126. [Google Scholar] [CrossRef] [PubMed]
- Fejes-Toth, K.; Sotirova, V.; Sachidanandam, R.; Assaf, G.; Hannon, G.J.; Kapranov, P.; Foissac, S.; Willingham, A.T.; Duttagupta, R.; Dumais, E.; et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 2009, 457, 1028–1032. [Google Scholar] [CrossRef]
- Kapranov, P.; Laurent, G.S.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H.B.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poynter, S.T.; Kadoch, C. Polycomb and trithorax opposition in development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 659–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Lee, J.T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 2011, 12, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Bousard, A.; Raposo, A.C.; Żylicz, J.J.; Picard, C.; Pires, V.B.; Qi, Y.; Gil, C.; Syx, L.; Chang, H.Y.; Heard, E.; et al. The role of Xist -mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019, 20, e48019. [Google Scholar] [CrossRef]
- Dixon-McDougall, T.; Brown, C.J. Independent domains for recruitment of PRC1 and PRC2 by human XIST. PLoS Genet. 2021, 17, e1009123. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Y.; Colognori, D.; Sunwoo, H.; Wang, D.; Lee, J.T. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 2019, 10, 2950. [Google Scholar] [CrossRef] [Green Version]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Shamovsky, I.; Ivannikov, M.; Kandel, E.S.; Gershon, D.; Nudler, E. RNA-mediated response to heat shock in mammalian cells. Nature 2006, 440, 556–560. [Google Scholar] [CrossRef]
- Feng, J.; Bi, C.; Clark, B.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006, 20, 1470–1484. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Turner, M.; Galloway, A.; Vigorito, E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 2014, 15, 484–491. [Google Scholar] [CrossRef]
- Sado, T.; Hoki, Y.; Sasaki, H. Tsix Silences Xist through Modification of Chromatin Structure. Dev. Cell 2005, 9, 159–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmer-Bensch, G. Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8, 1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARγ2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, R.; Mayeda, A.; Yoshida, M.; Nakagawa, S. MALAT1 long non-coding RNA in cancer. Biochim. Biophys. Acta 2016, 1859, 192–199. [Google Scholar] [CrossRef] [PubMed]
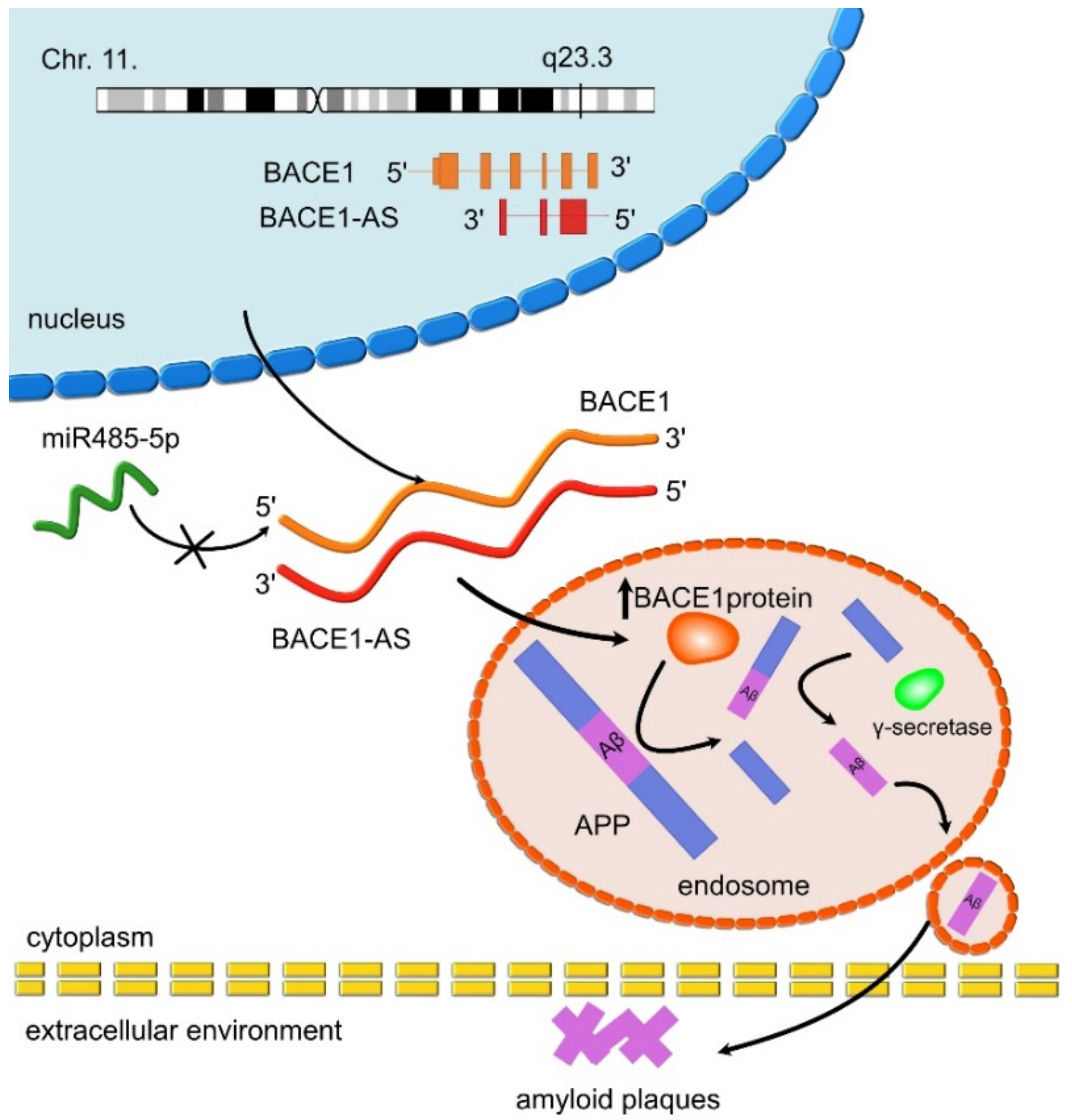
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St. Laurent, G., III; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Kallen, A.; Zhou, X.-B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.-S.; Zhang, H.; et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, X.; Zhou, J.; Hu, J.; Zhang, D.; Liu, J.; Qiao, Y.; Zhan, Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology 2017, 65, 1612–1627. [Google Scholar] [CrossRef] [Green Version]
- Thibaut, F. The role of sex and gender in neuropsychiatric disorders. Dialogues Clin. Neurosci. 2016, 18, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.M.; Clyne, A.M. Sex differences in the blood–brain barrier and neurodegenerative diseases. APL Bioeng. 2021, 5, 011509. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Shearwood, A.-M.J.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-Y.; Kuo, H.-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.-L.; Galasko, D.R.; Ringman, J.M.; Vinters, H.V.; Edland, S.D.; Pomakian, J.; Ubeda, O.J.; Rosario, E.R.; Teter, B.; Frautschy, S.A.; et al. Reduction of SorLA/LR11, a Sorting Protein Limiting β-Amyloid Production, in Alzheimer Disease Cerebrospinal Fluid. Arch. Neurol. 2009, 66, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Chen, Y. Long noncoding RNAs and Alzheimer’s disease. Clin. Interv. Aging 2016, 11, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef]
- Gavazzo, P.; Vassalli, M.; Costa, D.; Pagano, A. Novel ncRNAs transcribed by Pol III and elucidation of their functional relevance by biophysical approaches. Front. Cell. Neurosci. 2013, 7, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buggia-Prévot, V.; Thinakaran, G. Sorting the Role of SORLA in Alzheimer’s Disease. Sci. Transl. Med. 2014, 6, 223fs8. [Google Scholar] [CrossRef]
- Massone, S.; Ciarlo, E.; Vella, S.; Nizzari, M.; Florio, T.; Russo, C.; Cancedda, R.; Pagano, A. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim. Biophys. Acta 2012, 1823, 1170–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mus, E.; Hof, P.R.; Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 10679–10684. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Pestova, T.V.; Hellen, C.U.T.; Tiedge, H. Translational Control by a Small RNA: Dendritic BC1 RNA Targets the Eukaryotic Initiation Factor 4A Helicase Mechanism. Mol. Cell. Biol. 2008, 28, 3008–3019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacoangeli, A.; Bianchi, R.; Tiedge, H. Regulatory RNAs in brain function and disorders. Brain Res. 2010, 1338, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhao, Y.; Xu, N.; Zhang, S.; Wang, S.; Mao, Y.; Zhu, Y.; Li, B.; Jiang, Y.; Tan, Y.; et al. NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell. Mol. Life Sci. 2019, 76, 3005–3018. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.-Y.; Wang, G.-Q.; Wang, N.-N.; Yu, Q.-Y.; Liu, R.-L.; Shi, W.-Q. The long-non-coding RNA NEAT1 is a novel target for Alzheimer’s disease progression via miR-124/BACE1 axis. Neurol. Res. 2019, 41, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline–arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 746. [Google Scholar] [CrossRef] [Green Version]
- An, H.; Tan, J.T.; Shelkovnikova, T.A. Stress granules regulate stress-induced paraspeckle assembly. J. Cell Biol. 2019, 218, 4127–4140. [Google Scholar] [CrossRef] [Green Version]
- Mizielinska, S.; Grönke, S.; Niccoli, T.; Ridler, C.E.; Clayton, E.L.; Devoy, A.; Moens, T.; Norona, F.E.; Woollacott, I.O.C.; Pietrzyk, J.; et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 2014, 345, 1192–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, X.; Tan, W.; Westergard, T.; Krishnamurthy, K.; Markandaiah, S.S.; Shi, Y.; Lin, S.; Shneider, N.; Monaghan, J.; Pandey, U.B.; et al. Antisense Proline-Arginine RAN Dipeptides Linked to C9ORF72-ALS/FTD Form Toxic Nuclear Aggregates that Initiate In Vitro and In Vivo Neuronal Death. Neuron 2014, 84, 1213–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maharjan, N.; Künzli, C.; Buthey, K.; Saxena, S. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol. Neurobiol. 2016, 54, 3062–3077. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, B.; Bento-Abreu, A.; Gendron, T.F.; Boeynaems, S.; Bogaert, E.; Nuyts, R.; Timmers, M.; Scheveneels, W.; Hersmus, N.; Wang, J.; et al. A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. 2018, 135, 427–443. [Google Scholar] [CrossRef] [Green Version]
- Bampton, A.; Gittings, L.; Fratta, P.; Lashley, T.; Gatt, A. The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 2020, 140, 599–623. [Google Scholar] [CrossRef]
- Mizielinska, S.; Lashley, T.; Norona, F.E.; Clayton, E.L.; Ridler, C.E.; Fratta, P.; Isaacs, A.M. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013, 126, 845–857. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.-Y.; Berson, A.; Kennerdell, J.R.; Sartoris, A.; Unger, T.; Porta, S.; Kim, H.-J.; Smith, E.R.; Shilatifard, A.; Van Deerlin, V.; et al. Aberrant activation of non-coding RNA targets of transcriptional elongation complexes contributes to TDP-43 toxicity. Nat. Commun. 2018, 9, 4406. [Google Scholar] [CrossRef]
- Li, P.P.; Sun, X.; Xia, G.; Arbez, N.; Paul, S.; Zhu, S.; Peng, H.B.; Ross, C.A.; Koeppen, A.H.; Margolis, R.L.; et al. ATXN2-AS, a gene antisense toATXN2, is associated with spinocerebellar ataxia type 2 and amyotrophic lateral sclerosis. Ann. Neurol. 2016, 80, 600–615. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, S.; Pandini, C.; Garofalo, M.; Bordoni, M.; Pansarasa, O.; Cereda, C. Long non coding RNAs and ALS: Still much to do. Non-Coding RNA Res. 2018, 3, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Fan, L.; Mao, C.; Hu, X.; Zhang, S.; Yang, Z.; Hu, Z.; Sun, H.; Fan, Y.; Dong, Y.; Yang, J.; et al. New Insights Into the Pathogenesis of Alzheimer’s Disease. Front. Neurol. 2020, 10, 1312. [Google Scholar] [CrossRef]
- Maoz, R.; Garfinkel, B.; Soreq, H. Alzheimer’s Disease and ncRNAs. Adv. Exp. Med. Biol. 2017, 978, 337–361. [Google Scholar] [CrossRef] [PubMed]
- Millan, M.J. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: An integrative review. Prog. Neurobiol. 2017, 156, 1–68. [Google Scholar] [CrossRef]
- Cao, M.; Li, H.; Zhao, J.; Cui, J.; Hu, G. Identification of age- and gender-associated long noncoding RNAs in the human brain with Alzheimer’s disease. Neurobiol. Aging 2019, 81, 116–126. [Google Scholar] [CrossRef]
- Hong, H.; Mo, Y.; Li, D.; Xu, Z.; Liao, Y.; Yin, P.; Liu, X.; Xia, Y.; Fang, J.; Wang, Q.; et al. Aberrant Expression Profiles of lncRNAs and Their Associated Nearby Coding Genes in the Hippocampus of the SAMP8 Mouse Model with AD. Mol. Ther. Nucleic Acids 2020, 20, 140–154. [Google Scholar] [CrossRef]
- Wang, D.; Wang, P.; Bian, X.; Xu, S.; Zhou, Q.; Zhang, Y.; Ding, M.; Han, M.; Huang, L.; Bi, J.; et al. Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Tie, C.; Yu, B.; Zhang, W.; Wan, J. Identifying lncRNA–miRNA–mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging 2020, 12, 2897–2920. [Google Scholar] [CrossRef]
- Yang, B.; Xia, Z.-A.; Zhong, B.; Xiong, X.; Sheng, C.; Wang, Y.; Gong, W.; Cao, Y.; Wang, Z.; Peng, W. Distinct Hippocampal Expression Profiles of Long Non-coding RNAs in an Alzheimer’s Disease Model. Mol. Neurobiol. 2017, 54, 4833–4846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafi, O. Inverse relationship between Alzheimer’s disease and cancer, and other factors contributing to Alzheimer’s disease: A systematic review. BMC Neurol. 2016, 16, 236. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Liao, Y.-T.; He, J.-C.; Xie, C.-L.; Chen, S.-Y.; Fan, H.-H.; Su, Z.-P.; Wang, Z. Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 2018, 18, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, Z.; Yu, Y.; Cao, J.; Qiao, Y.; Qiao, H.; Suo, G. Comprehensive analysis of the lncRNA-associated ceRNA network identifies neuroinflammation biomarkers for Alzheimer’s disease. Mol. Omics 2019, 15, 459–469. [Google Scholar] [CrossRef]
- Ge, Y.; Song, X.; Liu, J.; Liu, C.; Xu, C. The Combined Therapy of Berberine Treatment with lncRNA BACE1-AS Depletion Attenuates Aβ25–35 Induced Neuronal Injury through Regulating the Expression of miR-132-3p in Neuronal Cells. Neurochem. Res. 2020, 45, 741–751. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Yang, H.; Xu, Y.; Zhou, X.; Zhang, X.; Xie, Z.; Bi, J. The effect of BACE1-AS on β-amyloid generation by regulating BACE1 mRNA expression. BMC Mol. Biol. 2019, 20, 23. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zheng, L.; Jiang, A.; Mo, Y.; Gong, Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. NeuroReport 2018, 29, 1061–1067. [Google Scholar] [CrossRef]
- Ahmadi, S.; Zobeiri, M.; Bradburn, S. Molecular mechanisms underlying actions of certain long noncoding RNAs in Alzheimer’s disease. Metab. Brain Dis. 2020, 35, 681–693. [Google Scholar] [CrossRef]
- Yin, R.-H.; Yu, J.-T.; Tan, L. The Role of SORL1 in Alzheimer’s Disease. Mol. Neurobiol. 2014, 51, 909–918. [Google Scholar] [CrossRef]
- Holstege, H.; Van Der Lee, S.J.; Hulsman, M.; Wong, T.H.; Van Rooij, J.G.; Weiss, M.M.; Louwersheimer, E.; Wolters, F.J.; Amin, N.; Uitterlinden, A.G.; et al. Characterization of pathogenic SORL1 genetic variants for association with Alzheimer’s disease: A clinical interpretation strategy. Eur. J. Hum. Genet. 2017, 25, 973–981. [Google Scholar] [CrossRef]
- Guo, C.-C.; Jiao, C.-H.; Gao, Z.-M. Silencing of LncRNA BDNF-AS attenuates Aβ25-35-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol. Res. 2018, 40, 795–804. [Google Scholar] [CrossRef]
- Bhattacharyya, N.; Pandey, V.; Bhattacharyya, M.; Dey, A. Regulatory role of long non coding RNAs (lncRNAs) in neurological disorders: From novel biomarkers to promising therapeutic strategies. Asian J. Pharm. Sci. 2021, in press. [Google Scholar] [CrossRef]
- Douglas, A.G. Non-coding RNA in C9orf72-related amyotrophic lateral sclerosis and frontotemporal dementia: A perfect storm of dysfunction. Non-Coding RNA Res. 2018, 3, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.; Dogini, D.; Lopes-Cendes, I. Role of non-coding RNAs in non-aging-related neurological disorders. Braz. J. Med Biol. Res. 2018, 51. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Masrori, P.; Van Damme, P. Amyotrophic lateral sclerosis: A clinical review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kabotyanski, E.B.; Reineke, L.C.; Shao, J.; Xiong, F.; Lee, J.-H.; Dubrulle, J.; Johnson, H.; Stossi, F.; Tsoi, P.S.; et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2020, 48, 2621–2642. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e7. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, N.; Eshleman, N.; Buchan, J.R. Stress Granules and ALS: A Case of Causation or Correlation? In RNA Metabolism in Neurodegenerative Diseases; Springer: Cham, Switzerland, 2018; pp. 173–212. [Google Scholar] [CrossRef]
- Van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 2018, 115, 2734–2739. [Google Scholar] [CrossRef] [Green Version]
- Farg, M.A.; Sundaramoorthy, V.; Sultana, J.M.; Yang, S.; Atkinson, R.A.; Levina, V.; Halloran, M.A.; Gleeson, P.A.; Blair, I.; Soo, K.Y.; et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014, 23, 3579–3595. [Google Scholar] [CrossRef]
- Webster, C.P.; Smith, E.F.; Bauer, C.S.; Moller, A.; Hautbergue, G.M.; Ferraiuolo, L.; Myszczynska, M.A.; Higginbottom, A.; Walsh, M.J.; Whitworth, A.J.; et al. The C9orf72 protein interacts with Rab1a and the ULK 1 complex to regulate initiation of autophagy. EMBO J. 2016, 35, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Vale, A.J.R.D. RNA phase transitions in repeat expansion disorders. Nature 2017, 546, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Česnik, A.B.; Darovic, S.; Mihevc, S.P.; Štalekar, M.; Malnar, M.; Motaln, H.; Lee, Y.; Mazej, J.; Pohleven, J.; Grosch, M.; et al. Nuclear RNA foci from C9ORF72 expansion mutation form paraspeckle-like bodies. J. Cell Sci. 2019, 132, jcs224303. [Google Scholar] [CrossRef] [Green Version]
- Davidson, Y.S.; Flood, L.; Robinson, A.C.; Nihei, Y.; Mori, K.; Rollinson, S.; Richardson, A.; Benson, B.C.; Jones, M.; Snowden, J.S.; et al. Heterogeneous ribonuclear protein A3 (hnRNP A3) is present in dipeptide repeat protein containing inclusions in Frontotemporal Lobar Degeneration and Motor Neurone disease associated with expansions in C9orf72 gene. Acta Neuropathol. Commun. 2017, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Piccolo, L.L.; Yamaguchi, M. RNAi of arcRNA hsrω affects sub-cellular localization of Drosophila FUS to drive neurodiseases. Exp. Neurol. 2017, 292, 125–134. [Google Scholar] [CrossRef]
- Muraoka, Y.; Nakamura, A.; Tanaka, R.; Suda, K.; Azuma, Y.; Kushimura, Y.; Piccolo, L.L.; Yoshida, H.; Mizuta, I.; Tokuda, T.; et al. Genetic screening of the genes interacting with Drosophila FIG4 identified a novel link between CMT-causing gene and long noncoding RNAs. Exp. Neurol. 2018, 310, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhang, L.; Qin, C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 2017, 132, 160–169. [Google Scholar] [CrossRef]
- Lipovich, L.; Tarca, A.L.; Cai, J.; Jia, H.; Chugani, H.T.; Sterner, K.N.; Grossman, L.I.; Uddin, M.; Hof, P.R.; Sherwood, C.C.; et al. Developmental Changes in the Transcriptome of Human Cerebral Cortex Tissue: Long Noncoding RNA Transcripts. Cereb. Cortex 2013, 24, 1451–1459. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, B.; Campbell, D.B. Contribution of Long Noncoding RNAs to Autism Spectrum Disorder Risk. Int. Rev. Neurobiol. 2013, 113, 35–59. [Google Scholar] [CrossRef]
- Tang, J.; Yu, Y.; Yang, W. Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci. Ther. 2017, 23, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, S.; Ohtsuki, T.; Yu, S.-Y.; Tanabe, E.-I.; Yara, K.; Kamioka, M.; Matsushima, E.; Matsuura, M.; Ishikawa, K.; Minowa, Y.; et al. Significant linkage to chromosome 22q for exploratory eye movement dysfunction in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003, 123B, 27–32. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ip, J.Y.; Sone, M.; Nashiki, C.; Pan, Q.; Kitaichi, K.; Yanaka, K.; Abe, T.; Takao, K.; Miyakawa, T.; Blencowe, B.J.; et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016, 6, 27204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [Green Version]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.-C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, J.K.; James, R.; Brandon, N.; A Thomson, P. DISC1 and DISC2: Discovering and dissecting molecular mechanisms underlying psychiatric illness. Ann. Med. 2004, 36, 367–378. [Google Scholar] [CrossRef]
- Chubb, J.E.; Bradshaw, N.J.; Soares, D.C.; Porteous, D.; Millar, J.K. The DISC locus in psychiatric illness. Mol. Psychiatry 2007, 13, 36–64. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, O.O.; Haroutunian, V.; Davis, K.L.; Hernandez, I.; Sokolov, B.P. Novel putative nonprotein-coding RNA gene from 11q14 displays decreased expression in brains of patients with schizophrenia. J. Neurosci. Res. 2003, 74, 111–122. [Google Scholar] [CrossRef]
- Walsh, T.; McClellan, J.M.; McCarthy, S.E.; Addington, A.M.; Pierce, S.B.; Cooper, G.M.; Nord, A.S.; Kusenda, M.; Malhotra, D.; Bhandari, A.; et al. Rare Structural Variants Disrupt Multiple Genes in Neurodevelopmental Pathways in Schizophrenia. Science 2008, 320, 539–543. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Bénard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, X.; Ju, W.; Flory, M.; Zhong, J.; Jiang, S.; Wang, P.; Dong, X.; Tao, X.; Chen, Q.; et al. Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl. Psychiatry 2015, 5, e660. [Google Scholar] [CrossRef] [Green Version]
- Noor, A.; Whibley, A.; Marshall, C.R.; Gianakopoulos, P.J.; Piton, A.; Carson, A.R.; Orlic-Milacic, M.; Lionel, A.C.; Sato, D.; Pinto, D.; et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism Spectrum Disorder and Intellectual Disability. Sci. Transl. Med. 2010, 2, 49ra68. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Person, R.E.; Beaudet, A.L. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 2012, 21, 3001–3012. [Google Scholar] [CrossRef]
- Meng, L.; Person, R.E.; Huang, W.; Zhu, P.J.; Costa-Mattioli, M.; Beaudet, A.L. Truncation of Ube3a-ATS Unsilences Paternal Ube3a and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model. PLoS Genet. 2013, 9, e1004039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Hosseini, E.; Bagheri-Hosseinabadi, Z.; De Toma, I.; Jafarisani, M.; Sadeghi, I. The importance of long non-coding RNAs in neuropsychiatric disorders. Mol. Asp. Med. 2019, 70, 127–140. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Y.; Zhang, F.; Xu, M.; Zhang, J.; Yan, J.; Ju, W.; Brown, W.T.; Zhong, N. Hypermethylation of the enolase gene (ENO2) in autism. Eur. J. Pediatr. 2014, 173, 1233–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegiel, J.; Kuchna, I.; Nowicki, K.; Imaki, H.; Wegiel, J.; Marchi, E.; Ma, S.Y.; Chauhan, A.; Chauhan, V.; Bobrowicz, T.W.; et al. The neuropathology of autism: Defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010, 119, 755–770. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Pedrosa, E.; Shah, A.; Hrabovsky, A.; Maqbool, S.; Zheng, D.; Lachman, H.M. RNA-Seq of Human Neurons Derived from iPS Cells Reveals Candidate Long Non-Coding RNAs Involved in Neurogenesis and Neuropsychiatric Disorders. PLoS ONE 2011, 6, e23356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusconi, F.; Battaglioli, E.; Venturin, M. Psychiatric Disorders and lncRNAs: A Synaptic Match. Int. J. Mol. Sci. 2020, 21, 3030. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.P.; Zhou, Y.-D.; Zhang, G.; Jin, Z.; Stoppel, D.C.; Anderson, M.P. Increased Gene Dosage of Ube3a Results in Autism Traits and Decreased Glutamate Synaptic Transmission in Mice. Sci. Transl. Med. 2011, 3, 103ra97. [Google Scholar] [CrossRef] [Green Version]
- Vatsa, N.; Jana, N.R. UBE3A and Its Link with Autism. Front. Mol. Neurosci. 2018, 11, 448. [Google Scholar] [CrossRef] [PubMed]
- Khatri, N.; Man, H.Y. The Autism and Angelman Syndrome Protein Ube3A/E6AP: The Gene, E3 Ligase Ubiquitination Targets and Neurobiological Functions. Front. Mol. Neurosci. 2019, 12, 109. [Google Scholar] [CrossRef] [Green Version]
- Velmeshev, D.; Magistri, M.; Faghihi, M. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol. Autism 2013, 4, 32. [Google Scholar] [CrossRef] [Green Version]
- Kerin, T.; Ramanathan, A.; Rivas, K.; Grepo, N.; Coetzee, G.A.; Campbell, D.B. A Noncoding RNA Antisense to Moesin at 5p14.1 in Autism. Sci. Transl. Med. 2012, 4, 128ra40. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, J.J.; Grepo, N.; Wilkinson, B.; Evgrafov, O.V.; Knowles, J.A.; Campbell, D.B. Impact of the Autism-Associated Long Noncoding RNA MSNP1AS on Neuronal Architecture and Gene Expression in Human Neural Progenitor Cells. Genes 2016, 7, 76. [Google Scholar] [CrossRef]
- Luo, T.; Ou, J.N.; Cao, L.F.; Peng, X.Q.; Li, Y.M.; Tian, Y.Q. The Autism-Related lncRNA MSNP1AS Regulates Moesin Protein to Influence the RhoA, Rac1, and PI3K/Akt Pathways and Regulate the Structure and Survival of Neurons. Autism Res. 2020, 13, 2073–2082. [Google Scholar] [CrossRef]
- Luo, T.; Liu, P.; Wang, X.Y.; Li, L.Z.; Zhao, L.P.; Huang, J.; Li, Y.M.; Ou, J.L.; Peng, X.Q. Effect of the autism-associated lncRNA Shank2-AS on architecture and growth of neurons. J. Cell. Biochem. 2018, 120, 1754–1762. [Google Scholar] [CrossRef]
- Zaslavsky, K.; Zhang, W.B.; McCready, F.P.; Rodrigues, D.C.; Deneault, E.; Loo, C.; Zhao, M.; Ross, P.J.; Hajjar, J.E.; Romm, A.; et al. SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat. Neurosci. 2019, 22, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Namvar, A.; Arsang-Jang, S.; Komaki, A.; Taheri, M. Expression Analysis of BDNF, BACE1, and Their Natural Occurring Antisenses in Autistic Patients. J. Mol. Neurosci. 2020, 70, 194–200. [Google Scholar] [CrossRef]
- Khavari, B.; Cairns, M.J. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells 2020, 9, 1837. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Ma, Y.; Wang, Y.X.; Di, Y. Expression profiles of long noncoding RNAs in retinopathy of prematurity. Neural Regen. Res. 2020, 15, 1962–1968. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.; Vanichkina, D.; Poth, E.; Beveridge, N.; Ratnu, V.; Nayler, S.; Nones, K.; Hu, J.; Bredy, T.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safa, A.; Badrlou, E.; Arsang-Jang, S.; Sayad, A.; Taheri, M.; Ghafouri-Fard, S. Expression of NF-κB associated lncRNAs in schizophrenia. Sci. Rep. 2020, 10, 18105. [Google Scholar] [CrossRef] [PubMed]
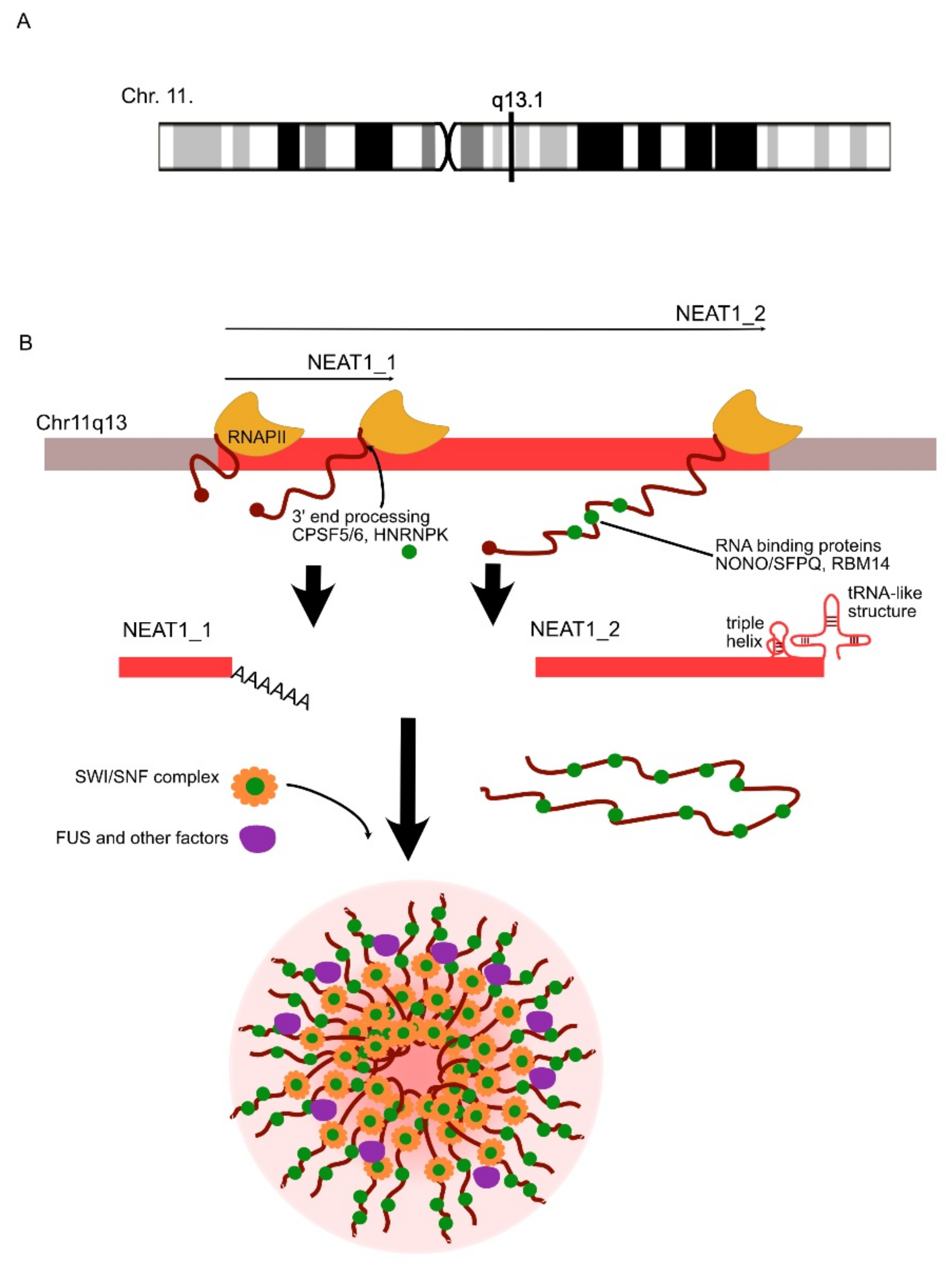
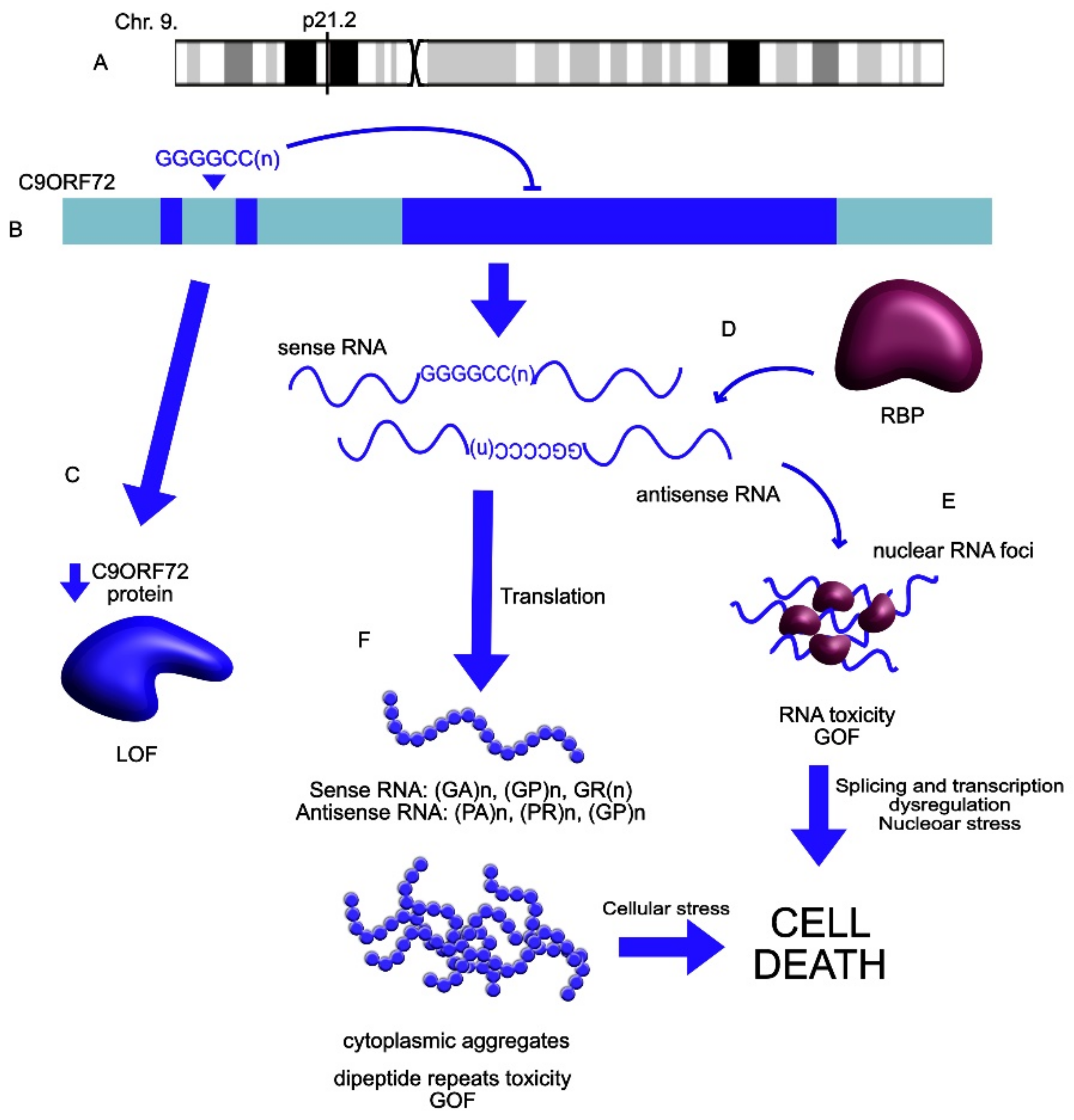
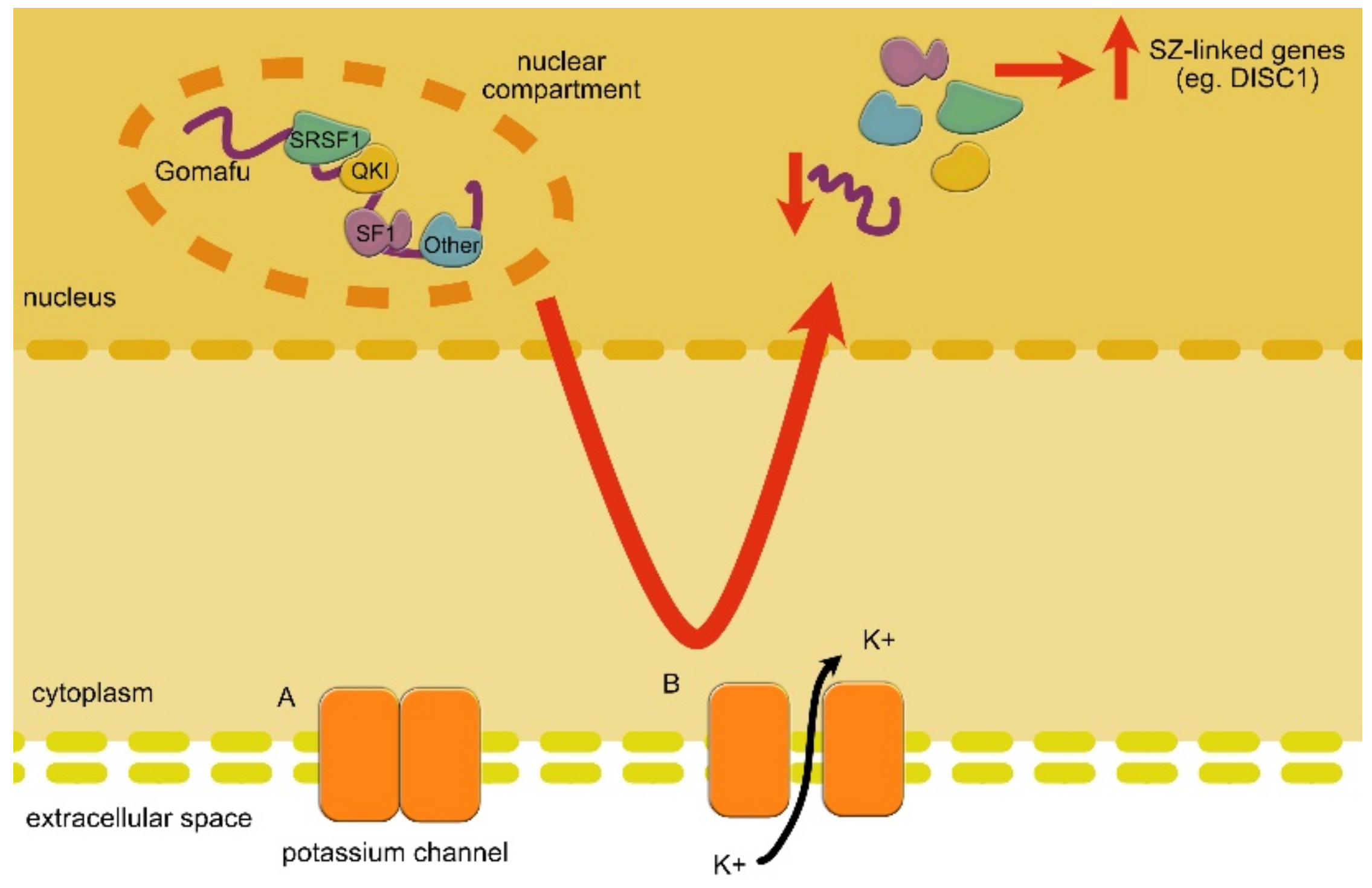
| LncRNA | Expression Change | Role | Linked Disorder | References |
|---|---|---|---|---|
| BACE1-AS | UP | BACE1-AS enhances mRNA BACE1 stability and activity. Leads to Aβ formation through the amyloid pathway. | AD | Faghihi et al., 2008 [97]; Faghihi et al., 2010 [71] |
| 51A | UP | 51A upregulates SORL1 variant A, which leads to Aβ accumulation through altered amyloid processing. | AD | Ma et al., 2009 [107]; Ciarlo et al., 2013 [108]; Luo and Chen 2016 [109] |
| 17A | UP | 17A disrupts GABAergic signalling (through the inhibition of GABAB R2 activity). This leads to an inflammation response and upregulation of Aβ formation and increases the Aβx-42/Aβx-40 ratio. | AD | Massone et al., 2011 [110]; Gavazzo et al., 2013 [111]; Buggia-Prevot and Thinakaran, 2014 [112] |
| NDM29 | UP | NDM29 promotes the cleavage activity of γ-secretase and BACE1 secretase, increasing the production of Aβ formation and the Aβx-42/Aβx-40 ratio. Moreover, it triggers an inflammatory response. | AD | Massone et al., 2012 [113] |
| BC200 | UP | BC200 takes part in the maintenance of long-term synaptic plasticity by targeting eIF4A and interacting with local proteins. In AD, it leads to the increased loss of synapses. | AD | Mus et al., 2007 [114]; Lin et al., 2008 [115] |
| NAT-Rad18 | UP | NAT-Rad18 increases the cell death rate in neurons, promoting the apoptotic processes. | AD | Iacoangeli et al., 2010 [116]; Massone et al., 2012 [113]; Luo and Chen, 2016 [109] |
| NEAT1 | UP/DOWN | NEAT1 is involved in the decreased clearance of Aβ. | AD | Wang et al., 2019 [117]; Zhao et al., 2019 [118]; |
| UP | NEAT1 regulates the assembly of paraspeckles and might trigger neurotoxic processes in ALS. | ALS | Clemson et al., 2009 [119]; Suzuki et al., 2019 [120] | |
| MALAT1 | UP | MALAT1 facilitates paraspeckle formation by binding with FUS and TDP-43. Moreover, it controls the phosphorylation of SR proteins and gene expression in cis. | ALS | Clark et al., 2014 [50]; An et al., 2019 [121]; Wu and Kuo, 2020 [106] |
| C9ORF72 | UP | C9ORF72 extended repeats mutation leads to the repeat-associated translation into neurotoxic misfolded proteins and dipeptides. Contributes to the SGs’ formation and cellular inclusions. | ALS | Mizielinska et al., 2014 [122]; Wen et al., 2014 [123]; Maharjan et al., 2017 [124]; Wan et al., 2017 [29]; Swinnen et al., 2018 [125] Bampton et al., 2020 [126] Mizielinska et al., 2013 [127] |
| SATIII(Hsrω) | UP | SAT III binds to TDP-43 and takes part in promoting its elongation (by binding to the ELL2 domain) during the transcription, which can affect TDP-43 neurotoxicity. | ALS | Chung et al., 2018 [128]; Chen, K. and Chen, 2020 [5]; Wu, et al., 2020 [106] |
| ATXN2-AS | DOWN | ATXN1-AS extended repeats form RNA foci and lead to an increase in apoptosis through interactions with caspase 3/7. | ALS | Li et al., 2016 [129] |
| SNAP25-AS | DOWN | SNAP25AS affects SNAP25 and processes controlled by it such as synaptic vesicle transport or axonal repair processes. | ALS | Gagliardi et al., 2018 [130]; Wu et al., 2020 [106] |
| LncRNA | Expression Change | Role | Linked Disorder | References |
|---|---|---|---|---|
| GOMAFU | DOWN | GOMAFU controls alternative splicing through interactions with splicing factors. Moreover, it affects the specification of amacrine cells and is involved in SZ-related eye movement disorder. | SZ | Takahashi et al., 2003 [172]; Rapicavoli et al., 2010 [173]; Tsuiji et al., 2011 [95]; Ip et al., 2016 [174] |
| MALAT1 | DOWN | MALAT1 controls the expression of genes linked to synaptogenesis through interactions with SR proteins. The downregulation of MALAT1 leads to impaired formation of synapses and reduced synaptic density. | SZ | Bernard et al., 2010 [175]; Madabhushi et al., 2015 [176] |
| DISC1-AS | DOWN | DISC1-AS affects cAMP signaling through interactions with DISC1 and DISC2. | SZ | Millar et al., 2004 [177]; Chubb et al., 2008 [178] |
| DISC2-AS | DOWN | DISC2-AS affects cAMP signaling (through interactions with DISC2), neuregulin signalling, axonal signalling and also long-term synaptic potentiation. | SZ | Polesskaya et al., 2003 [179]; Walsh et al., 2008 [180] |
| NEAT1 | DOWN | NEAT1 takes part in the unfolded proteins’ response under a condition of cellular stress. | SZ | Nakagawa et al., 2011 [181]; Hirose et al., 2014 [182] |
| SHANK2-AS | DOWN | SHANK2-AS affects the processes regulating post-synaptic density through interactions with SHANK2. | ASD | Wang et al., 2015 [183] |
| BDNF-AS | DOWN | BDNF-AS inhibits the BDNF transcript, which is a crucial transcription factor involved in neurite functioning. | ASD | Wang et al., 2015 [183] |
| PTCHD1AS1-3 | DOWN | PTCHD1AS1-3 is linked to the dysfunction of synapses and neurons in ASD. | ASD | Noor et al., 2010 [184] |
| NRON | UP | NRON inhibits NFAT (nuclear factor-activated T-cell) signaling. | Major Depressive Disorder | Willingham et al., 2005 [85] |
| AK081227 | UP | AK081227 downregulates GABAergic signalling through the inhibition of Gabrr2 expression. | Rett Syndrome | Petazzi et al., 2013 [54] |
| Ube3aATS | DOWN | Ube3aATS downregulation is associated with impaired contextual fear behavior (in Angelman syndrome). This is due to the impaired silencing of paternal Ube3a. | Angelman Syndrome | Meng et al., 2012 [185]; Meng et al., 2013 [186]; Meng et al., 2015 [187] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Non-Coding RNA 2021, 7, 36. https://doi.org/10.3390/ncrna7020036
Aliperti V, Skonieczna J, Cerase A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Non-Coding RNA. 2021; 7(2):36. https://doi.org/10.3390/ncrna7020036
Chicago/Turabian StyleAliperti, Vincenza, Justyna Skonieczna, and Andrea Cerase. 2021. "Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders" Non-Coding RNA 7, no. 2: 36. https://doi.org/10.3390/ncrna7020036






