Portuguese Neonatal Screening Program: A Cohort Study of 18 Years Using MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Subjects
2.2. Routine Biochemical Screening
2.3. Second Tier Tests (2TT)
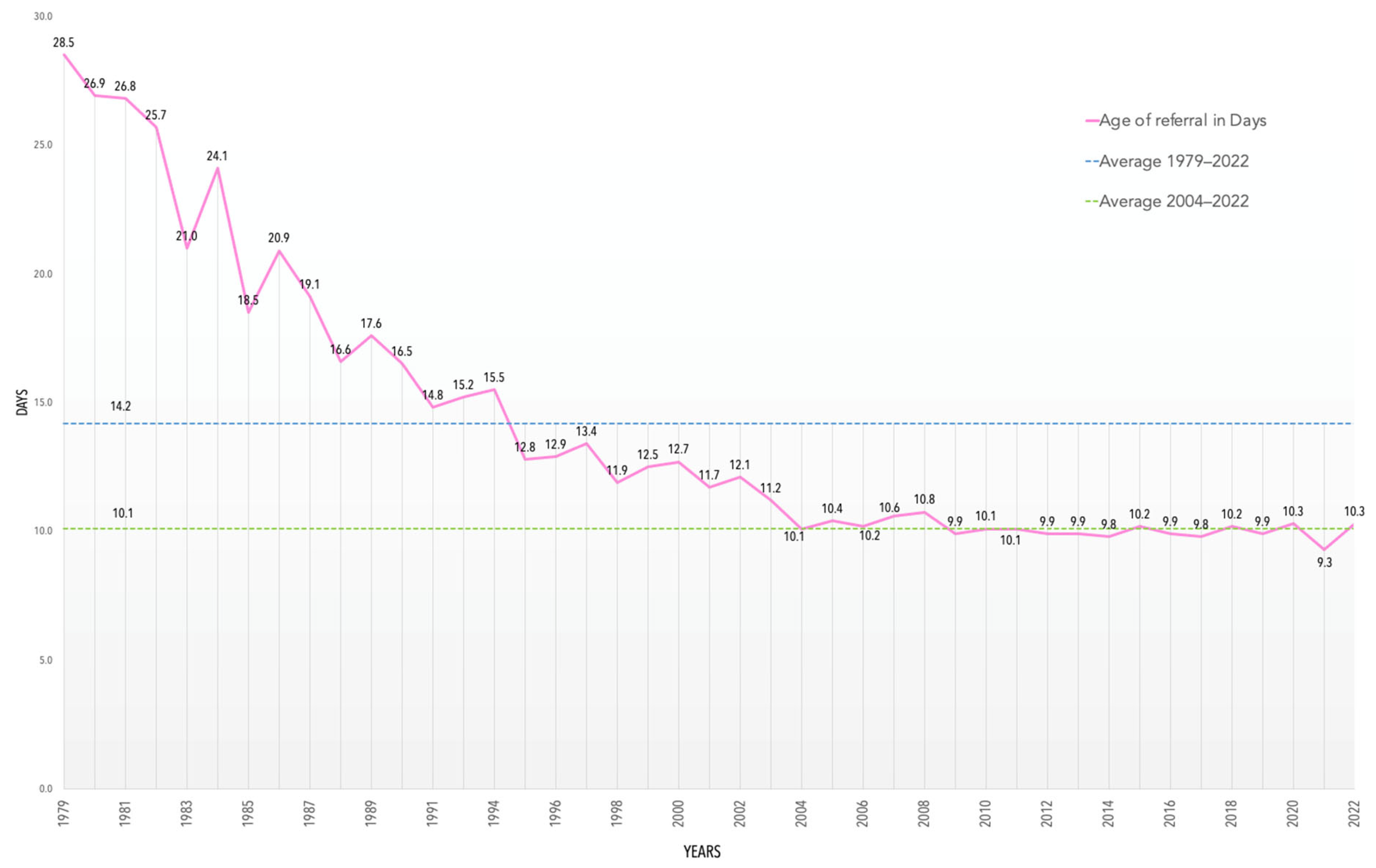
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Vilarinho, L.; Rocha, H.; De Sousa, C.M.M.; Marcão, A.; Fonseca, H.; Bogas, M.; Osório, R.V. Four Years of Expanded Newborn Screening in Portugal with Tandem Mass Spectrometry. J. Inherit. Metab. Dis. 2010, 33, 133–138. [Google Scholar] [CrossRef]
- Saudubray, J.; Baumgartner, M.; Walter, J.C. Inborn Metabolic Diseases; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Ferreira, C.R.; Van Karnebeek, C. Inborn Errors of Metabolism. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 449–481. [Google Scholar] [CrossRef]
- Rashed, M.S.; Özand, P.; Bucknall, M.P.; Little, D. Diagnosis of Inborn Errors of Metabolism from Blood Spots by Acylcarnitines and Amino Acids Profiling Using Automated Electrospray Tandem Mass Spectrometry. Pediatr. Res. 1995, 38, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Allard, P.; Grenier, A.; Korson, M.S.; Zytkovicz, T.H. Newborn Screening for Hepatorenal Tyrosinemia by Tandem Mass Spectrometry: Analysis of Succinylacetone Extracted from Dried Blood Spots. Clin. Biochem. 2004, 37, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- La Marca, G.; Malvagia, S.; Pasquini, E.; Innocenti, M.; Donati, M.A.; Zammarchi, E. Rapid 2nd-Tier Test for Measurement of 3-OH-Propionic and Methylmalonic Acids on Dried Blood Spots: Reducing the False-Positive Rate for Propionylcarnitine during Expanded Newborn Screening by Liquid Chromatography–Tandem Mass Spectrometry. Clin. Chem. 2007, 53, 1364–1369. [Google Scholar] [CrossRef]
- Turgeon, C.T.; Magera, M.J.; Cuthbert, C.; Loken, P.R.; Gavrilov, D.; Tortorelli, S.; Raymond, K.; Oglesbee, D.; Rinaldo, P.; Matern, D. Determination of Total Homocysteine, Methylmalonic Acid, and 2-Methylcitric Acid in Dried Blood Spots by Tandem Mass Spectrometry. Clin. Chem. 2010, 56, 1686–1695. [Google Scholar] [CrossRef]
- Forni, S.; Fu, X.; Palmer, S.; Sweetman, L. Rapid Determination of C4-Acylcarnitine and C5-Acylcarnitine Isomers in Plasma and Dried Blood Spots by UPLC–MS/MS as a Second Tier Test Following Flow-Injection MS/MS Acylcarnitine Profile Analysis. Mol. Genet. Metab. 2010, 101, 25–32. [Google Scholar] [CrossRef]
- DeArmond, P.D.; Dietzen, D.J.; Pyle-Eilola, A.L. Amino Acids Disorders; Elsevier eBooks: Amsterdam, The Netherlands, 2017; pp. 25–64. [Google Scholar] [CrossRef]
- Marcão, A.; Couce, M.L.; Nogueira, C.; Fonseca, H.; Ferreira, F.; Fraga, J.M.; Bóveda, M.D.; Vilarinho, L. Newborn Screening for Homocystinuria Revealed a High Frequency of MAT I/III Deficiency in Iberian Peninsula. In JIMD Reports; Springer: Berlin/Heidelberg, Germany, 2014; pp. 113–120. [Google Scholar] [CrossRef]
- Waters, D.; Rudan, I.; Woolham, D.; Wastnedge, E.; Patel, S.; Rudan, I. Global Birth Prevalence and Mortality from Inborn Errors of Metabolism: A Systematic Analysis of the Evidence. J. Glob. Health 2018, 8, 021102. [Google Scholar] [CrossRef]
- Posset, R.; Kölker, S.; Gleich, F.; Okun, J.G.; Gropman, A.; Nagamani, S.C.S.; Scharre, S.; Probst, J.; Walter, M.; Hoffmann, G.F.; et al. Severity-Adjusted Evaluation of Newborn Screening on the Metabolic Disease Course in Individuals with Cytosolic Urea Cycle Disorders. Mol. Genet. Metab. 2020, 131, 390–397. [Google Scholar] [CrossRef]
- Catsburg, C.; Anderson, S.; Upadhyaya, N.; Bechter, M. Arginase 1 Deficiency: Using Genetic Databases as a Tool to Establish Global Prevalence. Orphanet J. Rare Dis. 2022, 17, 94. [Google Scholar] [CrossRef]
- Almási, T.; Guey, L.T.; Lukacs, C.; Csetneki, K.; Vokó, Z.; Zelei, T. Systematic Literature Review and Meta-Analysis on the Epidemiology of Methylmalonic Acidemia (MMA) with a Focus on MMA Caused by Methylmalonyl-CoA Mutase (Mut) Deficiency. Orphanet J. Rare Dis. 2019, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yang, Z.; Yang, C.; Hu, H.; He, H.; Niu, T.; Liu, M.; Wang, D.; Sun, Y.; Shen, Y.; et al. C4OH Is a Potential Newborn Screening Marker—A Multicenter Retrospective Study of Patients with Beta-Ketothiolase Deficiency in China. Orphanet J. Rare Dis. 2021, 16, 224. [Google Scholar] [CrossRef] [PubMed]
- Rocha, H.; Castiñeiras, D.; Delgado, C.; Egea, J.M.O.; Yahyaoui, R.; González, Y.; Conde, M.; González, I.; Rueda, I.; Rello, L.; et al. Birth Prevalence of Fatty Acid Β-Oxidation Disorders in Iberia. In JIMD Reports; Springer: Berlin/Heidelberg, Germany, 2014; pp. 89–94. [Google Scholar] [CrossRef]
- Ventura, F.V.; Leandro, P.; Luz, A.C.; Rivera, I.; Silva, M.F.B.; Ramos, R.; Rocha, H.; Lopes, A.; Fonseca, H.; Gaspar, A.; et al. Retrospective Study of the Medium-Chain Acyl-CoA Dehydrogenase Deficiency in Portugal. Clin. Genet. 2013, 85, 555–561. [Google Scholar] [CrossRef]
- Fonseca, H.; Azevedo, L.; Serrano, C.; De Sousa, C.M.M.; Marcão, A.; Vilarinho, L. 3-Methylcrotonyl-CoA Carboxylase Deficiency: Mutational Spectrum Derived from Comprehensive Newborn Screening. Gene 2016, 594, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, C.; Marcão, A.; Rocha, H.; De Sousa, C.M.M.; Fonseca, H.; Valongo, C.; Vilarinho, L. Molecular Picture of Cobalamin C/D Defects before and after Newborn Screening Era. J. Med. Screen. 2016, 24, 6–11. [Google Scholar] [CrossRef]
- Fukao, T.; Yamaguchi, S.; Orii, T.; Hashimoto, T. Molecular Basis of β-Ketothiolase Deficiency: Mutations and Polymorphisms in the Human Mitochondrial Acetoacetyl-Coenzyme a Thiolase Gene. Hum. Mutat. 1995, 5, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Yamaguchi, S.; Wakazono, A.; Orii, T.; Hoganson, G.; Hashimoto, T. Identification of a Novel Exonic Mutation at -13 from 5′ Splice Site Causing Exon Skipping in a Girl with Mitochondrial Acetoacetyl-Coenzyme A Thiolase Deficiency. J. Clin. Investig. 1994, 93, 1035–1041. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grünert, S.C.; Sass, J.O. 3-Hydroxy-3-Methylglutaryl-Coenzyme A Lyase Deficiency: One Disease—Many Faces. Orphanet J. Rare Dis. 2020, 15, 48. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, S.H.; Liu, S.; Han, X.; Wang, Y.; Wang, L.L.; Yu, B. Two Infants with Beta-Ketothiolase Deficiency Identified by Newborn Screening in China. Front. Genet. 2019, 10, 451. [Google Scholar] [CrossRef]
- Farwell, K.D.; Shahmirzadi, L.; El-Khechen, D.; Powis, Z.; Chao, E.; Davis, B.T.; Baxter, R.; Zeng, W.; Mroske, C.; Parra, M.; et al. Enhanced Utility of Family-Centered Diagnostic Exome Sequencing with Inheritance Model–Based Analysis: Results from 500 Unselected Families with Undiagnosed Genetic Conditions. Genet. Med. 2015, 17, 578–586. [Google Scholar] [CrossRef]
- Boutron, A.; Acquaviva, C.; Vianey-Saban, C.; De Lonlay, P.; De Baulny, H.O.; Guffon, N.; Dobbelaere, D.; Feillet, F.; Labarthe, F.; Lamireau, D.; et al. Comprehensive cDNA Study and Quantitative Analysis of Mutant HADHA and HADHB Transcripts in a French Cohort of 52 Patients with Mitochondrial Trifunctional Protein Deficiency. Mol. Genet. Metab. 2011, 103, 341–348. [Google Scholar] [CrossRef]
- Ogawa, A.; Yamamoto, S.; Kanazawa, M.; Takayanagi, M.; Hasegawa, S.; Kohno, Y. Identification of Two Novel Mutations of the Carnitine/Acylcarnitine Translocase (CACT) Gene in a Patient with CACT Deficiency. J. Hum. Genet. 2000, 45, 52–55. [Google Scholar] [CrossRef]
- Ryder, B.; Inbar-Feigenberg, M.; Glamuzina, E.; Halligan, R.; Vara, R.; Elliot, A.; Coman, D.; Minto, T.; Lewis, K.; Schiff, M.; et al. New Insights into Carnitine-acylcarnitine Translocase Deficiency from 23 Cases: Management Challenges and Potential Therapeutic Approaches. J. Inherit. Metab. Dis. 2021, 44, 903–915. [Google Scholar] [CrossRef]
- Lipari Pinto, P.; Florindo, C.; Janeiro, P.; Santos, R.L.; Mexia, S.; Rocha, H.; De Almeida, I.T.; Vilarinho, L.; Gaspar, A.C. Acquired Vitamin B12 Deficiency in Newborns: Positive Impact on Newborn Health through Early Detection. Nutrients 2022, 14, 4397. [Google Scholar] [CrossRef]
- Wilcox, G. Impact of Pregnancy on Inborn Errors of Metabolism. Rev. Endocr. Metab. Disord. 2018, 19, 13–33. [Google Scholar] [CrossRef]
- Martins, E.; Marcão, A.; Bandeira, A.; Fonseca, H.; Nogueira, C.; Vilarinho, L. Methionine Adenosyltransferase I/III Deficiency in Portugal: High Frequency of a Dominantly Inherited Form in a Small Area of Douro High Lands. In JIMD Reports; Springer: Berlin/Heidelberg, Germany, 2012; pp. 107–112. [Google Scholar] [CrossRef]
- Huang, Y.; Sharma, R.; Feigenbaum, A.; Lee, C.; Sahai, I.; Russo, R.S.; Neira, J.; Brooks, S.S.; Jackson, K.E.; Wong, D.A.; et al. Arginine to Ornithine Ratio as a Diagnostic Marker in Patients with Positive Newborn Screening for Hyperargininemia. Mol. Genet. Metab. Rep. 2021, 27, 100735. [Google Scholar] [CrossRef] [PubMed]
- Rocha, H. Qué Hay de Nuevo En El Cribado Neonatal de Enfermedades Metabólicas. Acta Pediatr. Española 2015, (Suppl. S73), 12–13. [Google Scholar]
- Boemer, F.; Schoos, R.; De Halleux, V.; Kalenga, M.; Debray, F.-G. Surprising Causes of C5-Carnitine False Positive Results in Newborn Screening. Mol. Genet. Metab. 2014, 111, 52–54. [Google Scholar] [CrossRef]
- Bonham, J.R.; Carling, R.S.; Lindner, M.; Franzson, L.; Zetterström, R.; Boemer, F.; Cerone, R.; Eyskens, F.; Vilarinho, L.; Hougaard, D.M.; et al. Raising Awareness of False Positive Newborn Screening Results Arising from Pivalate-Containing Creams and Antibiotics in Europe When Screening for Isovaleric Acidemia. Int. J. Neonatal Screen. 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Özben, T. Expanded Newborn Screening and Confirmatory Follow-up Testing for Inborn Errors of Metabolism Detected by Tandem Mass Spectrometry. Clin. Chem. Lab. Med. 2012, 51, 157–176. [Google Scholar] [CrossRef]
- Peng, G.; Tang, Y.; Gandotra, N.; Enns, G.M.; Cowan, T.M.; Zhao, H.; Scharfe, C. Ethnic Variability in Newborn Metabolic Screening Markers Associated with False-positive Outcomes. J. Inherit. Metab. Dis. 2020, 43, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Estatística. Estatísticas Demográficas. Available online: https://www.ine.pt/xurl/pub/280978178 (accessed on 25 January 2024).
- Sampaleanu, L.M.; Vallée, F.; Thompson, G.D.; Howell, P.L. Three-Dimensional Structure of the Argininosuccinate Lyase Frequently Complementing Allele Q286R. Biochemistry 2001, 40, 15570–15580. [Google Scholar] [CrossRef] [PubMed]
- Taroni, F.; Verderio, E.; Dworzak, F.; Willems, P.J.; Cavadini, P.; DiDonato, S. Identification of a Common Mutation in the Carnitine Palmitoyltransferase II Gene in Familial Recurrent Myoglobinuria Patients. Nat. Genet. 1993, 4, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Morillo, E.; García, B.P.; Menéndez, F.V.Á. Challenges for Worldwide Harmonization of Newborn Screening Programs. Clin. Chem. 2016, 62, 689–698. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loeber, J.G.; Platis, D.; Zetterström, R.; Almashanu, S.; Boemer, F.; Bonham, J.R.; Borde, P.; Brincat, I.; Cheillan, D.; Dekkers, E.; et al. Neonatal Screening in Europe Revisited: An ISNS Perspective on the Current State and Developments since 2010. Int. J. Neonatal Screen. 2021, 7, 15. [Google Scholar] [CrossRef] [PubMed]


| Screened Disorders | OMIM | Cut-Offs and Ratios | |
|---|---|---|---|
| Amino Acid Disorders (AAD) | Phenylketonuria (PKU)/Hyperphenylalaninemia (HPhe) | 261600 | Phe (>150 μM) and Phe/Tyr (>1.5) |
| Maple syrup urine disease (MSUD) | 248600 | XLeu (>270 μM) and Val (>285 μM) Val/Phe > 4, Xleu/Phe > 5 | |
| Tyrosinemia type I (TYR1) | 276700 | Tyr (>210 μM) selects for 2TT (SA) | |
| Tyrosinemia type II (TYR2) | 276600 | Tyr (>500 μM) selects for 2TT | |
| Tyrosinemia type III (TYR3) | 276710 | Tyr (>500 μM) selects for 2TT | |
| Homocystinuria (CBS deficiency) | 236200 | Met (>45 μM) selects for 2TT | |
| Methionine adenosyltransferase deficiency (MATI/III deficiency) | 250850 | Met (>45 μM) selects for 2TT | |
| Organic Acidurias (OA) | 3-Methyl crotonyl-CoA carboxylase deficiency (3-MCCD) | 210200 | C5OH (>1.0 μM) |
| Isovaleric acidemia (IVA) | 243500 | C5 (>1.0 μM) selects for 2TT | |
| Propionic acidemia (PA) | 606054 | C3 (>5.25 μM) or C3/C2 (>0.2) selects for 2TT | |
| Methylmalonic acidemia (MMA Mut-/Mut0) | 251000 | C3 (>5.25 μM) or C3/C2 (>0.2) selects for 2TT | |
| Malonic acidemia (MAL) | 248360 | C3DC (>0.35 μM) | |
| Glutaric acidemia type I (GA1) | 231670 | C5DC (>0.2 μM) | |
| 3-Hydroxy-3-methylglutaryl CoA lyase deficiency (3HMGLD) | 246450 | C5OH (>1.0 μM) and C6DC (>0.07 μM) | |
| Methylmalonic acidemia type CblA/B (MAHCA or CblA and MAHCB or CblB) | 251100 251110 | C3 (>5.25 μM) and C3/C2 (>0.2) selects for 2TT | |
| Methylmalonic acidemia type CblC/D (MAHCC or CblC and MAHCD or CblD) | 277400 277410 | C3 (>5.25 μM) or C3/C2 (>0.2) selects for 2TT | |
| Urea Cycle Disorders (UCD) | Citrullinaemia type I (CTLN1) | 215700 | Cit (>200 μM) |
| Argininosuccinate lyase deficiency (ASL deficiency) | 207900 | ASA (>1 μM) | |
| Arginase deficiency (ARG deficiency) | 207800 | Arg (>50 μM) and Arg/Orn (>1.0) | |
| Fatty Acid Oxidation Disorders (FAOD) | Medium-chain acyl-CoA dehydrogenase deficiency (MCADD) | 201450 | C8 (>0.3 μM) and C8/C10 (>2.5) |
| Long-chain 3-OH acyl-CoA dehydrogenase deficiency (LCHADD)/Trifunctional Protein deficiency (TFP) | 609016 609015 | C16OH (>0.10 μM), C18:1OH (>0.07 μM), C18OH (>0.06 μM) and C16OH/C16 (>0.04) | |
| Multiple acyl-CoA dehydrogenase deficiency (MADD) | 231680 | Multiple elevations from C4 to C18 acyl carnitines | |
| Carnitine uptake defect (CUD) | 212140 | C0 (<6.8 μM) | |
| Very-long-chain acyl-CoA dehydrogenase deficiency (VLCADD) | 201475 | C14:1 (>0.46 μM), C14:2 (>0.17 μM) and C14:1/C12:1 (>6.0) | |
| Short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency (SCHADD) | 231530 | C4OH (>0.95 μM) | |
| Carnitine palmitoyl-transferase I deficiency (CPTIA) | 255120 | C0/(C16 + C18) (>30) | |
| Carnitine palmitoyl-transferase II deficiency (CPTII)/Carnitine-acylcarnitine translocase deficiency (CACT) | 255110 212138 | C0/(C16 + C18) (<3.0) | |
| Disorder | Primary Marker | Secondary Marker | Year of Implementation in the PNSP | References |
|---|---|---|---|---|
| Tyrosinemia | Tyrosine | SA | 2006 | [5] |
| Propionic/methylmalonic acidurias | Propionylcarnitine (C3) | MMA, 3OHprop, PropGly | 2017 | [6,7] |
| Cobalamin metabolism defects | Propionylcarnitine (C3) and ↓ methionine | MMA and tHcy | 2017 | [6,7] |
| Classic homocystinuria | Methionine | tHcy | 2017 | [7] |
| Isovaleric aciduria | Isovaleryl/2-methylbutyrylcarnitine (C5) | C5 and Piv-C5 | 2017 | [8] |
| Condition | 2TT Results | ||||||
|---|---|---|---|---|---|---|---|
| MMA | PropGly | 3OHprop | tHcy | Isovalerylcarnitine | 2-Methylbutyrylcarnitine | SA | |
| Propionic acidemia (PA) | N | ↑↑ | ↑↑ | N | |||
| Methylmalonyl- CoA mutase deficiency (Mut0 /Mut-) | ↑↑ | N | N | N | |||
| Cobalamin type A/B deficiency (CblA/B) | ↑ | N | N | N | |||
| Cobalamin type C/D deficiency (CblC/D) | ↑ | N | N | ↑ | |||
| Vitamin B12 deficiency (of maternal cause) a | N or ↑ | N | N | N or ↑ | |||
| Homocystinuria | ↑↑ | ||||||
| Methionine adenosyltransferase I/III deficiency | N or ↑ | ||||||
| Isovaleric aciduria | ↑↑ | ||||||
| 2-Methylbutyrylglycinuria b | ↑ | ||||||
| Tyrosinemia type I | ↑↑ | ||||||
| Detected Disorders | Positive Cases | Birth Prevalence | Estimated Worldwide Birth Prevalence |
|---|---|---|---|
| Amino acid disorders | 231 | 1:7640 | 1:6803 a |
| Phenylketonuria (PKU)/Hyperphenylalaninemia (HPhe) | 154 | 1:11,611 | 1:15,267 a |
| Maple syrup urine disease (MSUD) | 19 | 1:92,886 | 1:81,967 a |
| Tyrosinemia type I (TYR1) | 6 | 1:294,138 | 1:100,000 a |
| Tyrosinemia type II (TYR2) | 2 | 1:882,415 | <1:1,000,000 b |
| Tyrosinemia type III (TYR3) | 5 | 1:294,138 | <1:1,000,000 b |
| Homocystinuria (CBS deficiency) | 4 | 1:441,208 | 1:243,902 a |
| Methionine adenosyltransferase deficiency (MATI/III deficiency) | 41 | * | <1:1,000,000 b |
| Urea cycle disorders | 26 | 1:67,878 | 1:34,364 a |
| Citrullinemia type I (CTLN1) | 10 | 1:176,483 | 1:250,000 c |
| Argininosuccinate lyase deficiency (ASL deficiency) | 9 | 1:196,092 | 1:220,000 c |
| Arginase deficiency (ARG deficiency) | 7 | 1:252,119 | 1:35,700 d |
| Organic acid disorders | 116 | 1:15,214 | 1:11,481 a |
| 3-Methyl crotonyl-CoA carboxylase deficiency (3-MCCD) | 39 | 1:45,252 | Unknown b |
| Isovaleric acidemia (IVA) | 6 | 1:294,138 | 1:196,078 a |
| Propionic acidemia (PA) | 4 | 1:441,208 | 1:93,457 a |
| Methylmalonic acidemia (MMA Mut-/Mut 0) | 9 | 1:196,092 | <1:100,000 e |
| Cobalamin metabolism deficiency (CblA, B, C, and D)/Vitamin B12 deficiency | 22 | 1:80,220 | <1:100,000 b |
| Glutaric acidemia type 1 (GA 1) | 20 | 1:88,219 | 1:100,000 b |
| 3-Hydroxy-3-methylglutaryl CoA lyase deficiency (3HMGLD) | 11 | 1:160,439 | <1:1,000,000 b |
| ß-Ketothiolase deficiency (BKTD) | 1 | ** | Approximately 250 cases reported worldwide f |
| Holocarboxylase synthase deficiency (HLCS deficiency) | 2 | ** | 1:200,000 b |
| Malonic acidemia (MAL) | 2 | 1:882,415 | <1:1,000,000 b |
| Fatty acid oxidation disorders | 304 | 1:4819 | 1:15,360 a |
| Medium-chain acyl-CoA dehydrogenase deficiency (MCADD) | 240 | 1:6603 | 1:17,301 a |
| Long-chain 3-OH acyl-CoA dehydrogenase deficiency (LCHADD) | 16 | 1:110,302 | 1:250,000 b |
| Mitochondrial trifunctional protein deficiency (MTPD) | 1 | 1:1,764,830 | Less than 100 cases reported worldwide b |
| Short-chain 3-hydroxyacyl-CoA dehydrogenase Deficiency (SCHADD) | 2 | 1:882,415 | <1:1,000,000 b |
| Multiple acyl-CoA dehydrogenase deficiency (MADD) | 12 | 1:147,069 | 1:200,000 b |
| Brown–Vialetto–Van Laere Syndrome (BVVL) | 1 | ** | <1:1,000,000 b |
| Carnitine uptake defect (CUD) | 11 | 1:160,439 | Unknown b |
| Very-long-chain acyl-CoA dehydrogenase deficiency (VLCADD) | 12 | 1:147,069 | Over 400 cases reported worldwide b |
| Carnitine palmitoyl-transferase I (CPTIA) deficiency | 4 | 1:441,208 | <1:1,000,000 b |
| Carnitine palmitoyl-transferase II (CPTII) deficiency | 3 | 1:588,277 | <1:1,000,000 b |
| Carnitine-acylcarnitine translocase (CACT) deficiency | 2 | 1:882,415 | Approximately 60 cases reported worldwide b |
| Total | 677 | 1:2607 | 1:1964 a |
| Maternal Condition/Disorder | Number of Cases |
|---|---|
| Vitamin B12 deficiency | 27 |
| 3-MCCD | 18 |
| CUD | 8 |
| GA 1 | 5 |
| MCADD | 1 |
| Total | 59 |
| Year | Number of FPs for IVA |
|---|---|
| 2011 | 29 |
| 2012 | 33 |
| 2013 | 59 |
| 2014 | 35 |
| 2015 | 36 |
| 2016 1 | 5 |
| 2017 2 | 0 |
| Parameters | Value |
|---|---|
| Number of screened neonates | 1,764,830 |
| Global birth prevalence of IEM | 1:2607 |
| False positives (FPs) | 2636 |
| False negatives (FNs) | 8 |
| Positive predictive value (PPV) | 21% |
| False positive rate (%) | 0.15% |
| Sensitivity (%) | 98.89% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.M.; Marcão, A.; Sousa, C.; Nogueira, C.; Fonseca, H.; Rocha, H.; Vilarinho, L. Portuguese Neonatal Screening Program: A Cohort Study of 18 Years Using MS/MS. Int. J. Neonatal Screen. 2024, 10, 25. https://doi.org/10.3390/ijns10010025
Gonçalves MM, Marcão A, Sousa C, Nogueira C, Fonseca H, Rocha H, Vilarinho L. Portuguese Neonatal Screening Program: A Cohort Study of 18 Years Using MS/MS. International Journal of Neonatal Screening. 2024; 10(1):25. https://doi.org/10.3390/ijns10010025
Chicago/Turabian StyleGonçalves, Maria Miguel, Ana Marcão, Carmen Sousa, Célia Nogueira, Helena Fonseca, Hugo Rocha, and Laura Vilarinho. 2024. "Portuguese Neonatal Screening Program: A Cohort Study of 18 Years Using MS/MS" International Journal of Neonatal Screening 10, no. 1: 25. https://doi.org/10.3390/ijns10010025
APA StyleGonçalves, M. M., Marcão, A., Sousa, C., Nogueira, C., Fonseca, H., Rocha, H., & Vilarinho, L. (2024). Portuguese Neonatal Screening Program: A Cohort Study of 18 Years Using MS/MS. International Journal of Neonatal Screening, 10(1), 25. https://doi.org/10.3390/ijns10010025






