Estimation of the Total Nonstructural Carbohydrate Concentration in Apple Trees Using Hyperspectral Imaging
Abstract
:1. Introduction
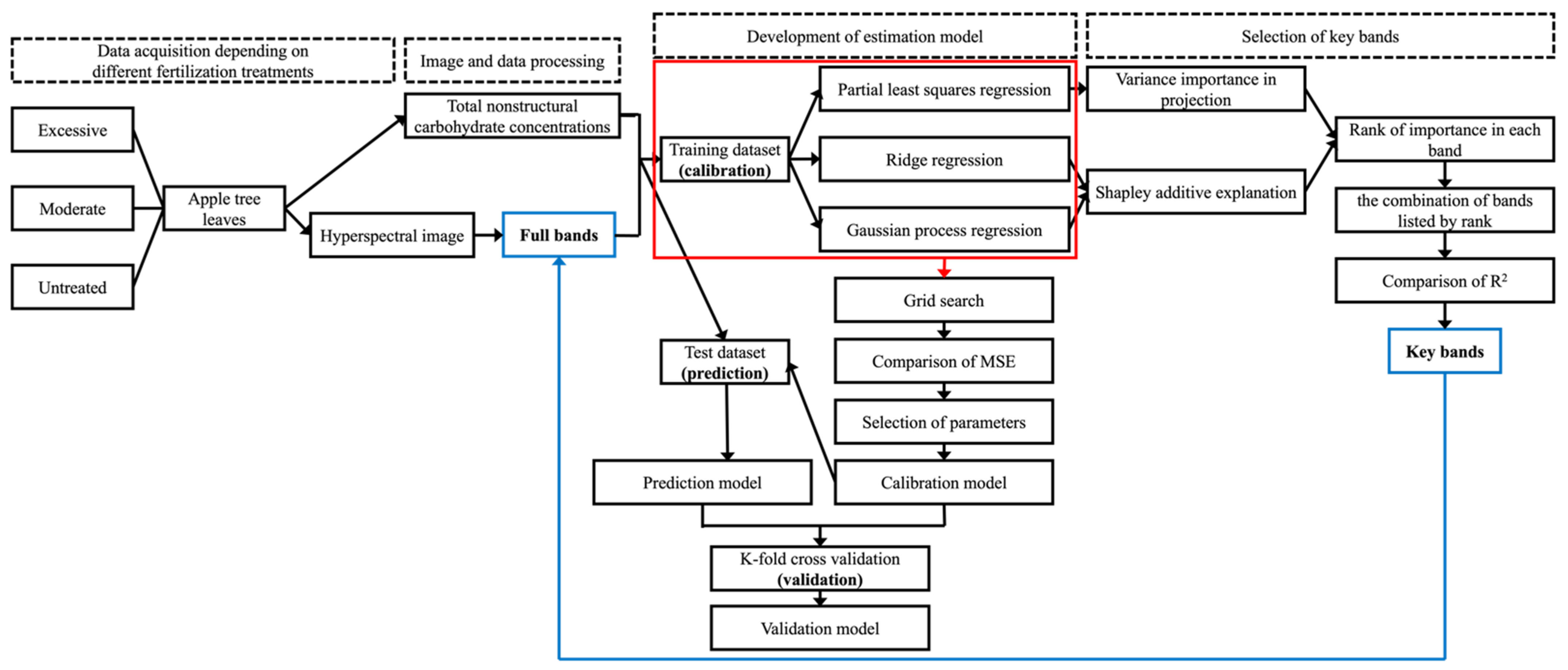
2. Materials and Methods
2.1. Experimental Design
2.2. Hyperspectral Image Acquisition and Processing
2.3. Total Nonstructural Carbohydrate Concentration
2.4. Analysis
2.4.1. Partial Least Squares Regression
2.4.2. Ridge Regression
2.4.3. Gaussian Process Regression
2.4.4. Variance Importance in Projection
2.4.5. Shapley Additive Explanation
3. Results
3.1. Basic Analysis of the Total Nonstructural Carbohydrate Concentration
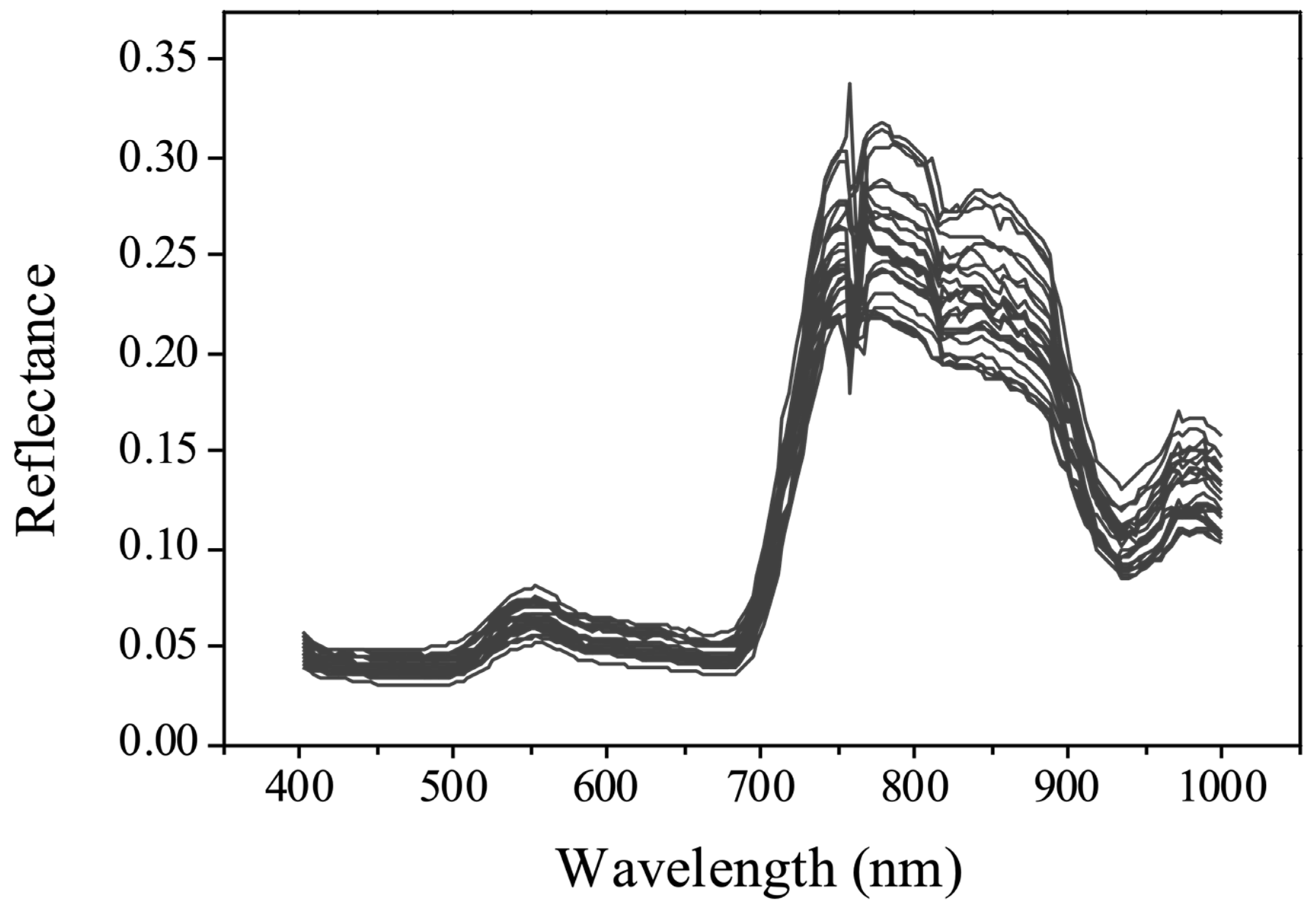
3.2. Reflectance Curves
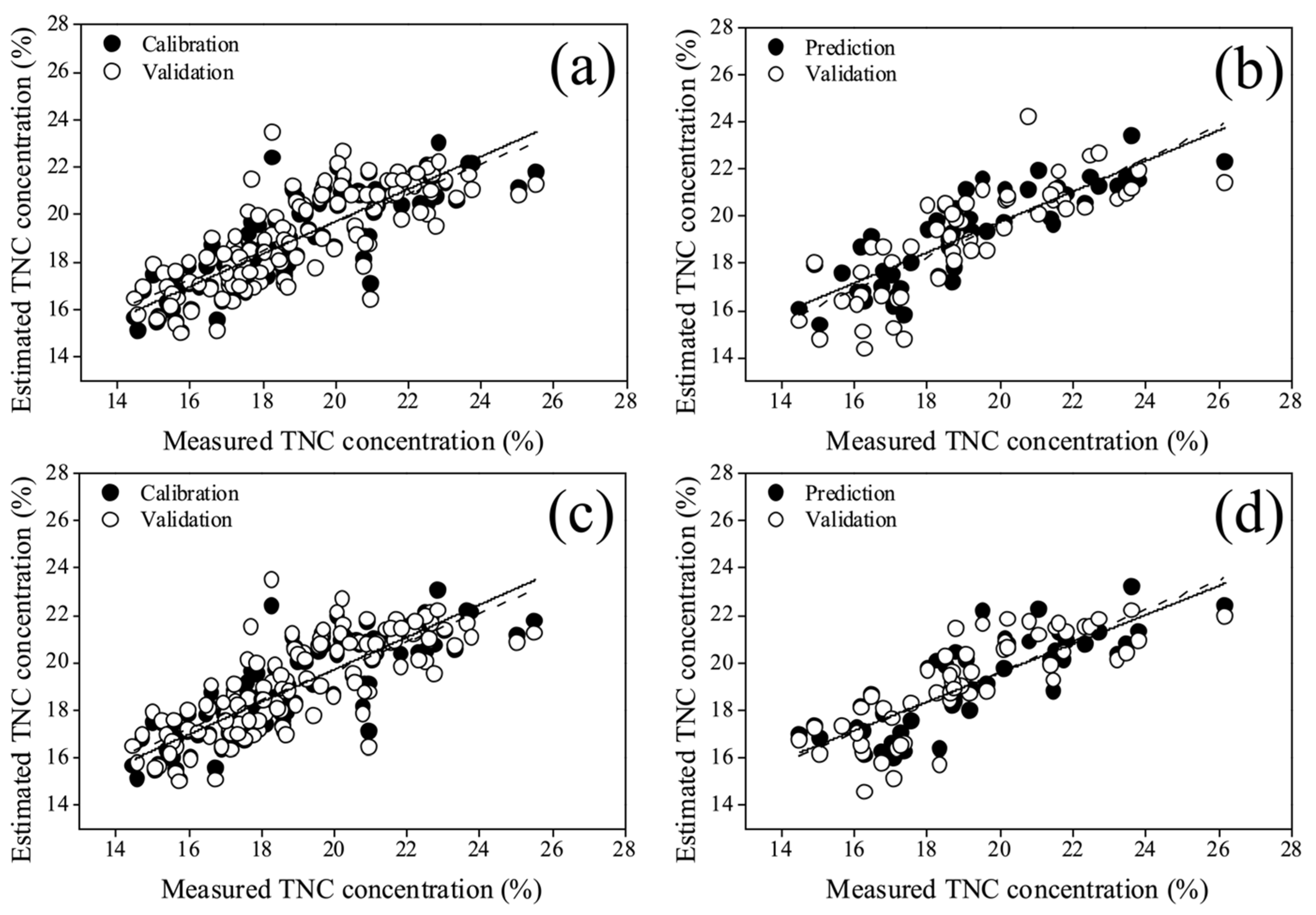
3.3. Estimation Performance of the Models with Full Bands
3.4. Estimation Performance of the Models with Key Bands
4. Discussion
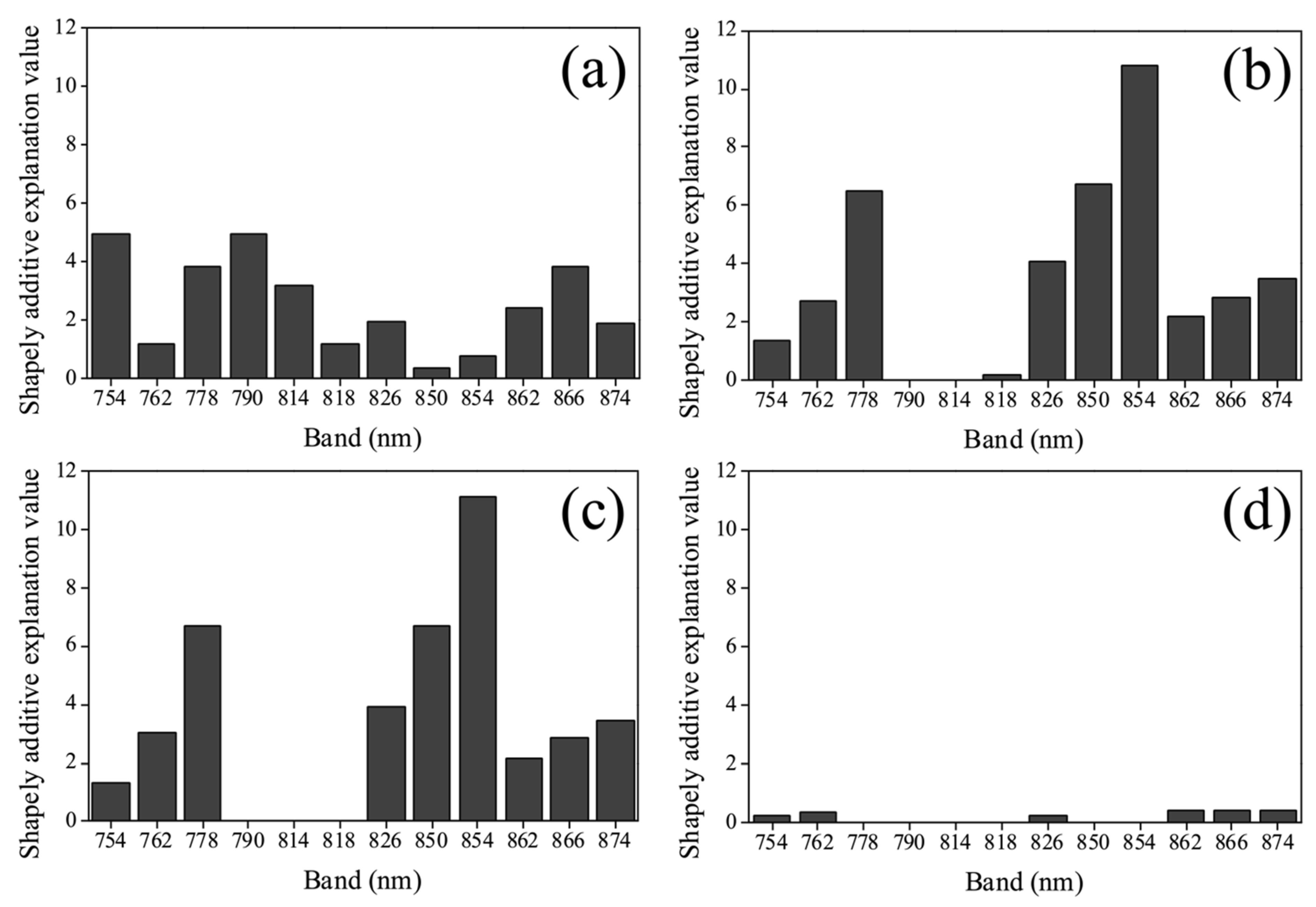
4.1. Relationship between Selected Bands and Total Nonstructural Carbohydrate Concentration
4.2. Comparison with Related Studies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aparadh, V.T.; Karadge, B.A. Comparative carbohydrates status in leaf developmental stages of Cleome species. Int. J. Pharm. Sci. Rev. Res. 2012, 14, 130–132. [Google Scholar]
- Larson, J.E.; Perkins-Veazie, P.; Ma, G.; Kon, T.M. Quantification and prediction with near infrared spectroscopy of carbohydrates throughout apple fruit development. Horticulturae 2023, 9, 279. [Google Scholar] [CrossRef]
- Haller, M.H.; Magness, J.R. Relation of Leaf Area and Position to Quality of Fruity and to Bud Differentiation in Apples (No. 1488-2016-123788). Ph.D. Thesis, University of Maryland, College Park, MD, USA, 1933. [Google Scholar]
- Rutkowski, K.; Łysiak, G.P. Effect of nitrogen fertilization on tree growth and nutrient content in soil and cherry leaves (Prunus cerasus L.). Agriculture 2023, 13, 578. [Google Scholar] [CrossRef]
- Shaolan, H.E.; Lie, D.E.N.G.; Yiqin, L.I.; Rong, L. Effects of floral promotion or inhibition treatments on flowering of citrus trees and protein fractions in buds. J. Trop. Subtrop. Bot. 1998, 6, 124–130. [Google Scholar]
- Budiarto, R.; Poerwanto, R.; Santosa, E.; Efendi, D. Shoot manipulations improve flushing and flowering of mandarin citrus in Indonesia. J. Appl. Hortic. 2018, 20, 112–118. [Google Scholar] [CrossRef]
- Goldschmidt, E.E.; Golomb, A. The carbohydrate balance of alternate-bearing citrus trees and the significance of reserves for flowering and Fruiting1. J. Am. Soc. Hortic. Sci. 1982, 107, 206–208. [Google Scholar] [CrossRef]
- Goldschmidt, E.E. Carbohydrate supply as a critical factor for citrus fruit development and productivity. HortScience 1999, 34, 1020–1024. [Google Scholar] [CrossRef]
- Zwieniecki, M.A.; Davidson, A.M.; Orozco, J.; Cooper, K.B.; Guzman-Delgado, P. The impact of non-structural carbohydrates (NSC) concentration on yield in Prunus dulcis, Pistacia vera, and Juglans regia. Sci. Rep. 2022, 12, 4360. [Google Scholar] [CrossRef]
- Bustan, A.; Avni, A.; Lavee, S.; Zipori, I.; Yeselson, Y.; Schaffer, A.A.; Riov, J.; Dag, A. Role of carbohydrate reserves in yield production of intensively cultivated oil olive (Olea europaea L.) trees. Tree Physiol. 2011, 31, 519–530. [Google Scholar] [CrossRef]
- Rossouw, G. Grapevine Carbohydrate and Nitrogen Allocation during Berry Maturation: Implications of Source-Sink Relations and Water Supply. Ph.D. Thesis, Charles Sturt University, Bathurst, Australia, 2017. [Google Scholar]
- Suárez, L.; Zarco-Tejada, P.J.; González-Dugo, V.; Berni, J.A.J.; Sagardoy, R.; Morales, F.; Fereres, E. Detecting water stress effects on fruit quality in orchards with time-series PRI airborne imagery. Remote Sens. Environ. 2010, 114, 286–298. [Google Scholar] [CrossRef]
- Skarpe, C.; Hester, A.J. Plant traits, browsing and gazing herbivores, and vegetation dynamics. Ecol. Stud. 2008, 195, 217–261. [Google Scholar]
- Demestihas, C.; Plénet, D.; Génard, M.; Raynal, C.; Lescourret, F. Ecosystem services in orchards. A review. Agron. Sustain. Dev. 2017, 37, 1–21. [Google Scholar] [CrossRef]
- Albrigo, L.G.; Buker, R.S.; Burn, J.K.; Castle, W.S.; Futch, S.; McCoy, C.W.; Muraro, R.P.; Rogers, M.E.; Syvertsen, J.P.; Timmer, L.W.; et al. The impact of four hurricanes in 2004 on the Florida citrus industry: Experiences and lessons learned. Proc. Fla. State Hortic. Soc. 2005, 118, 66–74. [Google Scholar]
- Sishodia, R.P.; Ray, R.L.; Singh, S.K. Applications of remote sensing in precision agriculture: A review. Remote Sens. 2020, 12, 3136. [Google Scholar] [CrossRef]
- Raj, R.; Kar, S.; Nandan, R.; Jagarlapudi, A. Precision agriculture and unmanned aerial Vehicles (UAVs). In Unmanned Aerial Vehicle: Applications in Agriculture and Environment; Springer: Berlin/Heidelberg, Germany, 2020; pp. 7–23. [Google Scholar]
- Ashapure, A.; Jung, J.; Chang, A.; Oh, S.; Yeom, J.; Maeda, M.; Maeda, A.; Dube, N.; Landivar, J.; Hague, S.; et al. Developing a machine learning based cotton yield estimation framework using multi-temporal UAS data. ISPRS J. Photogramm. 2020, 169, 180–194. [Google Scholar] [CrossRef]
- Hunt Jr, E.R.; Daughtry, C.S.T. What good are unmanned aircraft systems for agricultural remote sensing and precision agriculture? Int. J. Remote Sens. 2018, 39, 5345–5376. [Google Scholar] [CrossRef]
- Kumar, L.; Schmidt, K.; Dury, S.; Skidmore, A. Imaging spectrometry and vegetation science. In Imaging Spectrometry: Basic Principles and Prospective Applications; Springer: Berlin/Heidelberg, Germany, 2002; pp. 111–155. [Google Scholar]
- Rashwan, S.; Dobigeon, N. A split-and-merge approach for hyperspectral band selection. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1378–1382. [Google Scholar] [CrossRef]
- Sawant, S.; Manoharan, P. Hyperspectral band selection based on metaheuristic optimization approach. Infrared Phys. Technol. 2020, 107, 103295. [Google Scholar] [CrossRef]
- Li, M.; Zhu, X.; Li, W.; Tang, X.; Yu, X.; Jiang, Y. Retrieval of nitrogen content in apple canopy based on unmanned aerial vehicle hyperspectral images using a modified correlation coefficient method. Sustainability 2022, 14, 1992. [Google Scholar] [CrossRef]
- Huang, J.; Yan, X. Gaussian and non-Gaussian double subspace statistical process monitoring based on principal component analysis and independent component analysis. Ind. Eng. Chem. Res. 2015, 54, 1015–1027. [Google Scholar] [CrossRef]
- Malthouse, E.C. Limitations of nonlinear PCA as performed with generic neural networks. IEEE Trans. Neural Netw. 1998, 9, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.H.; Wang, L.Y.; Yang, Z.Q.; Zhang, Y.Y.; Li, X.; Song, L.; He, L.; Duan, J.Z.; Feng, W. Hyperspectral monitoring of powdery mildew disease severity in wheat based on machine learning. Front. Plant Sci. 2022, 13, 828454. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef]
- Li, W.; Zhu, X.; Yu, X.; Li, M.; Tang, X.; Zhang, J.; Xue, Y.; Zhang, C.; Jiang, Y. Inversion of nitrogen concentration in apple canopy based on UAV hyperspectral images. Sensors 2022, 22, 3503. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.S.; Jang, S.H.; Park, J.W.; Song, H.Y.; Ryu, C.S.; Jun, S.R.; Kim, S.H. Yield prediction and validation of onion (Allium cepa L.) using key variables in narrowband hyperspectral imagery and effective accumulated temperature. Comput. Electron. Agric. 2020, 178, 105667. [Google Scholar] [CrossRef]
- Mateos-Aparicio, G. Partial least squares (PLS) methods: Origins, evolution, and application to social sciences. Commun. Stat.-Theory Methods 2011, 40, 2305–2317. [Google Scholar] [CrossRef]
- Ji, S.; Gu, C.; Xi, X.; Zhang, Z.; Hong, Q.; Huo, Z.; Zhao, H.; Zhang, R.; Li, B.; Tan, C. Quantitative monitoring of leaf area index in rice based on hyperspectral feature bands and ridge regression algorithm. Remote Sens. 2022, 14, 2777. [Google Scholar] [CrossRef]
- Arefi, A.; Sturm, B.; von Gersdorff, G.; Nasirahmadi, A.; Hensel, O. Vis-NIR hyperspectral imaging along with Gaussian process regression to monitor quality attributes of apple slices during drying. LWT 2021, 152, 112297. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Gitelson, A.; Delegido, J.; Moreno, J.; Camps-Valls, G. Spectral band selection for vegetation properties retrieval using Gaussian processes regression. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 554–567. [Google Scholar] [CrossRef]
- Stellacci, A.M.; Castrignanò, A.; Troccoli, A.; Basso, B.; Buttafuoco, G. Selecting optimal hyperspectral bands to discriminate nitrogen status in durum wheat: A comparison of statistical approaches. Environ. Monit. Assess. 2016, 188, 1–15. [Google Scholar] [CrossRef]
- Kannangara, K.P.M.; Zhou, W.; Ding, Z.; Hong, Z. Investigation of feature contribution to shield tunneling-induced settlement using Shapley additive explanations method. J. Rock Mech. Geotech. Eng. 2022, 14, 1052–1063. [Google Scholar] [CrossRef]
- Leite, R.G.; Cardoso, A.D.S.; Fonseca, N.V.B.; Silva, M.L.C.; Tedeschi, L.O.; Delevatti, L.M.; Ruggieri, A.C.; Reis, R.A. Effects of nitrogen fertilization on protein and carbohydrate fractions of Marandu palisadegrass. Sci. Rep. 2021, 11, 14786. [Google Scholar] [CrossRef]
- Imada, S.; Tako, Y. Seasonal accumulation of photoassimilated carbon relates to growth rate and use for new aboveground organs of young apple trees in following spring. Tree Physiol. 2022, 42, 2294–2305. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Filella, I. Visible and near-infrared reflectance techniques for diagnosing plant physiological status. Trends Plant Sci. 1998, 3, 151–156. [Google Scholar] [CrossRef]
- Sun, W.; Du, Q. Hyperspectral band selection: A review. IEEE Geosci Remote Sens. Mag. 2019, 7, 118–139. [Google Scholar] [CrossRef]
- Hultquist, C.; Chen, G.; Zhao, K. A comparison of Gaussian process regression, random forests and support vector regression for burn severity assessment in diseased forests. Remote Sens. Lett. 2014, 5, 723–732. [Google Scholar] [CrossRef]
- Mutanen, T.; Sirro, L.; Rauste, Y. Tree height estimates in boreal forest using Gaussian process regression. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS) 2016, Beijing, China, 10–15 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1757–1760. [Google Scholar]
- Xie, R.; Darvishzadeh, R.; Skidmore, A.K.; Heurich, M.; Holzwarth, S.; Gara, T.W.; Reusen, I. Mapping leaf area index in a mixed temperate forest using Fenix airborne hyperspectral data and Gaussian processes regression. Int. J. Appl. Earth Obs. Geoinf. 2021, 95, 102242. [Google Scholar] [CrossRef]
- Ye, X.; Abe, S.; Zhang, S. Estimation and mapping of nitrogen content in apple trees at leaf and canopy levels using hyperspectral imaging. Precis Agric. 2020, 21, 198–225. [Google Scholar] [CrossRef]
- Ohyama, T. Nitrogen as a major essential element of plants. Nitrogen Assim. Plants 2010, 37, 1–17. [Google Scholar]
- Yu, R.; Zhu, X.; Cao, S.; Xiong, J.; Wen, X.; Jiang, Y.; Zhao, G. Estimation of chlorophyll content in apple leaves based on imaging spectroscopy. J. Appl. Spectrosc. 2019, 86, 457–464. [Google Scholar] [CrossRef]
- Streb, S.; Zeeman, S.C. Starch metabolism in Arabidopsis. Arab. Book Am. Soc. Plant Biol. 2012, 10, e0160. [Google Scholar] [CrossRef] [PubMed]
- Aluko, O.O.; Li, C.; Wang, Q.; Liu, H. Sucrose utilization for improved crop yields: A review article. Int. J. Mol. Sci. 2021, 22, 4704. [Google Scholar] [CrossRef]
- Giraldo, J.P.; Wu, H.; Newkirk, G.M.; Kruss, S. Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol. 2019, 14, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ying, Y.; Ping, J. Recent advances in plant nanoscience. Adv. Sci. 2022, 9, e2103414. [Google Scholar] [CrossRef] [PubMed]
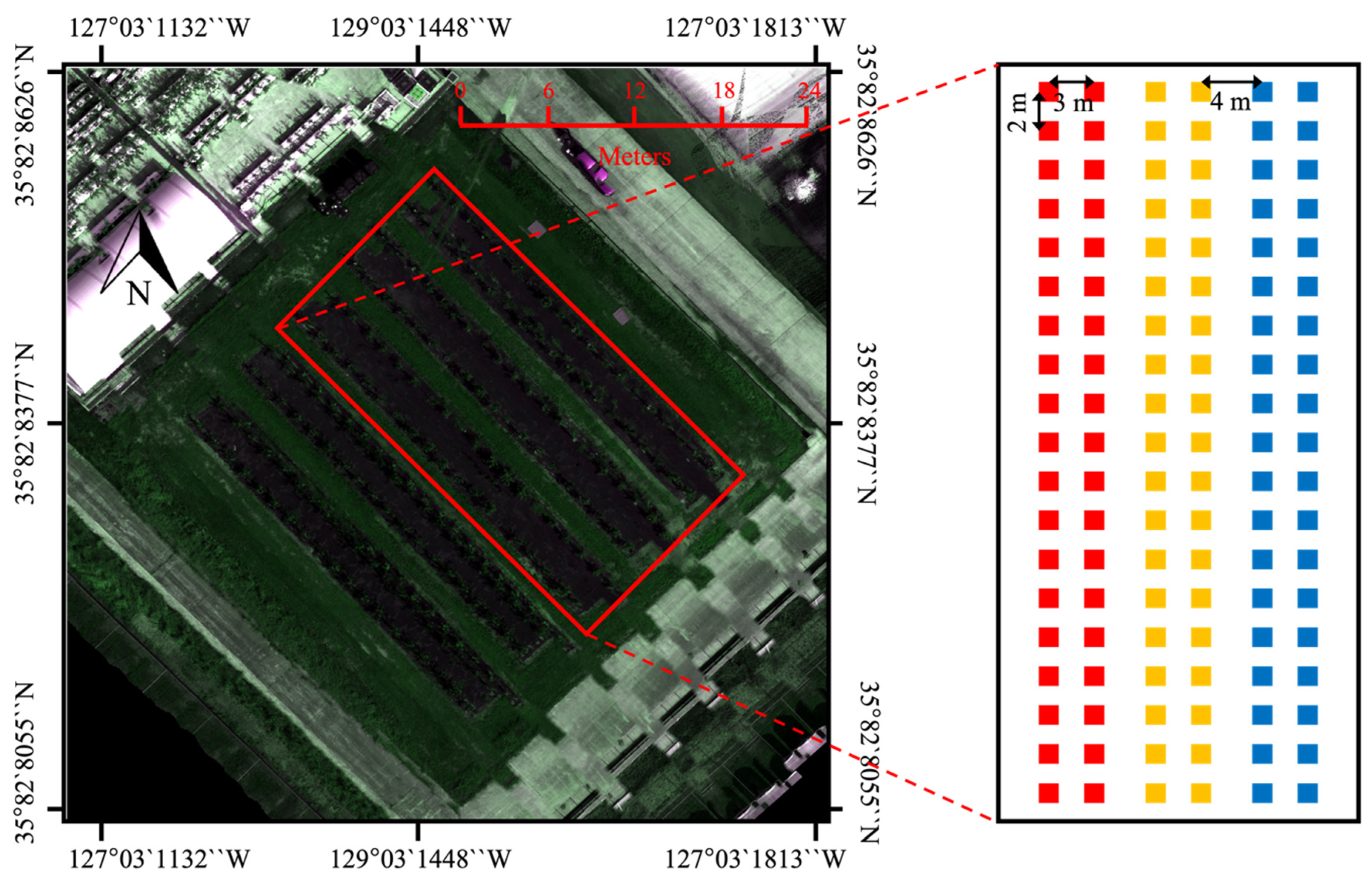


| Information | |
|---|---|
| Field coordinates | Coordinate 1: 35.828626, 127.031448 Coordinate 2: 35.828377, 127.031132 Coordinate 3: 35.828085, 127.031484 Coordinate 4: 35.828355, 127.031813 |
| Flight speed | 6 m/s |
| Overlap | 70% |
| Altitude | 60 m |
| Ground sample distance | 4.4 cm/pixel |
| May 23 | June 3 | June 17 | July 4 | July 19 | July 28 | August 16 | September 07 | September 21 | |
|---|---|---|---|---|---|---|---|---|---|
| E * | 1.04 a ** | 0.86 a | 1.27 a | 2.28 a | 2.47 a | 1.54 a | 2.29 a | 2.00 a | 1.12 a |
| M | 0.96 a | 1.04 a | 1.10 a | 1.20 a | 0.61 a | 1.44 b | 1.54 a | 1.45 a | 1.48 b |
| U | 1.20 a | 1.79 a | 0.46 a | 1.04 a | 0.62 a | 0.95 b | 1.84 a | 0.75 a | 1.36 b |
| All | 1.04 A | 1.38 BC | 1.05 A | 1.72 C | 1.43 B | 1.65 D | 1.96 EF | 1.45 DE | 1.77 F |
| PLS | RR | GP | ||
|---|---|---|---|---|
| n * | 127 | |||
| Standard deviation | 2.46 | |||
| (Training) Calibration | R2 | 0.71 | 0.73 | 0.73 |
| RMSE (%) | 1.32 | 1.29 | 1.29 | |
| RE (%) | 6.91 | 6.76 | 6.76 | |
| (Training) Validation | R2 | 0.61 | 0.59 | 0.59 |
| RMSE (%) | 1.54 | 1.57 | 1.57 | |
| RE (%) | 8.07 | 8.22 | 8.22 | |
| n | 55 | |||
| Standard deviation | 2.27 | |||
| (Test) Prediction | R2 | 0.70 | 0.72 | 0.72 |
| RMSEP (%) | 1.41 | 1.37 | 1.37 | |
| RE (%) | 7.43 | 7.22 | 7.22 | |
| (Test) Validation | R2 | 0.57 | 0.64 | 0.64 |
| RMSEP (%) | 1.68 | 1.55 | 1.55 | |
| RE (%) | 8.86 | 8.17 | 8.17 | |
| PLS | RR | GP | |
|---|---|---|---|
| Band 1 | 401 | 754 | 754 |
| Band 2 | 405 | 762 | 762 |
| Band 3 | 697 | 778 | 778 |
| Band 4 | 701 | 826 | 826 |
| Band 5 | 705 | 850 | 850 |
| Band 6 | 709 | 854 | 854 |
| Band 7 | 758 | 862 | 862 |
| Band 8 | 762 | 866 | 866 |
| Band 9 | 766 | 874 | 874 |
| PLS | RR | GP | ||
|---|---|---|---|---|
| n * | 127 | |||
| Stand ard deviation | 2.46 | |||
| (Training) Calibration | R2 | 0.71 | 0.73 | 0.73 |
| RMSE (%) | 1.32 | 1.29 | 1.29 | |
| RE (%) | 6.91 | 6.76 | 6.76 | |
| (Training) Validation | R2 | 0.61 | 0.59 | 0.59 |
| RMSE (%) | 1.54 | 1.57 | 1.57 | |
| RE (%) | 8.07 | 8.22 | 8.22 | |
| n | 55 | |||
| Standard deviation | 2.27 | |||
| (Test) Prediction | R2 | 0.30 | 0.63 | 0.68 |
| RMSEP (%) | 2.16 | 1.56 | 1.46 | |
| RE (%) | 11.4 | 8.23 | 7.70 | |
| (Test) Validation | R2 | 0.12 | 0.61 | 0.65 |
| RMSEP (%) | 2.41 | 1.62 | 1.52 | |
| RE (%) | 12.7 | 8.54 | 8.01 | |
| Number of Bands | Training Dataset | Test Dataset | ||
|---|---|---|---|---|
| 15 | Calibration | 0.73 | Prediction | 0.56 |
| Validation | 0.59 | Validation | 0.55 | |
| 14 | Calibration | 0.73 | Prediction | 0.56 |
| Validation | 0.59 | Validation | 0.56 | |
| 13 | Calibration | 0.73 | Prediction | 0.55 |
| Validation | 0.59 | Validation | 0.56 | |
| 12 | Calibration | 0.73 | Prediction | 0.55 |
| Validation | 0.59 | Validation | 0.55 | |
| 11 | Calibration | 0.73 | Prediction | 0.68 |
| Validation | 0.59 | Validation | 0.64 | |
| 10 | Calibration | 0.73 | Prediction | 0.68 |
| Validation | 0.59 | Validation | 0.63 | |
| 9 | Calibration | 0.73 | Prediction | 0.68 |
| Validation | 0.59 | Validation | 0.65 | |
| 8 | Calibration | 0.73 | Prediction | 0.62 |
| Validation | 0.59 | Validation | 0.58 | |
| 7 | Calibration | 0.73 | Prediction | 0.62 |
| Validation | 0.59 | Validation | 0.58 | |
| 6 | Calibration | 0.73 | Prediction | 0.42 |
| Validation | 0.59 | Validation | 0.45 | |
| 5 | Calibration | 0.73 | Prediction | 0.39 |
| Validation | 0.59 | Validation | 0.57 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Y.-S.; Park, K.-S.; Kim, E.-R.; Jeong, J.-C.; Ryu, C.-S. Estimation of the Total Nonstructural Carbohydrate Concentration in Apple Trees Using Hyperspectral Imaging. Horticulturae 2023, 9, 967. https://doi.org/10.3390/horticulturae9090967
Kang Y-S, Park K-S, Kim E-R, Jeong J-C, Ryu C-S. Estimation of the Total Nonstructural Carbohydrate Concentration in Apple Trees Using Hyperspectral Imaging. Horticulturae. 2023; 9(9):967. https://doi.org/10.3390/horticulturae9090967
Chicago/Turabian StyleKang, Ye-Seong, Ki-Su Park, Eun-Ri Kim, Jong-Chan Jeong, and Chan-Seok Ryu. 2023. "Estimation of the Total Nonstructural Carbohydrate Concentration in Apple Trees Using Hyperspectral Imaging" Horticulturae 9, no. 9: 967. https://doi.org/10.3390/horticulturae9090967






