You Do Not Mess with the Glia
Abstract
:1. Introduction
1.1. The More Some Things Change, the More Others Stay the Same
1.2. Quantitative Neuroanatomy: Counting Cells by Turning Brains into Soup
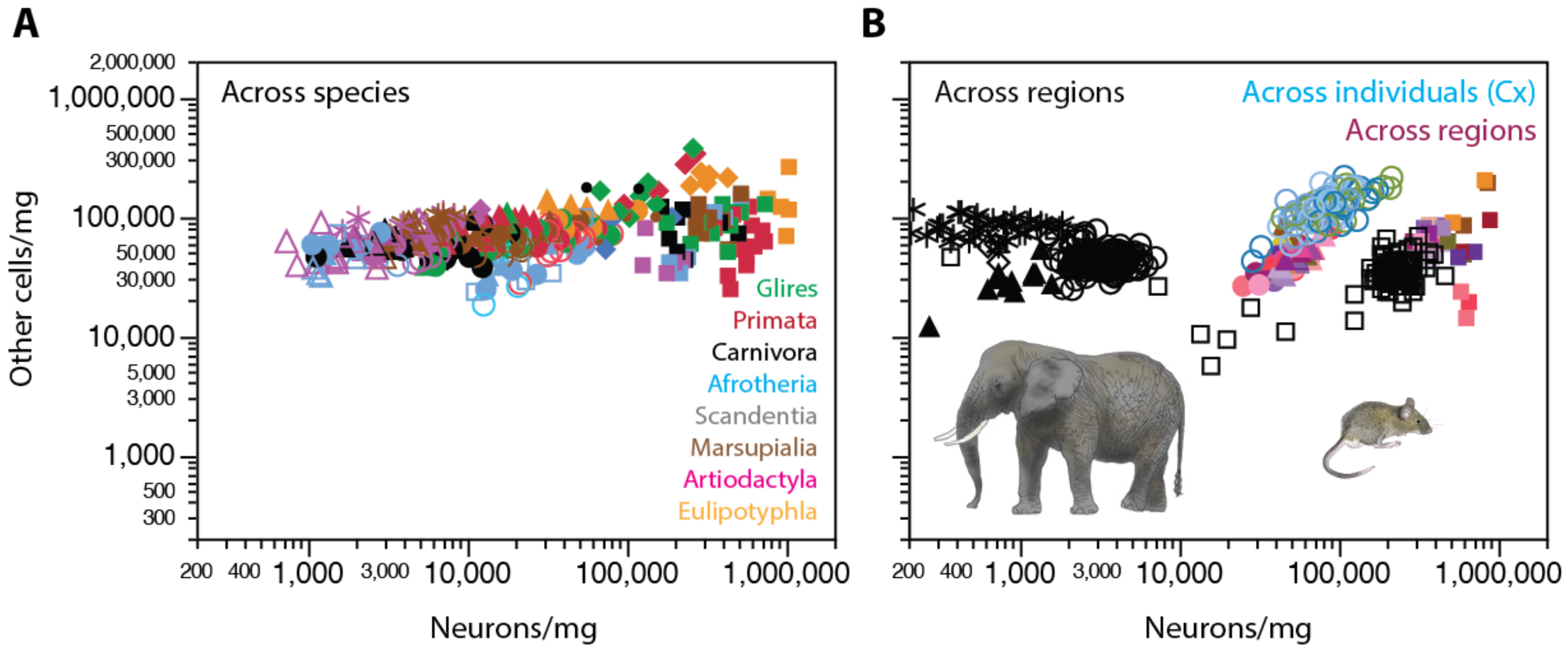
2. Neurons Are Highly Variable in Density; Other Cells, Not So Much
2.1. Relative Frequencies of Glial Cell Subtypes
2.2. Glia/Neuron Ratio
2.3. Why Should Glial Cell Size Be Constrained?
2.4. What Lies Ahead
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Virchow, R. Die Cellularpathologie in Ihrer Begründung auf Physiologische und Pathologische Gewebelehre; Hirschwald: Berlin, Germany, 1858. [Google Scholar]
- Allen, N.J.; Barres, B.A. Neuroscience: Glia—more than just brain glue. Nature 2009, 457, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Hartline, D.K. The evolutionary origins of glia. Glia 2011, 59, 1215–1236. [Google Scholar] [CrossRef] [PubMed]
- Umesono, Y.; Agata, K. Evolution and regeneration of the planarian central nervous system. Dev. Growth Differ. 2009, 51, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younossi-Hartenstein, A.; Ehlers, U.; Hartenstein, V. Embryonic development of the nervous system of the rhabdocoel flatworm Mesostoma lingua (Abilgaard, 1789). J. Comp. Neurol. 2000, 416, 461–474. [Google Scholar] [CrossRef]
- Zhu, B.; Pennack, J.A.; McQuilton, P.; Forero, M.G.; Mizuguchi, K.; Sutcliffe, B.; Gu, C.-J.; Fenton, J.C.; Hidalgo, A. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 2008, 6, e284. [Google Scholar] [CrossRef] [PubMed]
- Losada-Perez, M. The evolutionary origins of glia. Glia 2018, 59, 1215–1236. [Google Scholar] [CrossRef]
- De Robertis, E.M.; Sasai, Y. A common plan for dorsoventral patterning in Bilateria. Nature 1996, 380, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Hirth, F.; Kammermeier, L.; Frei, E.; Waldorf, U.; Noll, M.; Reichert, H. An urbilaterian origin of the tripartite brain: Developmental insights from Drosophila. Development 2003, 130, 2365–2373. [Google Scholar] [CrossRef] [PubMed]
- Hirth, F.; Reichert, H. Basic nervous system types: One or many? In Evolution of Nervous Systems; Kaas, J.H., Ed.; Academic Press: Amsterdam, The Netherlands, 2007; Volume 1, pp. 55–72. [Google Scholar]
- Moroz, L.L. On the independent origins of complex brains and neurons. Brain Behav. Evol. 2009, 74, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Northcutt, R.G. Cladistic analysis reveals brainless Urbilateria. Brain Behav. Evol. 2010, 76, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Ryan, T.J.; Grant, S.G.N. The origin and evolution of synapses. Nat. Rev. Neurosci. 2009, 10, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Helm, C.; Karl, A.; Beckers, P.; Kaul-Strehlow, S.; Ulbricht, E.; Kourtesis, I.; Kuhrt, H.; Hausen, H.; Bartolomaeus, T.; Reichenbach, A.; et al. Early evolution of radial glial cells in Bilateria. Proc. R. Soc. B 2017, 284, 20170743. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.J. Vertebrate Paleontology; Chapman and Hall: New York, NY, USA, 1997. [Google Scholar]
- Carroll, R.L. Patterns and Processes of Vertebrate Evolution; Cambridge Palaeobiology Series; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Cameron, C.B.; Garey, J.R.; Swalla, B.J. Evolution of the chordate body plan: New insights from phylogenetic analyses of deuterostome phyla. Proc. Natl. Acad. Sci. USA 2000, 97, 4469–4474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedges, S.B. Molecular evidence for the early history of living vertebrates in Major Events. In Early Vertebrate Evolution: Palaeontology, Phylogeny, Genetics and Development; Ahlberg, P.E., Ed.; Taylor & Francis: London, UK, 2001; pp. 119–134. [Google Scholar]
- Blair, J.E.; Hedges, S.B. Molecular clocks do not support the Cambrian Explosion. Mol. Biol. Evol. 2005, 22, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Greig, L.C.; Woodworth, M.B.; Galazo, M.J.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, A.F.; Kondo, S.; Abdel-Mannan, O.; Chodroff, R.A.; Sirey, T.M.; Bluy, L.E.; Webber, N.; DeProto, J.; Karlen, S.J.; Krubitzer, L.; et al. The subventricular zone is the developmental milestone of a 6-layered neocortex: Comparisons in metatherian and eutherian mammals. Cereb. Cortex 2010, 20, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lou, N.; Xu, Q.; Tian, G.F.; Peng, W.G.; Han, X.; Kang, J.; Takano, T.; Nedergaard, M. Astrocytic Ca2+ signaling evoked by sensory stimulation in vivo. Nat. Neurosci. 2006, 9, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Ramon, Y.; Cajal, S. Histologie du Système Nerveux de 1’Homme et des Vertébrés; Maloine: Paris, France, 1991; Volume 1, p. 986. [Google Scholar]
- Centro Virtual Cervantes Website. Available online: https://cvc.cervantes.es/ciencia/cajal/cajal_recuerdos/recuerdos/laminas.htm (accessed on 7 May 2018).
- Del Rio, H.P. La microglia y su transformacion en celulas en basoncito y cuerpos granulo-adiposos. Trab. Lab. Investig. Biol. Madrid. 1920, 18, 37–82. [Google Scholar]
- Somjen, G.G. Nervenkitt: Notes on the History of the Concept of Neuroglia. Glia 1988, 1, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Kamiya, T.; Tsuboi, T. Gliotransmitter release from astrocytes: Functional, developmental, and pathological implications in the brain. Front. Neurosci. 2016, 9, 499. [Google Scholar] [CrossRef] [PubMed]
- Halassa, M.M.; Fellin, T.; Takano, H.; Dong, J.H.; Haydon, P.G. Synaptic islands defined by the territory of a single astrocyte. J. Neurosci. 2007, 27, 6473–6477. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.G.; Kang, S.H.; Fukaya, M.; Bergles, D.E. Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 2013, 16, 668–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Sanes, J.R. Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 2017, 18, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Tasic, B.; Menon, V.; Nguyen, T.N.; Kim, T.K.; Jarsky, T.; Yao, Z.; Levi, B.; Gray, L.T.; Sorensen, S.A.; Dolbeare, T.; et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016, 19, 335–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeisel, A.; Muñoz-Machado, A.B.; Codeluppi, S.; Lönnerberg, P.; La Manno, G.; Juréus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Zhang, Y.; Enge, M.; Caneda, C.; Shuer, L.M.; Hayden, G.M.G.; Barres, B.A.; Quake, S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 2015, 112, 7285–7290. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 5, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Mullen, R.J.; Buck, C.R.; Smith, A.M. NeuN, a neuronal specific nuclear protein in vertebrates. Development 1992, 116, 201–211. [Google Scholar] [PubMed]
- Rothstein, J.D.; Martin, L.; Levey, A.I.; Dykes-Hoberg, M.; Jin, L.; Wu, D.; Nash, N.; Kuncl, R.W. Localization of neuronal and glial glutamate transporters. Neuron 1994, 13, 713–725. [Google Scholar] [CrossRef]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cornwell, A.; Li, J.; Peng, S.; Osorio, M.J.; Aalling, N.; Wang, S.; Benraiss, A.; Lou, N.; Goldman, S.A.; et al. SOX9 is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. J. Neurosci. 2017, 37, 4493–4507. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, S.E.; Botelho, L.; Medeiros, M.; Porfirio, J.; Herculano-Houzel, S. Invariant microglial cells densities suggest evolutionary conserved developmental mechanisms governing their addition to the brains of mammals. Unpublished work. 2018. [Google Scholar]
- Dos Santos, S.E.; Glassburn, L.; Hanflink, A.; Staub, M.; Palan, J.; Wimbiscus, M.; Herculano-Houzel, S. Similar densities of astrocytes found in different brain structures in a wide range of mammalian species. Unpublished work. 2018. [Google Scholar]
- Jiang, X.; Shen, S.; Cadwell, C.R.; Berns, P.; Sinz, F.; Ecker, A.S.; Patel, S.; Tolias, A.S. Principles of connectivity among morphologically defined cell types in adult neocortex. Science 2015, 350, aac9462. [Google Scholar] [CrossRef] [PubMed]
- Von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, D.W.; Peters, A. Neuroglial cells in the cerebral cortex of rats from young adulthood to old age: An electron microscope study. J. Neurocytol. 1974, 3, 405–429. [Google Scholar] [CrossRef] [PubMed]
- O’Kusky, J.; Colonnier, M. A laminar analysis of the number of neurons, glia, and synapses in the visual cortex (area 17) of adult macaque monkeys. J. Comp. Neurol. 1982, 210, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.; Josephson, K.; Vincent, S.L. Effects of aging on the neuroglial cells and pericytes within area 17 of the rhesus monkey cerebral cortex. Anat. Rec. 1991, 229, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Kettenmann, H.; Ransom, B.R. The Concept of Neuroglia: A Historical Perspective; University Press: Oxford, UK, 2005. [Google Scholar]
- Agulhon, C.; Petravicz, J.; McMullen, A.B.; Sweger, E.J.; Minton, S.K.; Taves, S.R.; Casper, K.B.; Fiacco, T.A.; McCarthy, K.D. What is the role of astrocyte calcium in neurophysiology. Neuron 2008, 59, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E.R.; Schwartz, J.H.; Jessell, T.M. Principles of Neural Science, 3rd ed.; Appleton & Lange: Norwalk, CT, USA, 1991. [Google Scholar]
- Kandel, E.R.; Schwartz, J.H.; Jessell, T.M.; Siegelbaum, S.A.; Hudspeth, A.J. Principles of Neural Science, 5th ed.; McGraw-Hill: New York, NY, USA, 2013. [Google Scholar]
- Haug, H. Brain sizes, surfaces, and neuronal sizes on the cortex cerebri: A stereological investigation of man and his variability and a comparison with some mammals (primates, whales, marsupials, insectivores, and one elephant). Am. J. Anat. 1987, 180, 126–142. [Google Scholar] [CrossRef] [PubMed]
- Tower, D.B.; Young, O.M. The activities of butyrylcholinesterase and carbonic anhydrase, the rate of anaerobic glycolysis, and the question of a constant density of glial cells in cerebral cortices of various mammalian species from mouse to whale. J. Neurochem. 1973, 20, 260–278. [Google Scholar] [CrossRef]
- Friede, R. Der quantitative Anteil der Glia an der Cortex Entwicklung. Acta Anat. 1954, 20, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Mota, B.; Lent, R. Cellular scaling rules for rodent brains. Proc. Natl. Acad. Sci. USA 2006, 103, 12138–12143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herculano-Houzel, S.; Collins, C.; Wong, P.; Kaas, J.H. Cellular scaling rules for primate brains. Proc. Natl. Acad. Sci. USA 2007, 104, 3562–3567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, F.A.C.; Carvalho, L.R.B.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.L.; Leite, R.E.P.; Jacob, F.W.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and non-neuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurocytol. 2009, 513, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Sarko, D.K.; Catania, K.C.; Leitch, D.B.; Kaas, J.H.; Herculano-Houzel, S. Cellular scaling rules of insectivore brains. Front. Neuroanat. 2009, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Gabi, M.; Collins, C.E.; Wong, P.; Kaas, J.H.; Herculano-Houzel, S. Cellular scaling rules for the brain of an extended number of primate species. Brain Behav. Evol. 2010, 76, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Kaas, J.H. Gorilla and orangutan brains conform to the primate scaling rules: Implications for hominin evolution. Brain Behav. Evol. 2011, 77, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Ribeiro, P.F.M.; Campos, L.; da Silva, A.V.; Torres, L.B.; Catania, K.C.; Kaas, J.H. Updated neuronal scaling rules for the brains of Glires (rodents/lagomorphs). Brain Behav. Evol. 2011, 78, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Neves, K., Jr.; Ferreira, F.M.; Tovar-Moll, F.; Gravett, N.; Bennett, N.C.; Kaswera, C.; Gilissen, E.; Manger, P.R.; Herculano-Houzel, S. Cellular scaling rules for the brains of Afrotheria. Front. Neuroanat. 2014, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Avelino-de-Souza, K.; Neves, K.; Porfírio, J.; Messeder, D.; Calazans, I.; Mattos, L.; Maldonado, J.; Manger, P.M. The elephant brain in numbers. Front. Neuroanat. 2014, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Kazu, R.S.; Maldonado, J.; Mota, B.; Manger, P.R.; Herculano-Houzel, S. Cellular scaling rules for the brains of Artiodactyla. Front. Neuroanat. 2014, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, S.E.; Porfirio, J.; Da Cunha, F.B.; Manger, P.R.; Tavares, W.; Pessoa, L.; Raghanti, M.A.; Sherwood, C.C.; Herculano-Houzel, S. Cellular scaling rules for the brain of marsupials: Not as “primitive” as expected. Brain Behav. Evol. 2017, 89, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Jardim-Messeder, D.; Lambert, K.; Noctor, S.; Marques Pestana, F.; DeCastro Leal, M.E.; Bertelsen, M.F.; Alagaili, A.N.; Mohammad, O.B.; Manger, P.R.; Herculano-Houzel, S. Dogs have the most neurons, though not the largest brain: Trade-off between body mass and number of neurons in the cerebral cortex of large carnivoran species. Front. Neuroanat. 2017, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Lent, R. Isotropic fractionator: A simple, rapid method for the quantification of total cell and neuron numbers in the brain. J. Neurosci. 2005, 25, 2518–2521. [Google Scholar] [CrossRef] [PubMed]
- Bahney, J.; von Bartheld, C.S. Validation of the isotropic fractionator: Comparison with unbiased stereology and DNA extraction for quantification of glial cells. J. Neurosci. Meth. 2014, 222, 165–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, D.J.; Balaram, P.; Young, N.A.; Kaas, J.H. Three counting methods agree on cell and neuron number in chimpanzee primary visual cortex. Front. Neuroanat. 2014, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Ngwenya, A.; Nahirney, J.; Brinkman, B.; Williams, L.; Iwaniuk, A.N. Comparison of estimates of neuronal number obtained using the isotropic fractionator method and unbiased stereology in day old chicks (Gallus domesticus). J. Neurosci. Meth. 2017, 287, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Messeder, D.; Fonseca-Azevedo, K.; Araujo Pantoja, N. When larger brains do not have more neurons: Intraspecific increase in numbers of cells is compensated by decreased average cell size. Front. Neuroanat. 2015, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Olkowicz, S.; Kocourek, M.; Lucan, R.K.; Portes, M.; Fitch, W.T.; Herculano-Houzel, S.; Nemec, P. Birds have primate-like numbers of neurons in the telencephalon. Proc. Natl. Acad. Sci. USA 2016, 113, 7255–7260. [Google Scholar] [CrossRef] [PubMed]
- Ngwenya, A.; Patzke, N.; Manger, P.R.; Herculano-Houzel, S. Continued growth of the central nervous system without mandatory addition of neurons in the Nile crocodile (Crocodylus niloticus). Brain Behav. Evol. 2016, 87, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S. The Isotropic Fractionator: A fast, reliable method to determine numbers of cells in the brain or other tissues. In Neuronal Network Analysis Concepts and Experimental Approaches; Fellin, T., Halassa, M., Eds.; Springer: Berlin, Germany, 2012; pp. 391–403. ISBN 978-1-61779-633-3. [Google Scholar]
- Mota, B.; Herculano-Houzel, S. All brains are made of this: A fundamental building block of brain matter with matching neuronal and glial masses. Front. Neuroanat. 2014, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Catania, K.; Manger, P.R.; Kaas, J.H. Mammalian brains are made of these: A dataset on the numbers and densities of neuronal and non-neuronal cells in the brain of glires, primates, scandentia, eulipotyphlans, afrotherians and artiodactyls, and their relationship with body mass. Brain Behav. Evol. 2015, 86, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Manger, P.R.; Kaas, J.H. Brain scaling in mammalian evolution as a consequence of concerted and mosaic changes in numbers of neurons and average neuronal cell size. Front. Neuroanat. 2014, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S.; Watson, C.; Paxinos, G. Distribution of neurons in functional areas of the mouse cerebral cortex reveals quantitatively different cortical zones. Front. Neuroanat. 2013, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, P.F.M.; Ventura-Antunes, L.; Gabi, M.; Mota, B.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.L.; Leite, R.E.P.; Jacob Filho, W.; Herculano-Houzel, S. The human cerebral cortex is neither one nor many: Neuronal distribution reveals two quantitatively different zones in the grey matter, three in the white matter, and explains local variations in cortical folding. Front. Neuroanat. 2013, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, F.; Cassot, F.; Lauwers-Cances, V.; Puwanarajah, P.; Duvernoy, H. Morphometry of the human cerebral cortex microcirculation: General characteristics and space-related profiles. Neuroimage 2008, 39, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Antunes, L.; Botelho, L.; Maldonado, J.; Herculano-Houzel, S. Smaller energy availability per neuron in sites with higher neuronal densities in the mouse brain. Unpublished work. 2018. [Google Scholar]
- Karbowski, J. Scaling of brain metabolism and blood flow in relation to capillary and neural scaling. PLoS ONE 2011, 6, e26709. [Google Scholar] [CrossRef] [PubMed]
- Pelvig, D.P.; Pakkenberg, H.; Stark, A.K.; Pakkenberg, B. Neocortical glial cell numbers in human brains. Neurobiol. Aging 2008, 29, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.-P.; Miyawaki, A.; Gage, F.H.; Jan, Y.N.; Han, L.Y. Local generation of glia is a major astrocyte source in postnatal cortex. Nature 2012, 484, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Bass, N.H.; Hess, H.H.; Pope, A.; Thalheimer, C. Quantitative cytoarchitectonic of neurons, glia, and DNA in rat cerebral cortex. J. Comp. Neurol. 1971, 143, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Mori, S. Light and electron microscopic features and frequencies of the glial cells present in the cerebral cortex of the rat brain. Arch. Histol. Jpn. 1972, 34, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Ling, E.-A.; Leblond, C.P. Investigations of glial cells in semithin sections. II Variation with age in the numbers of the various glial cell types in rat cortex and corpus callosum. J. Comp. Neurol. 1973, 149, 73–81. [Google Scholar] [CrossRef]
- Oberheim, N.A.; Takano, T.; Han, X.; He, W.; Lin, J.H.; Wang, F.; Xu, Q.; Wyatt, J.D.; Pilcher, W.; Ojemann, J.G.; et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009, 29, 3276–3287. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Han, D.; Efimova, O.; Guijarro, P.; Yu, Q.; Oleksiak, A.; Jiang, S.; Anokhin, K.; Velichkovsky, B.; Grünewald, S.; et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci. 2017, 20, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E.R.; Schwartz, J.H. Principles of Neural Science, 2nd ed.; Elsevier: New York, NY, USA; Amsterdam, The Netherlands; Ofxord, UK, 1985. [Google Scholar]
- Nedergaard, M.; Ransom, B.; Goldman, S.A. New roles for astrocytes: Redefining the functional architecture of the brain. Trends Neurosci. 2003, 26, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Ransom, B.; Behar, T.; Nedergaard, M. New roles for astrocytes (stars at last). Trends Neurosci. 2003, 26, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Yang, Z.; Butt, A. Astrocytes and NG2-glia: What’s in a name? J. Anat. 2005, 207, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, C.C.; Stimpson, C.D.; Raghanti, M.A.; Wildman, D.E.; Uddin, M.; Grossman, L.I.; Goodman, M.; Redmond, J.C.; Bonar, C.J.; Erwin, J.M.; et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc. Natl. Acad. Sci. USA 2006, 103, 13606–13611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, A.; Olszewski, J. Glia/nerve cell index of the cortex of the whale. Science 1957, 126, 76–77. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, J.; Matsumoto, Y.; Kreutzberg, G.W. Microglia: Intrinsic immunoeffector cell of the brain. Brain Res. Rev. 1995, 20, 269–287. [Google Scholar] [CrossRef]
- Cunningham, C.L.; Martínez-Cerdeño, V.; Noctor, S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013, 33, 4216–4233. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 2014, 62, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Kast, B. The best supporting actors. Nature 2001, 412, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C. The dark matter of the human brain. Discover 2009, 30, 30–31. [Google Scholar]
- Banhey, J.; von Bartheld, C.S. The cellular composition and glia-neuron ratio in the spinal cord of a human and a nonhuman primate: Comparison with other species and brain regions. Anat. Rec. 2018, 301, 697–710. [Google Scholar] [CrossRef]
- Von Bartheld, C.S. Myths and truths about the cellular composition of the human brain: A review of influential concepts. J. Chem. Neuroanat. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S. Scaling of brain metabolism with a fixed energy budget per neuron: Implications for neuronal activity, plasticity and evolution. PLoS ONE 2011, 6, e17514. [Google Scholar] [CrossRef] [PubMed]
- Mota, B.; Herculano-Houzel, S. Cortical folding scales universally with surface area and thickness, not number of neurons. Science 2015, 349, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D.; Smith, E.; Morowitz, H.J. A mechanism for the association of amino acids with their codons and the origins of the genetic code. Proc. Natl. Acad. Sci. USA 2005, 102, 4442–4447. [Google Scholar] [CrossRef] [PubMed]
- Perea, G.; Navarrete, M.; Araque, A. Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 2009, 32, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Morrison, B.M.; Li, Y.; Lengacher, S.; Farah, M.H.; Hoffman, P.N.; Liu, Y.; Tsingalia, A.; Jin, L.; Zhang, P.-W.; et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 2012, 487, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Ullian, E.M.; Sapperstein, S.K.; Chistopherson, K.S.; Barres, B.A. Control of synapse number by glia. Science 2001, 291, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Mauch, D.H.; Nägler, K.; Schumacher, S.; Göritz, C.; Müller, E.-C.; Otto, A.; Pfrieger, F.W. CNS synaptogenesis promoted by glia-derived cholesterol. Science 2001, 294, 1354–1357. [Google Scholar] [CrossRef] [PubMed]
- Magistretti, P.J. Neuron–glia metabolic coupling and plasticity. J. Exp. Biol. 2006, 209, 2304–2311. [Google Scholar] [CrossRef] [PubMed]
- Cragg, B.G. The density of synapses and neurons in the motor and visual areas of the cerebral cortex. J. Anat. 1967, 101, 639–654. [Google Scholar] [PubMed]
- Miki, T.; Fukui, Y.; Itoh, M.; Hisano, S.; Xie, Q.; Takeuchi, Y. Estimation of the numerical densities of neurons and synapses in cerebral cortex. Brain Res. Protoc. 1997, 2, 9–16. [Google Scholar] [CrossRef]
- Caplan, A.I.; Ordahl, C.P. Irreversible gene repression model for control of development. Science 1978, 201, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.; Bunce, C.M.; Lord, J.M.; McConnell, F.M. The development of cell lineages: A sequential model. Differentiation 1988, 39, 83–89. [Google Scholar] [CrossRef] [PubMed]
- McConnell, S.K. Constructing the cerebral cortex: Neurogenesis and fate determination. Neuron 1995, 15, 761–768. [Google Scholar] [CrossRef]
- Grimaldi, P.; Carletti, B.; Magrassi, L.; Rossi, F. Fate restriction and developmental potential of cerebellar progenitors. Transplantation studies in the developing CNS. Prog. Brain Res. 2005, 148, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Shen, Q.; Goderie, S.K.; He, W.; Capela, A.; Davis, A.A.; Temple, S. Timing of CNS cell generation: A programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 2000, 28, 69–80. [Google Scholar] [CrossRef]
- Bandeira, F.; Lent, R.; Herculano-Houzel, S. Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat. Proc. Natl. Acad. Sci. USA 2009, 106, 14108–14113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, S.E.; Noctor, S.C.; Herculano-Houzel, S. Discontinuous addition of cortical cells during embryonic and postnatal rat development. Unpublished work. 2018. [Google Scholar]
- Morterá, P.; Dos Santos, S.E.; Noctor, S.; Herculano-Houzel, S. Changing numbers of cells in pre- and postnatal development in the mouse. Unpublished work. 2018. [Google Scholar]
- Rowitch, D.H. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004, 5, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.Y.; Kohsaka, S.; Rezaie, P. The origin and cell lineage of microglia—New concepts. Brain Res. Rev. 2007, 53, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Kriegstein, A.; Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Ann. Rev. Neurosci. 2009, 32, 149–184. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Mildner, A. Microglia in the CNS: Immigrants from another world. Glia 2011, 59, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Lim, S.; Hoeffel, G.; Low, D.; Huber, T. Origin and differentiation of microglia. Front. Cell. Neurosci. 2013, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Prinz, M. Origin of microglia: Current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Golkaram, M.; Jang, J.; Hellander, S.; Kosik, K.S.; Petzold, L.R. The role of chromatin density in cell population heterogeneity during stem cell differentiation. Sci. Rep. 2017, 7, 13307. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Dent, S.Y.R. Chromatin modifiers and remodelers: Regulators of cellular differentiation. Nat. Rev. Gen. 2014, 15, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, F.; Caille, I.; Lim, D.A.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999, 97, 703–716. [Google Scholar] [CrossRef]
- Seri, B.; Garcia-Verdugo, J.M.; McEwen, B.S.; Alvarez-Buylla, A. Atrocytes give rise to new neurons in the adult mammalian hippocampus. J. Neurosci. 2001, 21, 7153–7160. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, F. The glial identity of neural stem cells. Nat. Neurosci. 2003, 6, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, O.A.; Fuentealba, L.C.; Alvarez-Buylla, A.; Rowitch, D.H. Astrocyte development and heterogeneity. Cold Spring Harb. Perspect. Biol. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Nielsen, K. Scaling: Why Is Animal Size So Important? Cambridge University Press: Cambridge, UK, 1984. [Google Scholar]
- Savage, V.M.; Allen, A.P.; Brown, L.H.; Gillooly, J.F.; Herman, A.B.; Woodruff, W.H.; West, G.B. Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 4718–4723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herculano-Houzel, S.; Avelino-de-Souza, K.; Manger, P.R. Constant densities of cells in the liver of reptile and mammalian species irrespective of body size. Unpublished work. 2018. [Google Scholar]
- Rosenberg, A.B.; Roco, C.M.; Muscat, R.A.; Kuchina, A.; Sample, P.; Yao, Z.; Graybuck, L.T.; Peeler, D.J.; Mukherjee, S.; Chen, W.; et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018, 360, 176–182. [Google Scholar] [CrossRef] [PubMed]











© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herculano-Houzel, S.; Dos Santos, S.E. You Do Not Mess with the Glia. Neuroglia 2018, 1, 193-219. https://doi.org/10.3390/neuroglia1010014
Herculano-Houzel S, Dos Santos SE. You Do Not Mess with the Glia. Neuroglia. 2018; 1(1):193-219. https://doi.org/10.3390/neuroglia1010014
Chicago/Turabian StyleHerculano-Houzel, Suzana, and Sandra E. Dos Santos. 2018. "You Do Not Mess with the Glia" Neuroglia 1, no. 1: 193-219. https://doi.org/10.3390/neuroglia1010014




