Magnetic Nylon 6 Nanocomposites for the Microextraction of Nucleic Acids from Biological Samples
Abstract
:1. Introduction
2. Results and Discussion
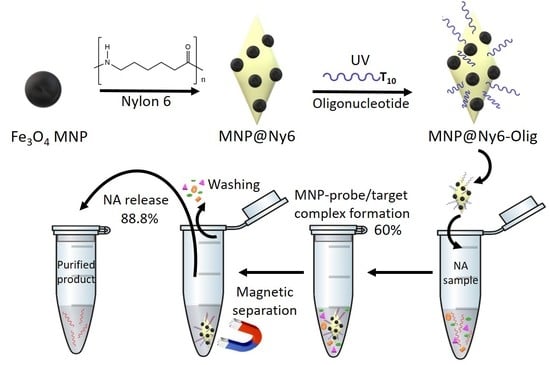
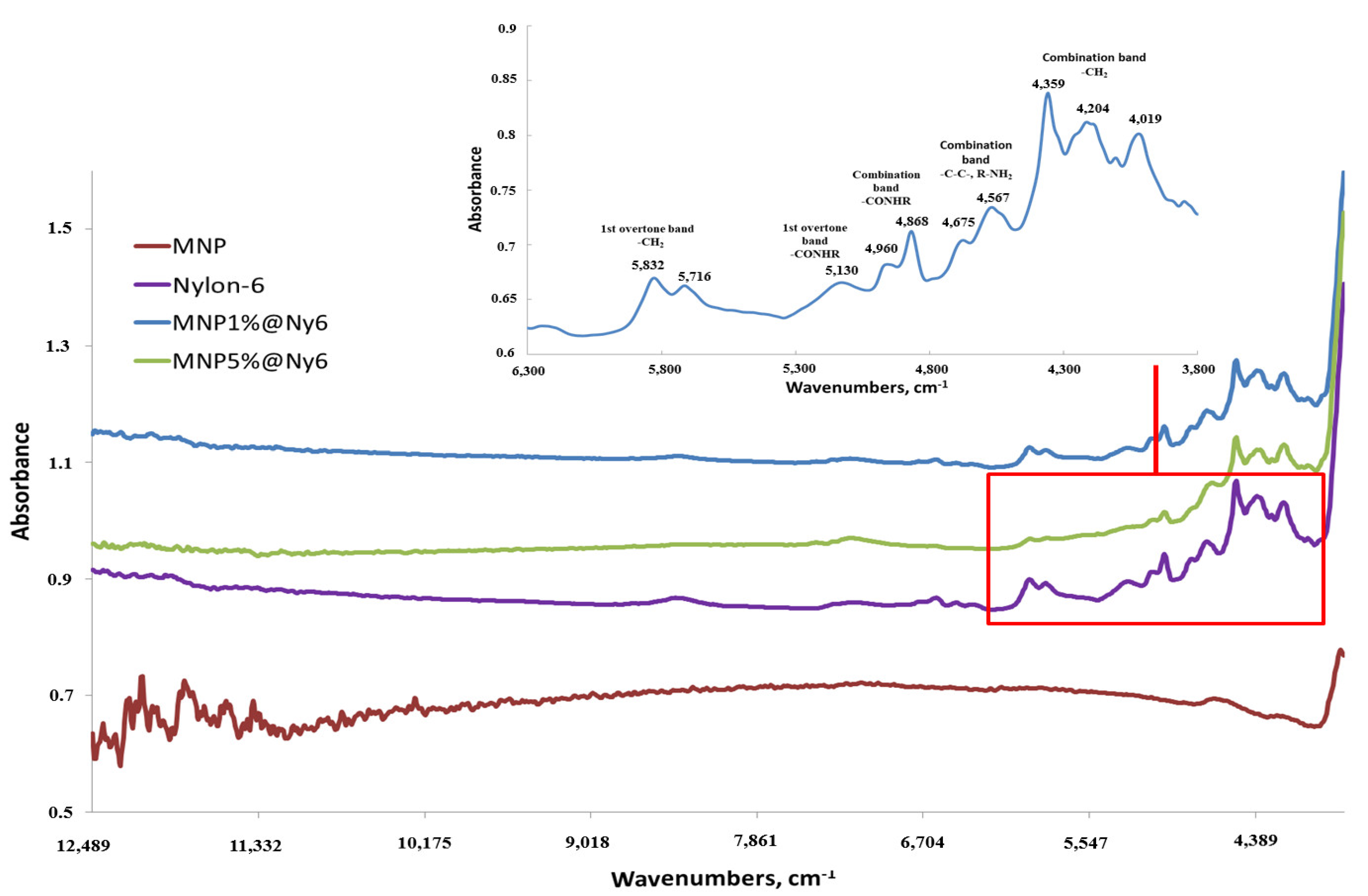
2.1. Synthesis and Characterization of MNP- and MNP@Ny6-Based Oligonucleotide Nanoprobes
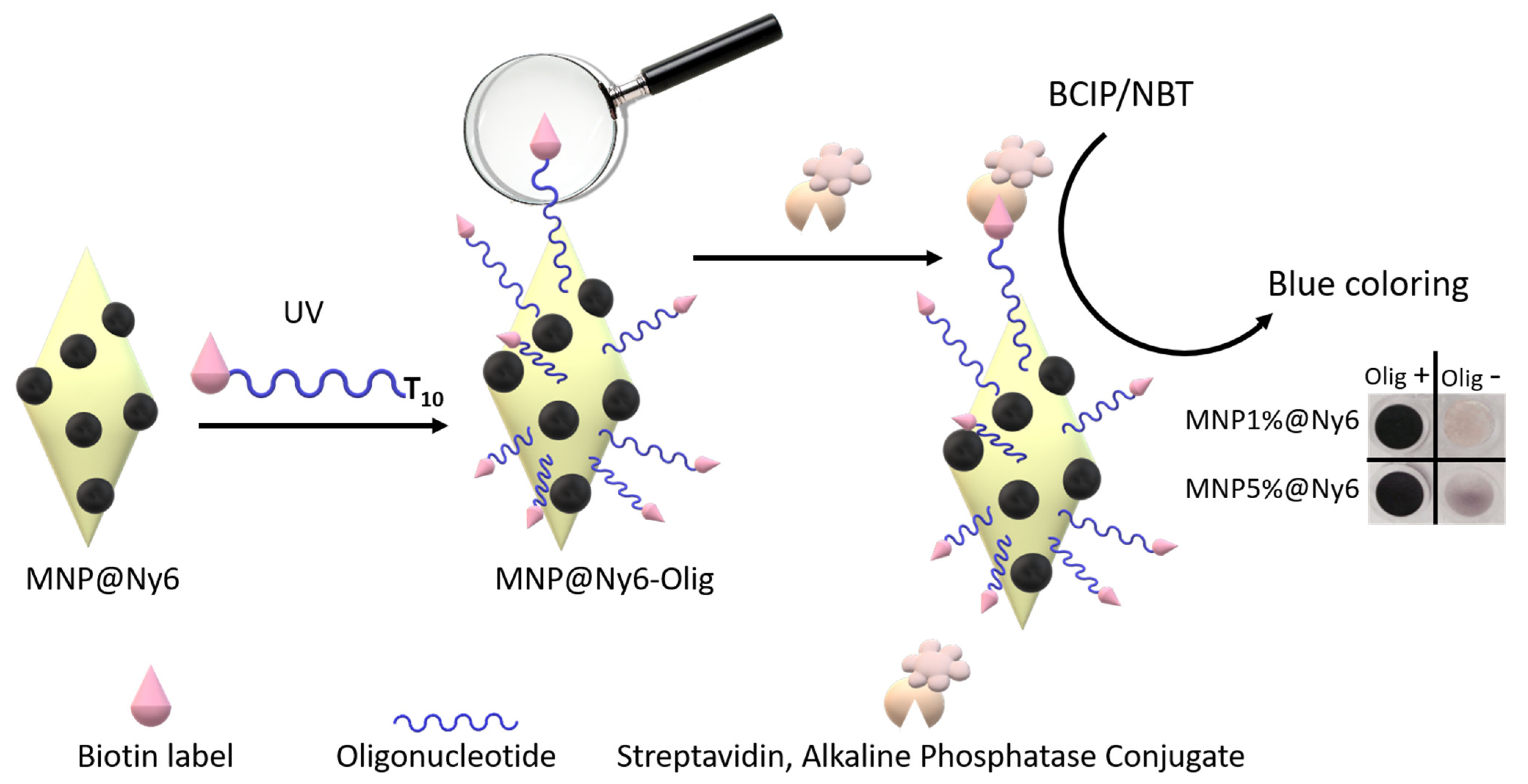
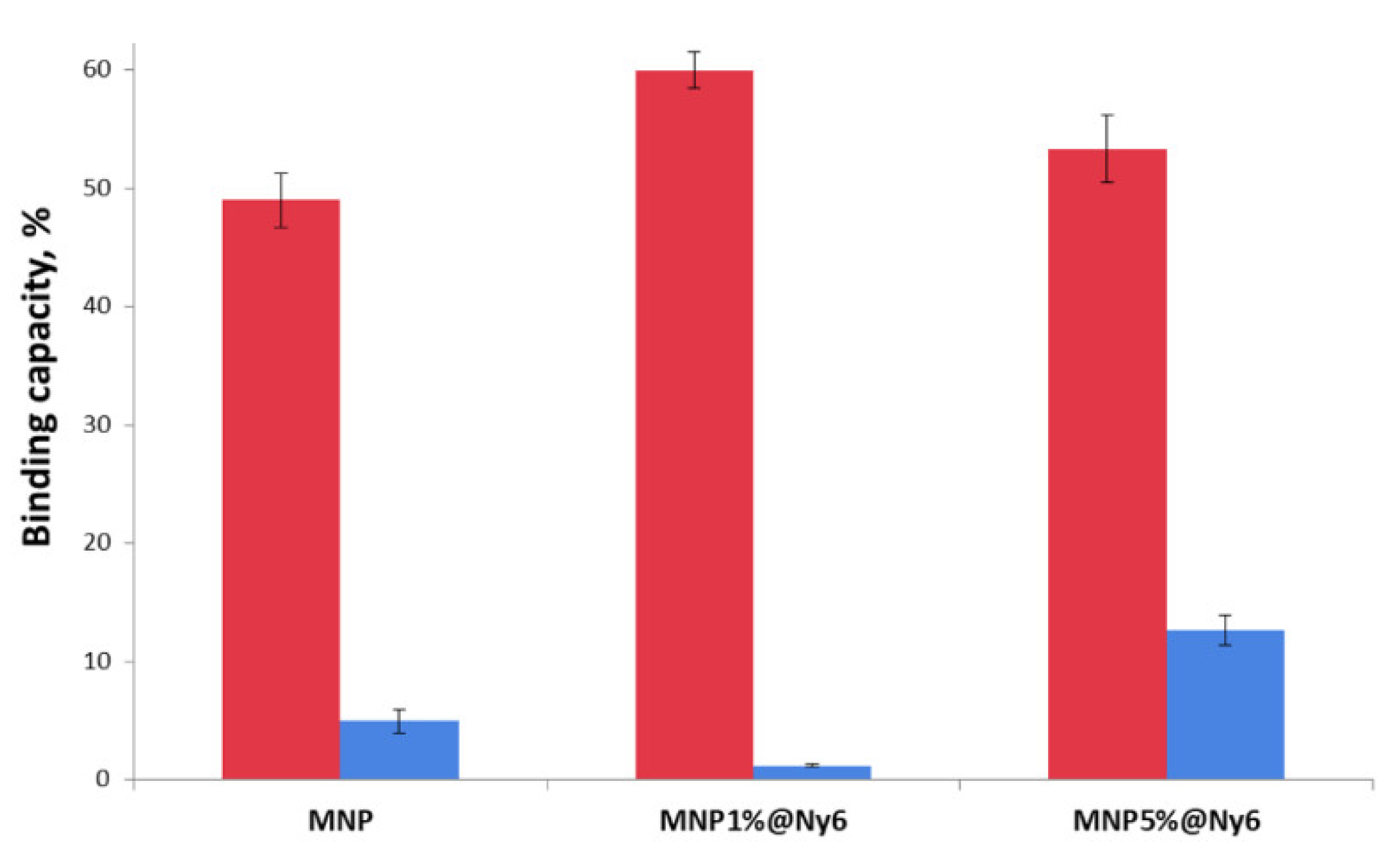
2.2. Oligonucleotides Microextraction from Model Samples by MNP-Olig and MNP@Ny6-Olig Nanocomposites
3. Materials and Methods
3.1. Materials
3.2. Magnetic Nanocomposites Characterization
3.3. Synthesis and Isolation of Oligonucleotides
3.4. Magnetic Nanoparticles (MNP) Synthesis
3.5. Synthesis of Oligonucleotide-Modified MNPs (MNP-Olig)
3.6. Synthesis of MNP and Nylon 6 Nanocomposites (MNP@Ny6)
3.7. Synthesis of Oligonucleotide-Modified MNP@Ny6 (MNP@Ny6-Olig)
3.8. Colorimetric Biotin Detection
3.9. RNA Binding on MNP-Olig or MNP@Ny6-Olig Nanocomposites
3.10. RNA Release
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reyes-Gallardo, E.M.; Lucena, R.; Cárdenas, S.; Valcárcel, M. Polymer-nanoparticles composites in bioanalytical sample preparation. Bioanalysis 2015, 7, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Bowers, A.N.; Trujillo-Rodríguez, M.J.; Farooq, M.Q.; Anderson, J.L. Extraction of DNA with magnetic ionic liquids using in situ dispersive liquid–liquid microextraction. Anal. Bioanal. Chem. 2019, 411, 7375–7385. [Google Scholar] [CrossRef] [PubMed]
- Kovrigina, E.; Chubarov, A.; Dmitrienko, E. High Drug Capacity Doxorubicin-Loaded Iron Oxide Nanocomposites for Cancer Therapy. Magnetochemistry 2022, 8, 54. [Google Scholar] [CrossRef]
- Anderson, S.D.; Gwenin, V.V.; Gwenin, C.D. Magnetic Functionalized Nanoparticles for Biomedical, Drug Delivery and Imaging Applications. Nanoscale Res. Lett. 2019, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lamichhane, N.; Sharma, S.; Verma, A.K.; Roy, I.; Sen, T. Iron oxide-based magneto-optical nanocomposites for in vivo biomedical applications. Biomedicines 2021, 9, 288. [Google Scholar] [CrossRef]
- Chouhan, R.S.; Horvat, M.; Ahmed, J.; Alhokbany, N.; Alshehri, S.M.; Gandhi, S. Magnetic nanoparticles—a multifunctional potential agent for diagnosis and therapy. Cancers 2021, 13, 2213. [Google Scholar] [CrossRef] [PubMed]
- Shabatina, T.I.; Vernaya, O.I.; Shabatin, V.P.; Melnikov, M.Y. Magnetic nanoparticles for biomedical purposes: Modern trends and prospects. Magnetochemistry 2020, 6, 30. [Google Scholar] [CrossRef]
- Ganapathe, L.S.; Mohamed, M.A.; Yunus, R.M.; Berhanuddin, D.D. Magnetite (Fe3O4) nanoparticles in biomedical application: From synthesis to surface functionalisation. Magnetochemistry 2020, 6, 68. [Google Scholar] [CrossRef]
- Hepel, M. Magnetic nanoparticles for nanomedicine. Magnetochemistry 2020, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Dulińska-Litewka, J.; Łazarczyk, A.; Hałubiec, P.; Szafrański, O.; Karnas, K.; Karewicz, A. Superparamagnetic iron oxide nanoparticles-current and prospective medical applications. Materials 2019, 12, 617. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Lu, F.; Chen, C.C.V.; Mo, K.C.; Hung, Y.; Guo, Z.X.; Lin, C.H.; Lin, M.H.; Lin, Y.H.; Chang, C.; et al. Manganese-enhanced MRI of rat brain based on slow cerebral delivery of manganese (II) with silica-encapsulated MnxFe1-xO nanoparticles. NMR Biomed. 2013, 26, 1176–1185. [Google Scholar] [CrossRef]
- Ray, S.; Cheng, C.A.; Chen, W.; Li, Z.; Zink, J.I.; Lin, Y.Y. Magnetic heating stimulated cargo release with dose control using multifunctional MR and thermosensitive liposome. Nanotheranostics 2019, 3, 166–178. [Google Scholar] [CrossRef]
- Ghambari, H.; Reyes-Gallardo, E.M.; Lucena, R.; Saraji, M.; Cárdenas, S. Magnetic polyamide nanocomposites for the microextraction of benzophenones from water samples. Molecules 2019, 24, 953. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Chen, T.; Iqbal, M.Z.; Yang, F.; Hampp, N.; Wu, A.; Luo, L. Applications of magnetic materials separation in biological nanomedicine. Electrophoresis 2019, 40, 2011–2028. [Google Scholar] [CrossRef]
- Marengo, A.; Cagliero, C.; Sgorbini, B.; Anderson, J.L.; Emaus, M.N.; Bicchi, C.; Bertea, C.M.; Rubiolo, P. Development of an innovative and sustainable one-step method for rapid plant DNA isolation for targeted PCR using magnetic ionic liquids. Plant Methods 2019, 15, 23. [Google Scholar] [CrossRef]
- Wang, L.; He, K.; Sadak, O.; Wang, X.; Wang, Q.; Xu, X. Visual detection of in vitro nucleic acid replication by submicro- and nano-sized materials. Biosens. Bioelectron. 2020, 169, 112602. [Google Scholar] [CrossRef]
- Sosa-Acosta, J.R.; Iriarte-Mesa, C.; Ortega, G.A.; Díaz-García, A.M. DNA–Iron Oxide Nanoparticles Conjugates: Functional Magnetic Nanoplatforms in Biomedical Applications. Top. Curr. Chem. 2020, 378, 1–29. [Google Scholar] [CrossRef]
- Li, P.; Li, M.; Yue, D.; Chen, H. Solid-phase extraction methods for nucleic acid separation. A review. J. Sep. Sci. 2022, 45, 172–184. [Google Scholar] [CrossRef]
- Chubarov, A.S. Serum Albumin for Magnetic Nanoparticles Coating. Magnetochemistry 2022, 8, 13. [Google Scholar]
- Bobrikova, E.; Chubarov, A.; Dmitrienko, E. The Effect of pH and Buffer on Oligonucleotide Affinity for Iron Oxide Nanoparticles. Magnetochemistry 2021, 7, 128. [Google Scholar] [CrossRef]
- Vanyorek, L.; Ilosvai, Á.M.; Szőri-Dorogházi, E.; Váradi, C.; Kristály, F.; Prekob, Á.; Fiser, B.; Varga, T.; Kónya, Z.; Viskolcz, B. Synthesis of iron oxide nanoparticles for DNA purification. J. Dispers. Sci. Technol. 2021, 42, 693–700. [Google Scholar] [CrossRef]
- Wang, J.; Ali, Z.; Si, J.; Wang, N.; He, N.; Li, Z. Simultaneous extraction of DNA and RNA from hepatocellular carcinoma (Hep G2) based on silica-coated magnetic nanoparticles. J. Nanosci. Nanotechnol. 2017, 17, 802–806. [Google Scholar] [CrossRef]
- Danthanarayana, A.N.; Manatunga, D.C.; De Silva, R.M.; Chandrasekharan, N.V.; De Silva, K.M.N. Magnetofection and isolation of DNA using polyethyleneimine functionalized magnetic iron oxide nanoparticles. R. Soc. Open Sci. 2018, 5, 181369. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.; He, Z.; Liu, H.; Xu, Y.; Huang, H.; Yang, G.; Xiao, Z.; Li, S.; Liu, H.; Deng, Y.; et al. Application of magnetic nanoparticles in nucleic acid detection. J. Nanobiotechnol. 2020, 18, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Chacón-Torres, J.C.; Reinoso, C.; Navas-León, D.G.; Briceño, S.; González, G. Optimized and scalable synthesis of magnetic nanoparticles for RNA extraction in response to developing countries’ needs in the detection and control of SARS-CoV-2. Sci. Rep. 2020, 10, 19004. [Google Scholar] [CrossRef]
- Ali, T.H.; Mandal, A.M.; Heidelberg, T.; Hussen, R.S.D. Sugar based cationic magnetic core–shell silica nanoparticles for nucleic acid extraction. RSC Adv. 2022, 12, 13566–13579. [Google Scholar] [CrossRef]
- Frenea-Robin, M.; Marchalot, J. Basic Principles and Recent Advances in Magnetic Cell Separation. Magnetochemistry 2022, 8, 11. [Google Scholar] [CrossRef]
- Gessner, I.; Fries, J.W.U.; Brune, V.; Mathur, S. Magnetic nanoparticle-based amplification of microRNA detection in body fluids for early disease diagnosis. J. Mater. Chem. B 2021, 9, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Min, J.H.; Woo, M.K.; Yoon, H.Y.; Jang, J.W.; Wu, J.H.; Lim, C.S.; Kim, Y.K. Isolation of DNA using magnetic nanoparticles coated with dimercaptosuccinic acid. Anal. Biochem. 2014, 447, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Acosta, J.R.; Silva, J.A.; Fernández-Izquierdo, L.; Díaz-Castañón, S.; Ortiz, M.; Zuaznabar-Gardona, J.C.; Díaz-García, A.M. Iron Oxide Nanoparticles (IONPs) with potential applications in plasmid DNA isolation. Colloids Surf. A Physicochem. Eng. Asp. 2018, 545, 167–178. [Google Scholar] [CrossRef]
- Berensmeier, S. Magnetic particles for the separation and purification of nucleic acids. Appl. Microbiol. Biotechnol. 2006, 73, 495–504. [Google Scholar] [CrossRef]
- Hossain, S.; Rahman, M.; Nahar, Y.; Rahman, A.; Sharafat, M.K.; Hossain, M.; Ochiai, B.; Elaissari, A.; Ahmad, H. A simple in situ synthesis of iron oxide magnetic nanoparticles embedded in thermosensitive polymer for DNA capture. J. Mater. Res. 2020, 35, 2441–2450. [Google Scholar] [CrossRef]
- Damavandi, F.; Wang, W.; Shen, W.Z.; Cetinel, S.; Jordan, T.; Jovel, J.; Montemagno, C.; Wong, G.K.S. Enrichment of low abundance DNA/RNA by oligonucleotide-clicked iron oxide nanoparticles. Sci. Rep. 2021, 11, 13053. [Google Scholar] [CrossRef]
- Lim, S.H.; Bestvater, F.; Buchy, P.; Mardy, S.; Yu, A.D.C. Quantitative analysis of nucleic acid hybridization on magnetic particles and quantum dot-based probes. Sensors 2009, 9, 5590–5599. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Wang, C.; Wang, J.; Sun, Z.; Xiao, R.; Wang, S. Fe3O4@Ag magnetic nanoparticles for microRNA capture and duplex-specific nuclease signal amplification based SERS detection in cancer cells. Biosens. Bioelectron. 2016, 79, 574–580. [Google Scholar] [CrossRef]
- Li, B.; Mou, X.; Chen, Z.; Chen, H.; Deng, Y.; Li, S.; Su, E.; He, L.; He, N. The development of a rapid high-quality universal nucleic acid extraction kit based on magnetic separation. Sci. China Chem. 2017, 60, 1602–1608. [Google Scholar] [CrossRef]
- Pinchon, E.; Leon, F.; Temurok, N.; Morvan, F.; Vasseur, J.J.; Clot, M.; Foulongne, V.; Cantaloube, J.F.; Perre, P.V.; Daynès, A.; et al. Rapid and specific DNA detection by magnetic field-enhanced agglutination assay. Talanta 2020, 219, 121344. [Google Scholar] [CrossRef]
- Yang, S.Y.; Chieh, J.J.; Wang, W.C.; Yu, C.Y.; Hing, N.S.; Horng, H.E.; Hong, C.Y.; Yang, H.C.; Chang, C.F.; Lin, H.Y. Magnetic nanoparticles for high-sensitivity detection on nucleic acids via superconducting-quantum-interference-device-based immunomagnetic reduction assay. J. Magn. Magn. Mater. 2011, 323, 681–685. [Google Scholar] [CrossRef]
- Camacho-fernández, J.C.; Jin, M.; Liu, X.; Berg, A.V.D.; Zhou, G. Ultrasensitive DNA detection based on two-step quantitative amplification on magnetic nanoparticles. Nanotechnology 2016, 27, 335102. [Google Scholar]
- Pivetal, J.; Frénéa-Robin, M.; Haddour, N.; Vézy, C.; Zanini, L.F.; Ciuta, G.; Dempsey, N.M.; Dumas-Bouchiat, F.; Reyne, G.; Bégin-Colin, S.; et al. Development and applications of a DNA labeling method with magnetic nanoparticles to study the role of horizontal gene transfer events between bacteria in soil pollutant bioremediation processes. Environ. Sci. Pollut. Res. 2015, 22, 20322–20327. [Google Scholar] [CrossRef] [Green Version]
- Bordelon, H.; Russ, P.K.; Wright, D.W.; Haselton, F.R. A Magnetic Bead-Based Method for Concentrating DNA from Human Urine for Downstream Detection. PLoS ONE 2013, 8, e68369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Tjiu, W.W.; Liu, T.; Lui, W.Y.; Phang, I.Y.; Zhang, W. De Dramatically enhanced mechanical performance of nylon-6 magnetic composites with nanostructured hybrid one-dimensional carbon nanotube—Two-dimensional clay nanoplatelet heterostructures. J. Phys. Chem. B 2011, 115, 3392–3399. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.Z.; Safari, Z.; Madady, N. Synthesis of Co3O4@SiO2 Core/Shell–Nylon 6 Magnetic Nanocomposite as an Adsorbent for Removal of Congo Red from Wastewater. J. Inorg. Organomet. Polym. Mater. 2020, 30, 3199–3212. [Google Scholar] [CrossRef]
- Choi, E.Y.; Kim, K.; Kim, C.K.; Kang, E. Reinforcement of nylon 6,6/nylon 6,6 grafted nanodiamond composites by in situ reactive extrusion. Sci. Rep. 2016, 6, 37010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, K.; Khan, I.; Ahad, M.; Shah, T.; Sadiq, M.; Zada, A.; Zada, N. Preparation of ZnO/Nylon 6/6 nanocomposites, their characterization and application in dye decolorization. Appl. Water Sci. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Dmitrienko, E.V.; Bulushev, R.D.; Haupt, K.; Kosolobov, S.S.; Latyshev, A.V.; Pyshnaya, I.A.; Pyshnyi, D.V. A simple approach to prepare molecularly imprinted polymers from nylon-6. J. Mol. Recognit. 2013, 26, 368–375. [Google Scholar] [CrossRef]
- Shakiba, M.; Rezvani Ghomi, E.; Khosravi, F.; Jouybar, S.; Bigham, A.; Zare, M.; Abdouss, M.; Moaref, R.; Ramakrishna, S. Nylon—A material introduction and overview for biomedical applications. Polym. Adv. Technol. 2021, 32, 3368–3383. [Google Scholar] [CrossRef]
- Kirillov, V.L.; Balaev, D.A.; Semenov, S.V.; Shaikhutdinov, K.A.; Martyanov, O.N. Size control in the formation of magnetite nanoparticles in the presence of citrate ions. Mater. Chem. Phys. 2014, 145, 75–81. [Google Scholar] [CrossRef]
- Kirillov, V.L.; Yakushkin, S.S.; Balaev, D.A.; Dubrovskiy, A.A.; Semenov, S.V.; Knyazev, Y.V.; Bayukov, O.A.; Velikanov, D.A.; Yatsenko, D.A.; Martyanov, O.N. Dimethylsulfoxide as a media for one-stage synthesis of the Fe3O4-Based ferro fluids with a controllable size distribution. Mater. Chem. Phys. 2019, 225, 292–297. [Google Scholar] [CrossRef]
- Clavijo, C.; Osma, J.F. Functionalized leather: A novel and effective hazardous solid waste adsorbent for the removal of the diazo dye congo red from aqueous solution. Water 2019, 11, 1906. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sun, Y.; Wang, J.; Yang, Y.; Li, Y.; Yuan, Y.; Liu, C. Charge-Reversal APTES-Modified Mesoporous Silica Nanoparticles with High Drug Loading and Release Controllability. Appl. Mater. Interfaces 2016, 8, 17166–17175. [Google Scholar] [CrossRef]
- Digigow, R.G.; Dechézelles, J.; Dietsch, H.; Geissbühler, I.; Vanhecke, D.; Geers, C.; Hirt, A.M.; Rothen-rutishauser, B. Preparation and characterization of functional silica hybrid magnetic nanoparticles. J. Magn. Magn. Mater. 2014, 362, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Chandra, S.; Beaune, G.; Shirahata, N.; Winnik, F.M. A one-pot synthesis of water soluble highly fluorescent silica nanoparticles. J. Mater. Chem. B 2017, 5, 1363. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.F.; Goh, P.; Rezaei, M.; Arzhandi, D.; Ismail, N. Aptes and teos modified binary recyclable hybrid Fe3O4@GO nanocomposite for photocatalytic dye removal. J. Teknol. 2018, 80, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Hao, N.; Jayawardana, K.W.; Chen, X.; Zoysa, T.D.; Yan, M. One-step synthesis of amine-functionalized hollow mesoporous silica nanoparticles as efficient antibacterial and anticancer materials. ACS Appl. Mater. Interfaces 2015, 7, 1040–1045. [Google Scholar] [CrossRef] [Green Version]
- Dmitrienko, E.V.; Pyshnaya, I.A.; Pyshnyi, D.V. Oligonucleotide Derivatives in the Hybridization Analysis of Nucleic Acids. I. Covalent Immobilization of Oligonucleotide Probes on Nylon. Russ. J. Bioorgan. Chem. 2010, 36, 645–656. [Google Scholar] [CrossRef]
- Gebeyehu, G.; Rao, P.Y.; Soochan, P.; Simms, D.A.; Klevan, L. Novel biotinylated nucleotide-analogs for labeling and colorimetric detection of DNA. Nucleic Acids Res. 1987, 15, 4513–4534. [Google Scholar] [CrossRef] [Green Version]
- Juthani, N.; Doyle, P.S. A Platform for Multiplexed Colorimetric microRNA Detection using Shape-encoded Hydrogel Particles. Analyst 2020, 145, 5134. [Google Scholar] [CrossRef]
- Dmitrienlo, E.; Naumova, O.; Fomin, B.; Kupryushkin, M.; Volkova, A.; Amirkhanov, N.; Semenov, D.; Pyshnaya, I.; Pyshnyi, D. Surface modification of SOI-FET sensors for label-free and specific detection of short RNA analyte. Nanomedicine 2016, 11, 2073–2082. [Google Scholar] [CrossRef]
- Lomzov, A.A.; Kupryushkin, M.S.; Shernyukov, A.V.; Nekrasov, M.D.; Dovydenko, I.S.; Stetsenko, D.A.; Pyshnyi, D.V. Diastereomers of a mono-substituted phosphoryl guanidine trideoxyribonucleotide: Isolation and properties. Biochem. Biophys. Res. Commun. 2019, 513, 807–811. [Google Scholar] [CrossRef]



| Description | 5′-Sequence-3′ | NA Type, Function |
|---|---|---|
| oligonucleotide-NH2 | GTCTTCCTTCTCCGCTT-Sp1-NH2 | DNA, capture probe |
| FAM-oligonucleotide-NH2 | FAM-Sp2-GTCTTCCTTCTCCGCTT-Sp1-NH2 | |
| T10-oligonucleotide | (T)10-GTCTTCCTTCTCCGCTT | |
| T10-oligonucleotide-FAM * | (T)10-GTCTTCCTTCTCCGCTT-Sp2-FAM | |
| T10-oligonucleotide-Biotin | (T)10-GTCTTCCTTCTCCGCTT-Sp3-Biotin | |
Sp1 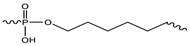 Sp2 Sp2 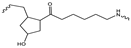 Sp3 Sp3 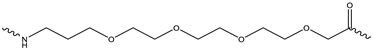 | ||
| Description | 5′-Sequence-3′ | NA Type, Function |
|---|---|---|
| oligonucleotide-NH2 No. 1 specific for RNA | GTCTTCCTTCTCCGCTT-Sp1-NH2 * | DNA, capture probe for MNP |
| oligonucleotide-NH2 No. 2 non-specific for RNA | TAACCGATTTCAGATTT-Sp1-NH2 | |
| T10-oligonucleotide No. 1 specific for RNA | (T)10-GTCTTCCTTCTCCGCTT | DNA, capture probe for MNP@Ny6 |
| T10-oligonucleotide No. 2 non-specific for RNA | (T)10-TAACCGATTTCAGATTT | |
| Target | Fam- GAGGCGAAGCGGAGAAGGAAGAC | RNA, target |
| MNP Type | Capture DNA Probe Immobilization | Initial Target RNA Amount | Target RNA Binding | Target RNA Release | RNA Capture Efficiency |
|---|---|---|---|---|---|
| MNP | 50.0% | 0.30 nmol | 0.147 ± 0.015 nmol 49.0% a | 0.033 ± 0.010 nmol 22.4% b | 11.0% c |
| MNP1%Ny6 | 73.3% | 0.180 ± 0.010 nmol 60.0% | 0.160 ± 0.013 nmol 88.9% | 53.3% | |
| MNP5%Ny6 | 66.7% | 0.160 ± 0.018 nmol 53.3% | 0.087 ± 0.013 nmol 54.4% | 29.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulgakova, A.; Chubarov, A.; Dmitrienko, E. Magnetic Nylon 6 Nanocomposites for the Microextraction of Nucleic Acids from Biological Samples. Magnetochemistry 2022, 8, 85. https://doi.org/10.3390/magnetochemistry8080085
Bulgakova A, Chubarov A, Dmitrienko E. Magnetic Nylon 6 Nanocomposites for the Microextraction of Nucleic Acids from Biological Samples. Magnetochemistry. 2022; 8(8):85. https://doi.org/10.3390/magnetochemistry8080085
Chicago/Turabian StyleBulgakova, Anastasia, Alexey Chubarov, and Elena Dmitrienko. 2022. "Magnetic Nylon 6 Nanocomposites for the Microextraction of Nucleic Acids from Biological Samples" Magnetochemistry 8, no. 8: 85. https://doi.org/10.3390/magnetochemistry8080085